

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.5 + phase: 0
(370 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
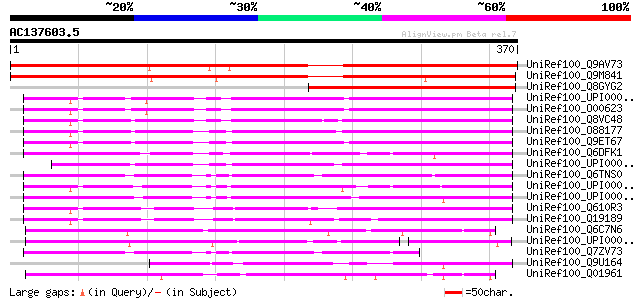
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9AV73 Putative peroxin [Oryza sativa] 489 e-137
UniRef100_Q9M841 Putative peroxisome assembly protein 12 [Arabid... 483 e-135
UniRef100_Q8GYG2 Hypothetical protein At3g04460/T27C4_11 [Arabid... 270 6e-71
UniRef100_UPI000036AC22 UPI000036AC22 UniRef100 entry 182 1e-44
UniRef100_O00623 Peroxisome assembly protein 12 [Homo sapiens] 179 1e-43
UniRef100_Q8VC48 Peroxisome assembly protein 12 [Mus musculus] 175 2e-42
UniRef100_O88177 Peroxisome assembly protein 12 [Rattus norvegicus] 172 1e-41
UniRef100_Q9ET67 Peroxisome assembly protein 12 [Cricetulus long... 172 1e-41
UniRef100_Q6DFK1 Pex12-prov protein [Xenopus laevis] 158 2e-37
UniRef100_UPI00003AB629 UPI00003AB629 UniRef100 entry 155 2e-36
UniRef100_Q6TNS0 Peroxisomal biogenesis factor 12 [Brachydanio r... 150 5e-35
UniRef100_UPI000033819C UPI000033819C UniRef100 entry 149 9e-35
UniRef100_UPI00003609A7 UPI00003609A7 UniRef100 entry 136 8e-31
UniRef100_Q610R3 Hypothetical protein CBG17349 [Caenorhabditis b... 136 8e-31
UniRef100_Q19189 Putative peroxisome assembly protein 12 [Caenor... 135 2e-30
UniRef100_Q6C7N6 Similar to sp|Q01961 Pichia pastoris Peroxisome... 118 3e-25
UniRef100_UPI00003C10E5 UPI00003C10E5 UniRef100 entry 114 6e-24
UniRef100_Q7ZV73 Hypothetical protein zgc:56182 [Brachydanio rerio] 109 1e-22
UniRef100_Q9U164 Possible peroxisome assembly factor-3 [Leishman... 97 7e-19
UniRef100_Q01961 Peroxisome assembly protein 12 [Pichia pastoris] 88 4e-16
>UniRef100_Q9AV73 Putative peroxin [Oryza sativa]
Length = 369
Score = 489 bits (1260), Expect = e-137
Identities = 249/394 (63%), Positives = 292/394 (73%), Gaps = 49/394 (12%)
Query: 1 MLFQVGGQGSRPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEHESFSLLM 60
MLFQVGGQG+RPTFFEM+AA+QLP SLRAAL+YS+GV ALR P LHK+LDYE E F+LLM
Sbjct: 1 MLFQVGGQGARPTFFEMSAAQQLPASLRAALSYSLGVFALRRPLLHKVLDYEDEFFALLM 60
Query: 61 LVLEAHSLRTTDASFSESLYGLRRRPANIKLNDNDSSSSS------SQLRRRQKLLSLLF 114
VLE+HSLRTTD SFSESLYGLRRRP + + + S + S S LR+RQK+LS++F
Sbjct: 61 AVLESHSLRTTDGSFSESLYGLRRRPVKVSVKRSSSGAESNDKAYDSVLRKRQKVLSVVF 120
Query: 115 LVVLPYLKSKLHSIYNKEREARIQATIWGD-----------ENESYTFNARASVTT---- 159
LVVLPY KSKL SIYNKEREAR+QA++WG ++ T A+ TT
Sbjct: 121 LVVLPYFKSKLQSIYNKEREARLQASLWGQGDVRFDEADLVSDQGETSQAQVEATTGEVS 180
Query: 160 ---LITKRFQKIVGLCYPLLHAGTEGFQFAYQLLYLLDATGYYSLALHALGIHVCRATGQ 216
I K F ++G+CYP +HA EG FAYQLLYLLD T +YS ALHALG+HVCRATGQ
Sbjct: 181 NMARIKKNFAALIGVCYPWIHATNEGLSFAYQLLYLLDGTAFYSPALHALGLHVCRATGQ 240
Query: 217 ELMDASSRISKIRSRERERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEW 276
EL T+Q LL+C YT LDYAQTGLIAAVFFFKMMEW
Sbjct: 241 EL-------------------------TMQRVLLNCMYTSLDYAQTGLIAAVFFFKMMEW 275
Query: 277 WYQSAEERMSAPTVYPPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVF 336
WYQSAEERMSAPTVYPPPPPPP PKVAK+G+ LP DRT+CPLC QKR NPSV++ SGFVF
Sbjct: 276 WYQSAEERMSAPTVYPPPPPPPLPKVAKDGLPLPPDRTLCPLCCQKRNNPSVLSASGFVF 335
Query: 337 CYACIFKFLTQYKRCPATMVPATVDQIRRLFHDV 370
CY+CIFK ++Q+KRCP T++PATV+QIRRLFHD+
Sbjct: 336 CYSCIFKSVSQHKRCPITLMPATVEQIRRLFHDL 369
>UniRef100_Q9M841 Putative peroxisome assembly protein 12 [Arabidopsis thaliana]
Length = 372
Score = 483 bits (1244), Expect = e-135
Identities = 241/396 (60%), Positives = 293/396 (73%), Gaps = 52/396 (13%)
Query: 1 MLFQVGGQGSRPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEHESFSLLM 60
MLFQVGG+G+RPTFFEMAAA+QLP SLRAALTYS+GV ALR FLHK+LDYE E F+ LM
Sbjct: 1 MLFQVGGEGTRPTFFEMAAAQQLPASLRAALTYSLGVFALRRSFLHKILDYEDEFFAALM 60
Query: 61 LVLEAHSLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQ------LRRRQKLLSLLF 114
L+LE HSLRTTD SF+ESLYGLRR+ A ++L + + SS+ L +RQ++LS++F
Sbjct: 61 LILEGHSLRTTDGSFAESLYGLRRKSARLRLRKDSARKDSSEEVQHSGLEKRQRILSVVF 120
Query: 115 LVVLPYLKSKLHSIYNKEREARIQATIWGDENESY-----------------TFNARASV 157
LVVLPY KSKLH+IYNKEREAR++ ++WG E++ + + N SV
Sbjct: 121 LVVLPYFKSKLHAIYNKEREARLRESLWGAEDQGFDEADFFTGDDSIVSREPSGNEELSV 180
Query: 158 TTLITKRFQKIVGLCYPLLHAGTEGFQFAYQLLYLLDATGYYSLALHALGIHVCRATGQE 217
+ + +K + +CYP +HA +EG F YQLLYLLDATG+YSL L ALGI VCRATGQE
Sbjct: 181 RVQLATKIKKFIAVCYPWIHASSEGLSFTYQLLYLLDATGFYSLGLQALGIQVCRATGQE 240
Query: 218 LMDASSRISKIRSRERERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWW 277
L T+QGALLSC+Y VLDYAQTGLIAAVF FKMMEWW
Sbjct: 241 L-------------------------TVQGALLSCSYAVLDYAQTGLIAAVFIFKMMEWW 275
Query: 278 YQSAEERMSAPTVYPPPPPPPPPKV----AKEGVQLPSDRTICPLCLQKRVNPSVMTVSG 333
YQSAEER+SAPTVYPPPPPPP PKV AKEG+ LP DR++C LCLQKR NPSV+TVSG
Sbjct: 276 YQSAEERLSAPTVYPPPPPPPAPKVINQMAKEGIPLPPDRSLCALCLQKRANPSVVTVSG 335
Query: 334 FVFCYACIFKFLTQYKRCPATMVPATVDQIRRLFHD 369
FVFCY+C+FK++++YKRCP T++PA+VDQIRRLF D
Sbjct: 336 FVFCYSCVFKYVSKYKRCPVTLIPASVDQIRRLFQD 371
>UniRef100_Q8GYG2 Hypothetical protein At3g04460/T27C4_11 [Arabidopsis thaliana]
Length = 152
Score = 270 bits (689), Expect = 6e-71
Identities = 120/151 (79%), Positives = 139/151 (91%)
Query: 219 MDASSRISKIRSRERERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWY 278
MD SSRISKIR+ ERERLRGP W+KT+QGALLSC+Y VLDYAQTGLIAAVF FKMMEWWY
Sbjct: 1 MDTSSRISKIRNHERERLRGPPWLKTVQGALLSCSYAVLDYAQTGLIAAVFIFKMMEWWY 60
Query: 279 QSAEERMSAPTVYPPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCY 338
QSAEER+SAPTVYPPPPPPP PK+AKEG+ LP DR++C LCLQKR NPSV+TVSGFVFCY
Sbjct: 61 QSAEERLSAPTVYPPPPPPPAPKMAKEGIPLPPDRSLCALCLQKRANPSVVTVSGFVFCY 120
Query: 339 ACIFKFLTQYKRCPATMVPATVDQIRRLFHD 369
+C+FK++++YKRCP T++PA+VDQIRRLF D
Sbjct: 121 SCVFKYVSKYKRCPVTLIPASVDQIRRLFQD 151
>UniRef100_UPI000036AC22 UPI000036AC22 UniRef100 entry
Length = 359
Score = 182 bits (463), Expect = 1e-44
Identities = 121/364 (33%), Positives = 185/364 (50%), Gaps = 32/364 (8%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A + L ++R AL + + VLA P FL + D E F+LL L+L+ H
Sbjct: 17 QPSIFEVVAQDSLMTAVRPALQHVVKVLAESNPTHYGFLWRWFD---EIFTLLDLLLQQH 73
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSS--SSSQLRRRQKLLSLLFLVVLPYLKSK 124
L T ASFSE+ YGL+R I + D S +S+ L ++Q S++FLV+LPYLK K
Sbjct: 74 YLSRTSASFSENFYGLKR----IVMGDTHKSQRLASAGLPKQQLWKSIMFLVLLPYLKVK 129
Query: 125 LHSIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQ 184
L + + RE DE + ++R KRF + YP ++ EG+
Sbjct: 130 LEKLVSSLREE--------DEYSIHPPSSR-------WKRFYRAFLAAYPFVNMAWEGWF 174
Query: 185 FAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKT 244
QL Y+L ++S L G+ + R T Q++ + +K +R + IK+
Sbjct: 175 LVQQLRYILGKAQHHSPLLRLAGVQLGRLTVQDIQALEHKPAKASMMQRPARSVSEKIKS 234
Query: 245 LQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVA 303
AL V TGL VFF + ++WWY S +E + + T P PPPP
Sbjct: 235 ---ALKKAVGGVALSLSTGLSVGVFFLQFLDWWYSSENQETIKSLTALPTPPPPVHLDYN 291
Query: 304 KEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQI 363
+ LP +T+CPLC + RVN +V+ SG+VFCY C+F ++ ++ CP T P V +
Sbjct: 292 SDSPLLPKMKTVCPLCRKTRVNDTVLATSGYVFCYRCVFHYVRSHQACPITGYPTEVQHL 351
Query: 364 RRLF 367
+L+
Sbjct: 352 IKLY 355
>UniRef100_O00623 Peroxisome assembly protein 12 [Homo sapiens]
Length = 359
Score = 179 bits (454), Expect = 1e-43
Identities = 119/364 (32%), Positives = 184/364 (49%), Gaps = 32/364 (8%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A + L ++R AL + + VLA P FL + D E F+LL L+L+ H
Sbjct: 17 QPSIFEVVAQDSLMTAVRPALQHVVKVLAESNPTHYGFLWRWFD---EIFTLLDLLLQQH 73
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSS--SSSQLRRRQKLLSLLFLVVLPYLKSK 124
L T ASFSE+ YGL+R I + D S +S+ L ++Q S++FLV+LPYLK K
Sbjct: 74 YLSRTSASFSENFYGLKR----IVMGDTHKSQRLASAGLPKQQLWKSIMFLVLLPYLKVK 129
Query: 125 LHSIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQ 184
L + + RE DE + ++R KRF + YP ++ EG+
Sbjct: 130 LEKLVSSLREE--------DEYSIHPPSSR-------WKRFYRAFLAAYPFVNMAWEGWF 174
Query: 185 FAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKT 244
QL Y+L ++S L G+ + R T Q++ + +K ++ + I +
Sbjct: 175 LVQQLRYILGKAQHHSPLLRLAGVQLGRLTVQDIQALEHKPAKASMMQQPARSVSEKINS 234
Query: 245 LQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVA 303
AL V TGL VFF + ++WWY S +E + + T P PPPP
Sbjct: 235 ---ALKKAVGGVALSLSTGLSVGVFFLQFLDWWYSSENQETIKSLTALPTPPPPVHLDYN 291
Query: 304 KEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQI 363
+ LP +T+CPLC + RVN +V+ SG+VFCY C+F ++ ++ CP T P V +
Sbjct: 292 SDSPLLPKMKTVCPLCRKTRVNDTVLATSGYVFCYRCVFHYVRSHQACPITGYPTEVQHL 351
Query: 364 RRLF 367
+L+
Sbjct: 352 IKLY 355
>UniRef100_Q8VC48 Peroxisome assembly protein 12 [Mus musculus]
Length = 359
Score = 175 bits (444), Expect = 2e-42
Identities = 117/362 (32%), Positives = 181/362 (49%), Gaps = 28/362 (7%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A + L ++R AL + + VLA P FL + D E F+LL +L+ H
Sbjct: 17 QPSIFEVVAQDSLMTAVRPALQHVVKVLAESNPAHYGFLWRWFD---EIFTLLDFLLQQH 73
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
L T ASFSE YGL+R A + + +S+ L + S +FLV+LPYLK KL
Sbjct: 74 YLSRTSASFSEHFYGLKRIVAGS--SPHLQRPASAGLPKEHLWKSAMFLVLLPYLKVKLE 131
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
+ + RE DE + ++R KRF + YP ++ EG+
Sbjct: 132 KLASSLREE--------DEYSIHPPSSR-------WKRFYRAFLAAYPFVNMAWEGWFLT 176
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQ 246
QL Y+L ++S L G+ + R T Q++ R+ + S +E +R + ++
Sbjct: 177 QQLRYILGKAEHHSPLLKLAGVRLARLTAQDMQAIKQRLVEA-SAMQEPVRSVG--EKIK 233
Query: 247 GALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVAKE 305
AL V TGL VFF + ++WWY S +E + + T P PPPP +
Sbjct: 234 SALKKAVGGVALSLSTGLSVGVFFLQFLDWWYSSENQEAIKSLTALPTPPPPVHLDYNSD 293
Query: 306 GVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRR 365
LP +T+CPLC + RVN +V+ SG+VFCY C+F ++ ++ CP T P V + +
Sbjct: 294 SPLLPKMKTVCPLCRKTRVNDTVLATSGYVFCYRCVFNYVRSHQACPITGYPTEVQHLIK 353
Query: 366 LF 367
L+
Sbjct: 354 LY 355
>UniRef100_O88177 Peroxisome assembly protein 12 [Rattus norvegicus]
Length = 359
Score = 172 bits (436), Expect = 1e-41
Identities = 116/363 (31%), Positives = 179/363 (48%), Gaps = 30/363 (8%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A + L ++R AL + + VLA P F + D E F+LL +L+ H
Sbjct: 17 QPSIFEVVAQDSLMTAVRPALQHVVKVLAESNPAHYGFFWRWFD---EIFTLLDFLLQQH 73
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
L T ASFSE YGL+R A + +S+ L + S +FLV+LPYLK KL
Sbjct: 74 YLSRTSASFSEHFYGLKRIVAGS--SPQLQRPASAGLPKEHLWKSAMFLVLLPYLKVKLE 131
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
+ + RE + Y+ + +S KRF ++ YP + EG+
Sbjct: 132 KLASTLRE-----------EDEYSIHPPSSHW----KRFYRVFLAAYPFVTMTWEGWFLT 176
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRS-RERERLRGPQWIKTL 245
QL Y+L ++S L G+ + R T Q++ R+ + + +E R G K +
Sbjct: 177 QQLRYILGKAEHHSPLLKLAGVRLGRLTAQDIQAMEHRLVEASAMQEPVRSIG----KKI 232
Query: 246 QGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVAK 304
+ AL V TGL VFF + ++WWY S +E + + T P PPPP
Sbjct: 233 KSALKKAVGGVALSLSTGLSVGVFFLQFLDWWYSSENQETIKSLTALPTPPPPVHLDYNS 292
Query: 305 EGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIR 364
+ LP +T+CPLC + RVN +V+ SG+VFCY C+F ++ ++ CP T P V +
Sbjct: 293 DSPLLPKMKTVCPLCRKARVNDTVLATSGYVFCYRCVFNYVRSHQACPITGYPTEVQHLI 352
Query: 365 RLF 367
+L+
Sbjct: 353 KLY 355
>UniRef100_Q9ET67 Peroxisome assembly protein 12 [Cricetulus longicaudatus]
Length = 359
Score = 172 bits (436), Expect = 1e-41
Identities = 114/362 (31%), Positives = 176/362 (48%), Gaps = 28/362 (7%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A + L ++R AL + + VLA P F + D E F+LL +L+ H
Sbjct: 17 QPSIFEVVAQDSLMTAVRPALQHVVKVLAESNPAHYGFFWRWFD---EIFTLLDFLLQQH 73
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
L T ASFSE YGL+R A + +S+ L + S +FLV+LPYLK KL
Sbjct: 74 YLSRTSASFSEHFYGLKRIVAGS--SQQPQRPASAGLPKEHLWKSTMFLVLLPYLKVKLE 131
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
+ + RE + Y+ + +S KRF + YP ++ EG+
Sbjct: 132 KLASSLRE-----------EDEYSIHPPSSHW----KRFYRAFLAAYPFVNMAWEGWFLT 176
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQ 246
QL Y+L ++S L G+ + R T Q++ R+S+ + + IK
Sbjct: 177 QQLRYILGKAEHHSPLLKLAGVRLGRLTAQDIQAIEHRLSEASVMQDPVRSVGEKIKL-- 234
Query: 247 GALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVAKE 305
AL + TGL VFF + ++WWY S +E + + T P PPPP +
Sbjct: 235 -ALKKAVGGIALSLSTGLSVGVFFLQFLDWWYSSENQETIKSLTALPTPPPPVHLDYNSD 293
Query: 306 GVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRR 365
LP +T+CPLC + RVN +V+ SG+VFCY C+F ++ ++ CP T P V + +
Sbjct: 294 SPLLPKMKTVCPLCRKTRVNDTVLATSGYVFCYRCVFNYVRSHQACPITGYPTEVQHLIK 353
Query: 366 LF 367
L+
Sbjct: 354 LY 355
>UniRef100_Q6DFK1 Pex12-prov protein [Xenopus laevis]
Length = 353
Score = 158 bits (400), Expect = 2e-37
Identities = 115/361 (31%), Positives = 179/361 (48%), Gaps = 32/361 (8%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLL-DYEHESFSLLMLVLEAHSLR 69
RP+ FE+ A E L + R AL + + VLA P + L + E ++LL +L+ H L
Sbjct: 17 RPSIFEVVAQESLMAAARPALHHIVKVLAESNPARYGTLWRWFDELYTLLECLLQQHYLS 76
Query: 70 TTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLHSIY 129
ASFSE+ YGL+R ++ + L R++ SLL LV++PYL+ KL +
Sbjct: 77 WASASFSENFYGLKRVTLGKQVGQRN-------LARKEYWKSLLLLVLIPYLRIKLEKLV 129
Query: 130 NKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFAYQL 189
N RE E Y+ S KR K + YP L G E + YQL
Sbjct: 130 NSLRE-----------EEDYSIQNPTS----FHKRCYKAILASYPFLKLGWEAWFLFYQL 174
Query: 190 LYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKT-LQGA 248
Y+L ++S L G+ + R T ++L A + ++ + + Q I++ L+ A
Sbjct: 175 RYILWNGKHHSPLLRLAGVQLARLTMEDLR-AMEKQEELTNTVSNVVSISQHIRSILKKA 233
Query: 249 LLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKEGVQ 308
L + T +V + L VFF + ++WWY SAE R + ++ P PPPP E
Sbjct: 234 LGAVTLSV----SSSLSLGVFFLQFLDWWY-SAENRETLKSLGNLPVPPPPIHFDLETYS 288
Query: 309 --LPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRRL 366
LP RT+CPLC + RVN + + SG+VFCY C + ++ ++RCP + P + + +L
Sbjct: 289 PLLPKLRTVCPLCRKVRVNDTALGTSGYVFCYRCAYYYVKTHQRCPVSGYPTELQHLIKL 348
Query: 367 F 367
+
Sbjct: 349 Y 349
>UniRef100_UPI00003AB629 UPI00003AB629 UniRef100 entry
Length = 322
Score = 155 bits (391), Expect = 2e-36
Identities = 97/339 (28%), Positives = 165/339 (48%), Gaps = 25/339 (7%)
Query: 31 LTYSIGVLALRTPFLHKLLDYEH-ESFSLLMLVLEAHSLRTTDASFSESLYGLRRRPANI 89
++ S+ VLA P + LL E + LL +L+ H L ASFSE+ Y L+R P
Sbjct: 3 VSLSMQVLAESNPGQYGLLWRRFDEIYVLLDFLLQQHYLARCSASFSENFYSLKRIPTG- 61
Query: 90 KLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLHSIYNKEREARIQATIWGDENESY 149
+ +++ L ++Q SLL L+++PYLK KL + + RE + Y
Sbjct: 62 --DCKQQPLATAGLPKKQHWKSLLLLILVPYLKGKLEKLVSSLRE-----------EDEY 108
Query: 150 TFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFAYQLLYLLDATGYYSLALHALGIH 209
+ + +S KRF + YP ++ EG+ QL Y+L ++S L G+
Sbjct: 109 SIHPPSSSW----KRFYRAFLAAYPFINMAWEGWFLIQQLCYILGKAQHHSPLLRLAGVR 164
Query: 210 VCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVF 269
+ R T +++ +++ S + ++ +Q A+ + TGL VF
Sbjct: 165 LVRLTAEDIQALEKKLAVAASSQTHSIK-----TQVQSAVRKALGGIAFSLSTGLSVCVF 219
Query: 270 FFKMMEWWYQSA-EERMSAPTVYPPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSV 328
F + ++WWY S +E + + T P PPPP + LP +T+CPLC + RVN +
Sbjct: 220 FLQFLDWWYSSENQETIKSLTALPTPPPPVHLDHGVDSALLPKLKTVCPLCRKVRVNATA 279
Query: 329 MTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRRLF 367
++ SGFVFCY C++ ++ ++RCP T + + +L+
Sbjct: 280 LSTSGFVFCYRCVYNYVKTHQRCPITGYATELQHLVKLY 318
>UniRef100_Q6TNS0 Peroxisomal biogenesis factor 12 [Brachydanio rerio]
Length = 350
Score = 150 bits (379), Expect = 5e-35
Identities = 100/358 (27%), Positives = 173/358 (47%), Gaps = 29/358 (8%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEH-ESFSLLMLVLEAHSLR 69
RP+ FE+ A + L +++ AL +++ +LA P + +L E + +L ++L+ H L
Sbjct: 17 RPSVFEVLAQDSLMTAVKPALLHAVKILATSNPARYGVLWRRFDEIYVILDMLLQHHFLS 76
Query: 70 TTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLHSIY 129
T ASFSE+ YGL+R + + + ++ LRRRQ SLL L ++PYL +KL I
Sbjct: 77 RTSASFSENFYGLKR------VGSHSARAAHLGLRRRQHWRSLLLLALIPYLHTKLEKIL 130
Query: 130 NKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFAYQL 189
++R DE++ F+ R + L ++ + YP + +G+ F QL
Sbjct: 131 ARQR----------DEDD---FSIRLPQSFL--QKMYRAFLAAYPFVCMAWDGWVFCQQL 175
Query: 190 LYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQGAL 249
LY+ T +S L G+ + T ++ ++ E + G + + LQ +
Sbjct: 176 LYVFGKTRTHSPLLWLAGVKLSYLTANDIHSLD-----LKPSGPELISGQSFGEKLQRVV 230
Query: 250 LSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKEGVQL 309
+ V T L VFF + +EWWY S + P PPPP ++E
Sbjct: 231 STAVGGVAVSLSTSLSIGVFFLQFLEWWYSSENQSTVKSLTSLPTPPPPLHLHSQETTH- 289
Query: 310 PSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRRLF 367
+ +CP+C + R N + + SG+VFCY CI+ ++ RCP T P+ + + +++
Sbjct: 290 -THIKVCPICRKVRTNDTALATSGYVFCYRCIYVYVKANHRCPLTSYPSELQHLIKIY 346
>UniRef100_UPI000033819C UPI000033819C UniRef100 entry
Length = 348
Score = 149 bits (377), Expect = 9e-35
Identities = 104/364 (28%), Positives = 181/364 (49%), Gaps = 41/364 (11%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ FE+ A E L ++R AL +++ VLA P FL + D E + LL L+L+ H
Sbjct: 16 QPSIFEVLAQESLMEAVRPALRHAVKVLAESNPSRFGFLWRRFD---ELYLLLDLLLQNH 72
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
L ASFSE+ YGL+R PA + + L R+ SLL L ++PYL++KL
Sbjct: 73 FLSHCSASFSENFYGLKRVPAGQR------PAVRLGLNRKSLWRSLLLLCLVPYLRAKLE 126
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
+ ++R DE + F+ R + + ++R + YP + + + + F
Sbjct: 127 ATLARQR----------DEED---FSIRLAQSR--SQRLYRAAVAAYPYISSAWQLWAFF 171
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQW---IK 243
+QLL++ +S L + + R + ++ D + ++ + W +
Sbjct: 172 HQLLFVFGVAKGHSPLLWLARVRLARLSAADIRDMELKAARSGTPAGGSFGRRAWQLASR 231
Query: 244 TLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVA 303
+G +S + T L VFF + +EWWY S++ R + + P PPPP +
Sbjct: 232 AARGVAVSLS--------TSLSLGVFFLQFLEWWY-SSDNRDTVKALTSTPVPPPPLHLQ 282
Query: 304 KEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQI 363
++ PS+RT CPLC + R N +V++ SGFVFCY CI+ ++ +RCP + P+ + +
Sbjct: 283 EDDQSSPSNRT-CPLCRRLRANATVLSTSGFVFCYRCIYTYVKANRRCPVSGYPSELQHL 341
Query: 364 RRLF 367
+++
Sbjct: 342 IKIY 345
>UniRef100_UPI00003609A7 UPI00003609A7 UniRef100 entry
Length = 355
Score = 136 bits (343), Expect = 8e-31
Identities = 100/364 (27%), Positives = 168/364 (45%), Gaps = 33/364 (9%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEHESFSLLM-LVLEAHSLR 69
+P+ FE+ A E L +++ AL +++ VLA P +L + LL+ L+L+ H L
Sbjct: 15 QPSIFEVLAQESLMEAVKPALRHAVKVLAESHPSRFGVLWQRFDELYLLLDLLLQNHFLS 74
Query: 70 TTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLHSIY 129
ASFSE+ YGL+R PA L L R+ SLL L ++PYL++KL +
Sbjct: 75 HCSASFSENFYGLKRVPAGRGL------PVRLGLNRKSHWCSLLLLCLVPYLRAKLEATL 128
Query: 130 NKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFAYQL 189
++R DE + F+ R + + ++R + YP + + + F +QL
Sbjct: 129 ARQR----------DEED---FSIRLAQSR--SQRLYRAAVAAYPYISSAWHLWAFLHQL 173
Query: 190 LYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQGAL 249
L++ + +S L + + R T +++ D + + W Q A
Sbjct: 174 LFVFGVSKNHSPLLWVARVRLARLTARDIQDMELMAGRPEMPAKGSFGQRAWQLMSQAAR 233
Query: 250 LSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEER-MSAPTVYPPPPPPPPPKVAKEGVQ 308
V T L VFF + +EWWY S + + A T P PP P + +
Sbjct: 234 -----GVAVSLSTSLSLGVFFLQFLEWWYSSDNQSTVKALTSMPVPPAPLHLQEDQSSPD 288
Query: 309 LPSDRTI-----CPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQI 363
P T+ CPLC R N ++++ SG+VFCY CI+ ++ +RCP T P+ + +
Sbjct: 289 SPRATTVHHKRTCPLCHCLRANATILSTSGYVFCYRCIYTYVKANRRCPVTGYPSELQHL 348
Query: 364 RRLF 367
+++
Sbjct: 349 IKIY 352
>UniRef100_Q610R3 Hypothetical protein CBG17349 [Caenorhabditis briggsae]
Length = 342
Score = 136 bits (343), Expect = 8e-31
Identities = 101/362 (27%), Positives = 178/362 (48%), Gaps = 45/362 (12%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ F++ A E L S+R AL + + LA P +HK D E + + L+L+ H
Sbjct: 20 QPSVFDIIAQENLATSIRPALQHLVKYLAFFKPKTFLSVHKNFD---EYYLIFDLILQNH 76
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
L+ ASF+E+ Y ++R ND R+++LSLL LV PY++ KL+
Sbjct: 77 YLKNYGASFTENFYSMKRVVTKNGSMPNDG---------RERILSLLTLVGWPYVEDKLN 127
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
++++ +E Y + +S+T + +K QK+ + +P + + +
Sbjct: 128 QLHDRLKEV-------------YEIRSWSSITDIKSK-CQKMFVVIWPYIKTVLKAVKTV 173
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQ 246
QL Y+L+ + +S L+ G+ + T ++L DA + + P ++T
Sbjct: 174 LQLAYILNRSSIHSPWLYFSGVVLKHLTPEDL-DAFNAV-------------PLHLQTGF 219
Query: 247 GALLSCTYTVLDYAQTGLIA-AVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKE 305
+ + L + L A +FF + +++ Y + +++ + PPPP + KE
Sbjct: 220 FNRIWRFFLGLPGIISRLFAYGLFFVQFLDYMYNTDLAKLTKTGLNSAIPPPPHKMIIKE 279
Query: 306 GVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRR 365
L D CP+CL+KRVN + + VSG+VFCY CI +++ Y++CP T PA V + R
Sbjct: 280 SEVLSLDTNKCPICLKKRVNDTALFVSGYVFCYTCINQYVNTYQKCPVTGCPANVQHLIR 339
Query: 366 LF 367
LF
Sbjct: 340 LF 341
>UniRef100_Q19189 Putative peroxisome assembly protein 12 [Caenorhabditis elegans]
Length = 359
Score = 135 bits (339), Expect = 2e-30
Identities = 100/370 (27%), Positives = 176/370 (47%), Gaps = 46/370 (12%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTP----FLHKLLDYEHESFSLLMLVLEAH 66
+P+ F++ A E L S+R AL + + LA P +H+ D E + + L+L+ H
Sbjct: 22 QPSVFDIIAQENLATSIRPALQHLVKYLAFFKPKTFLSVHRNFD---EYYIIFDLILQNH 78
Query: 67 SLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLH 126
LR ASF+E+ Y ++R + ND R++++SL+ LV PY+++KL+
Sbjct: 79 YLRNYGASFTENFYSMKRIASGTGNPPNDG---------RERIMSLITLVGWPYVENKLN 129
Query: 127 SIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFA 186
+Y++ +E Y + +S+ + K QK+ + +P + + + A
Sbjct: 130 QLYDRLKEV-------------YECRSWSSINGMKAK-CQKMFVIIWPYIKTALKAVKSA 175
Query: 187 YQLLYLLDATGYYSLALHALGIHVCRATGQEL---------MDASSRISKIRSRERERLR 237
QL Y+L+ + +S L+ G+ + T ++L + +IS+ E LR
Sbjct: 176 LQLAYILNRSSIHSPWLYFSGVILKHLTPEDLEAFNAVPLHLQTGYQISRGTLNEHIHLR 235
Query: 238 GPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPP 297
+ + +L V GL FF + +++ Y + +++ + P P
Sbjct: 236 ---FFNRIWRFILGLPGIVSRLFAYGL----FFVQFLDYMYNTDLAKLTKTGLDGAIPSP 288
Query: 298 PPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVP 357
P + E L D CP+CL+KRVN + + VSG+VFCY CI +++ Y +CP T P
Sbjct: 289 PHKMIISESEILSLDTNKCPICLKKRVNDTALFVSGYVFCYTCINQYVNTYNKCPVTGCP 348
Query: 358 ATVDQIRRLF 367
A V + RLF
Sbjct: 349 ANVQHLIRLF 358
>UniRef100_Q6C7N6 Similar to sp|Q01961 Pichia pastoris Peroxisome assembly protein
PAS10 [Yarrowia lipolytica]
Length = 408
Score = 118 bits (295), Expect = 3e-25
Identities = 100/375 (26%), Positives = 166/375 (43%), Gaps = 39/375 (10%)
Query: 12 PTFFEMAAAEQLPRSLRAALTYSIGVLALRTP-FLHKLLDYEHESFSLLMLVLEAHSLRT 70
PT FE+ +A+QL + ++ Y + A R P +L ++++ E ++L M ++E ++L+T
Sbjct: 17 PTLFELLSAKQLEGLIAPSVRYILAFYAQRHPRYLLRIVNRYDELYALFMGLVEYYNLKT 76
Query: 71 TDASFSESLYGLRR-----RPA-NIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSK 124
+ASF+E YGL+R PA + D + +L +++ SL FL+V+PY+K K
Sbjct: 77 WNASFTEKFYGLKRTQILTNPALRTRQAVPDLVEAEKRLSKKKIWGSLFFLIVVPYVKEK 136
Query: 125 LHSIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQ 184
L + Y + + + I E + + F + YP++ G
Sbjct: 137 LDARYERLKGRYLARDI---NEERIEIKRTGTAQQIAVFEFDYWLLKLYPIVTMGCTTAT 193
Query: 185 FAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSR------------E 232
A+ +L+L T YS+ L I R T + + R S+ +
Sbjct: 194 LAFHMLFLFSVTRAYSIDDFLLNIQFSRMTRYDYQMETQRDSRNAANVAHTMKSISEYPV 253
Query: 233 RERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERM------- 285
ER+ K A+ S + L Y L ++F K +EWWY S R
Sbjct: 254 AERVMLLLTTKAGANAMRSAALSGLSYV---LPTSIFALKFLEWWYASDFARQLNQKRRG 310
Query: 286 -SAPTVYPPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKF 344
+ P K+A+ + D + CPLC ++ VNP+V+ SG+VFCY CI++
Sbjct: 311 DLEDNLPVPDKVKGADKLAESVAKWKEDTSKCPLCSKELVNPTVIE-SGYVFCYTCIYRH 369
Query: 345 LTQYK-----RCPAT 354
L RCP T
Sbjct: 370 LEDGDEETGGRCPVT 384
>UniRef100_UPI00003C10E5 UPI00003C10E5 UniRef100 entry
Length = 447
Score = 114 bits (284), Expect = 6e-24
Identities = 86/284 (30%), Positives = 141/284 (49%), Gaps = 18/284 (6%)
Query: 12 PTFFEMAAAEQLPRSLRAALTYSIGVLALRTP-FLHKLLDYEHESFSLLMLVLEAHSLRT 70
P+FFE+AA QL ++ A+ Y + VLA R P +L +++++ E ++LLM +E+H LR
Sbjct: 22 PSFFELAAQSQLSELIKPAVRYVVAVLAQRNPRYLLRIVNWFDEVYALLMTAVESHYLRV 81
Query: 71 TDASFSESLYGLRRRP------ANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSK 124
+SFSE+ YGLRRR +++ + S + + L +R+ LSLLFLV LPYL++K
Sbjct: 82 WGSSFSENFYGLRRRKRPGLSRSSVTRATSLSIAKTESLGKREIRLSLLFLVGLPYLRAK 141
Query: 125 LHSIYNKEREARIQ-ATIWGDEN---ESYTFNARASVTTLITKRFQKIVGLCYPLLHAGT 180
L I+ + A ++GD+ S+ +++ + L T Q + YP
Sbjct: 142 LDDIWERNGGGLTNTANLFGDDRTSAPSFQDSSQPLMKRLETTLMQ-VFKTAYPYFKTLY 200
Query: 181 EGFQFAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQ 240
+ + Y + YL D T Y+ L + + V R + R + +R P
Sbjct: 201 QFWLLTYNVRYLFDKTPYWRPWLSLMRVDVRRVGPND----GPRRKMLPKNMPSLVRQP- 255
Query: 241 WIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEER 284
K L L + + + GL A++F FK +EWWY + R
Sbjct: 256 -TKFLAVVLKLVPGMLFEALKYGLPASIFLFKFLEWWYGADNPR 298
Score = 48.1 bits (113), Expect = 4e-04
Identities = 23/77 (29%), Positives = 36/77 (45%), Gaps = 2/77 (2%)
Query: 292 PPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRC 351
P P P ++ + CPLC +N + SG+VFCY C F ++ Q+ C
Sbjct: 369 PTDAPTHPKLLSASANERKLIHNSCPLCGAMPINNPAVLPSGYVFCYTCAFNYVEQHATC 428
Query: 352 PAT--MVPATVDQIRRL 366
P + VP D +R++
Sbjct: 429 PVSSISVPERKDALRKV 445
>UniRef100_Q7ZV73 Hypothetical protein zgc:56182 [Brachydanio rerio]
Length = 303
Score = 109 bits (272), Expect = 1e-22
Identities = 82/290 (28%), Positives = 141/290 (48%), Gaps = 28/290 (9%)
Query: 11 RPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEH-ESFSLLMLVLEAHSLR 69
RP+ FE+ A + L +++ AL +++ +LA P + +L E + +L ++L+ H L
Sbjct: 17 RPSVFEVLAQDSLMTAVKPALLHAVKILATSNPARYGVLWRRFDEIYVILDMLLQHHFLS 76
Query: 70 TTDASFSESLYGLRRRPANIKLNDNDSSSSSSQLRRRQKLLSLLFLVVLPYLKSKLHSIY 129
T ASFSE+ YGL+R + + + ++ LRRRQ SLL L ++PYL +KL I
Sbjct: 77 RTSASFSENFYGLKR------VGSHSARAAHLGLRRRQHWRSLLLLALIPYLHTKLEKIL 130
Query: 130 NKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGFQFAYQL 189
++R DE++ F+ R + + L ++ + YP + +G+ F QL
Sbjct: 131 ARQR----------DEDD---FSIRLAQSFL--QKMYRAFLAAYPFVCMAWDGWVFCQQL 175
Query: 190 LYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIKTLQGAL 249
LY+ T +S L G+ + T ++ ++ E G + + LQ +
Sbjct: 176 LYVFGKTRTHSPLLWLAGVKLSYLTANDIHSLD-----LKPSGPELSSGQSFGEKLQRVV 230
Query: 250 LSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPP 299
+ V T L VFF + +EWWY S+E + + ++ P PPPP
Sbjct: 231 SAAVGGVAVSLSTSLSIGVFFLQFLEWWY-SSENQSTVKSLTSLPTPPPP 279
>UniRef100_Q9U164 Possible peroxisome assembly factor-3 [Leishmania major]
Length = 461
Score = 97.1 bits (240), Expect = 7e-19
Identities = 73/271 (26%), Positives = 117/271 (42%), Gaps = 43/271 (15%)
Query: 103 LRRRQKLLSLLFLVVLPYLKSKLHSIYNKEREARIQATIWGDENESYTFNARASVTTLIT 162
L RR K++SLL L + PYL+ K Y + + + A + +Y + RA++ +
Sbjct: 226 LSRRNKVISLLLLTLKPYLERKAEQWYVRNTDTSVDAVAMRNAY-AYRYPHRAALLHFLA 284
Query: 163 KRFQKIVGLCYPLLHAGTEGFQFAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDAS 222
K YP+ H +G +F Y + +LL+ T Y S GI + R+T ++ +
Sbjct: 285 K-------YVYPIYHVMKQGSRFLYMMCFLLEMTPYTSPLHRVFGIALRRSTLEDSLATG 337
Query: 223 SRISKIRSRERERLRGPQWIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAE 282
R + R L L+ C + +LD+ + +A+
Sbjct: 338 PRAQRALLVARVVL-----------ILVFCGFRLLDFTR------------------NAD 368
Query: 283 ERMSAPTVYPPPPPPPPPKVAKEGVQLPSDRTI------CPLCLQKRVNPSVMTVSGFVF 336
S + P PPP V V LP DR+ CP+C + N +V SG V
Sbjct: 369 GASSPRQIREEGLPIPPPPVLGSDVPLPEDRSSLPKAGECPVCRRHVTNAAVCLTSGIVG 428
Query: 337 CYACIFKFLTQYKRCPATMVPATVDQIRRLF 367
CY C+ ++ + + CPAT V+QIRR+F
Sbjct: 429 CYPCLQGYVREQRTCPATHQSMGVEQIRRVF 459
Score = 37.0 bits (84), Expect = 0.88
Identities = 28/101 (27%), Positives = 46/101 (44%)
Query: 1 MLFQVGGQGSRPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEHESFSLLM 60
+L Q+ PT E+ + +SL A ++ +LA ++ LL + E + +
Sbjct: 6 LLSQINTASPLPTVVEVQFTTSINQSLYKAFQFAHTLLAEKSDRAAVLLPWSDEIYLAVS 65
Query: 61 LVLEAHSLRTTDASFSESLYGLRRRPANIKLNDNDSSSSSS 101
+LE L D +FSE ++ LRR + L SSSS
Sbjct: 66 GMLERLMLEHADTTFSEMIFALRRAEVDGPLAAAPPLSSSS 106
>UniRef100_Q01961 Peroxisome assembly protein 12 [Pichia pastoris]
Length = 409
Score = 87.8 bits (216), Expect = 4e-16
Identities = 95/385 (24%), Positives = 166/385 (42%), Gaps = 60/385 (15%)
Query: 12 PTFFEMAAAEQLPRSLRAALTYSIGVLALRTP-FLHKLLDYEHESFSLLMLVLEAHSLRT 70
PT FE+ +A++L + L ++ Y + R P +L K+ ++ E + +E L
Sbjct: 17 PTLFEIISAQELEKLLTPSIRYILVHYTQRYPRYLLKVANHFDELNLAIRGFIEFRQLSH 76
Query: 71 TDASFSESLYGLRR-RPANIKLNDNDSSSSSSQLRRRQKL------LSLLFLVVLPYLKS 123
+++F + YGL++ R + S + L +R++L +SL +V +PYL+
Sbjct: 77 WNSTFIDKFYGLKKVRNHQTISTERLQSQVPTLLEQRRRLSKTQIAVSLFEIVGVPYLRD 136
Query: 124 KLHSIYNKEREARIQATIWGDENESYTFNARASVTTLITKRFQKIVGLCYPLLHAGTEGF 183
KL +Y+K + + + + S+ T + F K+ YP+L +
Sbjct: 137 KLDHLYDKLYPKLMMNNL----------DPKESLKTFVQYYFLKL----YPILLSVLTTI 182
Query: 184 QFAYQLLYLLDATGYYSLALHALGIHVCRATGQELMDASSRISKIRSRERERLRGPQWIK 243
Q Q+LYL S+ + + R + R++K ++ G +
Sbjct: 183 QVLLQVLYLSGTFKSPSIIMWLFKMKYARLNSYDYTLDEQRVNKFLNKTSPGKLGTGNNR 242
Query: 244 ----TLQGALLSCTYTVLDYAQTGLI--------AAVFFFKMMEWWYQSA-EERMSAP-T 289
TL +L + + GL+ A++F K +EWW S +M+ P
Sbjct: 243 IRPITLTESLYLLYSDLTRPLKKGLLITGGTLFPASIFLLKFLEWWNSSDFATKMNKPRN 302
Query: 290 VYPPPPPPPPPKVAKEGVQLPSDRTI-------------CPLCLQKRVNPSVMTVSGFVF 336
+ PPP ++K+ L +DR I CPLC ++ NP+V+ +G+VF
Sbjct: 303 PFSDSELPPPINLSKD---LLADRKIKKLLKKSQSNDGTCPLCHKQITNPAVIE-TGYVF 358
Query: 337 CYACIFKFLTQYK-------RCPAT 354
CY CIFK LT + RCP T
Sbjct: 359 CYTCIFKHLTSSELDEETGGRCPIT 383
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 601,658,906
Number of Sequences: 2790947
Number of extensions: 24773562
Number of successful extensions: 290480
Number of sequences better than 10.0: 530
Number of HSP's better than 10.0 without gapping: 275
Number of HSP's successfully gapped in prelim test: 265
Number of HSP's that attempted gapping in prelim test: 283624
Number of HSP's gapped (non-prelim): 3061
length of query: 370
length of database: 848,049,833
effective HSP length: 129
effective length of query: 241
effective length of database: 488,017,670
effective search space: 117612258470
effective search space used: 117612258470
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137603.5


