

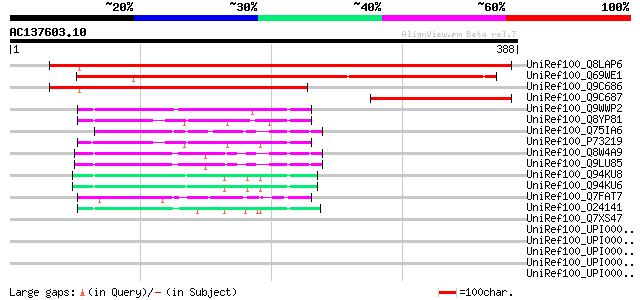
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.10 + phase: 0
(388 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LAP6 Hypothetical protein At1g51110/F23H24_8 [Arabid... 477 e-133
UniRef100_Q69WE1 Plastid-lipid associated protein PAP/fibrillin ... 350 3e-95
UniRef100_Q9C686 Hypothetical protein F23H24.16 [Arabidopsis tha... 279 1e-73
UniRef100_Q9C687 Hypothetical protein F23H24.17 [Arabidopsis tha... 140 6e-32
UniRef100_Q9WWP2 Fibrillin [Nostoc sp.] 66 1e-09
UniRef100_Q8YP81 Fibrillin [Anabaena sp.] 63 2e-08
UniRef100_Q75IA6 Putative PAP_fibrillin [Oryza sativa] 62 4e-08
UniRef100_P73219 Fibrillin [Synechocystis sp.] 59 2e-07
UniRef100_Q8W4A9 Hypothetical protein At3g26090; MPE11.24 [Arabi... 56 2e-06
UniRef100_Q9LU85 Arabidopsis thaliana genomic DNA, chromosome 3,... 56 2e-06
UniRef100_Q94KU8 Plastid-lipid associated protein PAP2 [Brassica... 54 7e-06
UniRef100_Q94KU6 Plastid-lipid associated protein PAP2 [Brassica... 54 7e-06
UniRef100_Q7FAT7 OSJNBa0010H02.5 protein [Oryza sativa] 50 1e-04
UniRef100_O24141 Plastid-lipid-Associated Protein [Nicotiana tab... 50 1e-04
UniRef100_Q7XS47 OSJNBa0035M09.17 protein [Oryza sativa] 47 0.001
UniRef100_UPI00002A2AA1 UPI00002A2AA1 UniRef100 entry 45 0.005
UniRef100_UPI00003294C9 UPI00003294C9 UniRef100 entry 43 0.013
UniRef100_UPI0000311B17 UPI0000311B17 UniRef100 entry 43 0.013
UniRef100_UPI000029F438 UPI000029F438 UniRef100 entry 43 0.013
UniRef100_UPI000029F0EC UPI000029F0EC UniRef100 entry 43 0.013
>UniRef100_Q8LAP6 Hypothetical protein At1g51110/F23H24_8 [Arabidopsis thaliana]
Length = 409
Score = 477 bits (1227), Expect = e-133
Identities = 239/360 (66%), Positives = 286/360 (79%), Gaps = 6/360 (1%)
Query: 31 NSRFVSAGCSKVEQISIVTEES-----ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVL 85
N R + CS ++ T++S E LI AL+GIQGRG+S+SP+QLN +E A++VL
Sbjct: 48 NCRTLRISCSSSSTVTDQTQQSSFNDAELKLIDALIGIQGRGKSASPKQLNDVESAVKVL 107
Query: 86 EHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQ-TNDPRVT 144
E + G+ +PT+S LIEGRW+L+FTTRPGTASPIQRTF GVD F+VFQ+VYL+ TNDPRV+
Sbjct: 108 EGLEGIQNPTDSDLIEGRWRLMFTTRPGTASPIQRTFTGVDVFTVFQDVYLKATNDPRVS 167
Query: 145 NIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGW 204
NIV FSD IGELKVEA ASI DGKR+LFRFDRAAF KFLPFKVPYPVPF+LLGDEAKGW
Sbjct: 168 NIVKFSDFIGELKVEAVASIKDGKRVLFRFDRAAFDLKFLPFKVPYPVPFRLLGDEAKGW 227
Query: 205 LDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKNSG 264
LDTTYLS SGNLRISRGNKGTTFVLQK+T PRQKLL IS GV EAID+ ++ N NS
Sbjct: 228 LDTTYLSPSGNLRISRGNKGTTFVLQKETVPRQKLLATISQDKGVAEAIDEFLASNSNSA 287
Query: 265 EEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFSMSG 324
E++ EL EG WQMIW+SQ TDSW+ENAANGLMG+QI+EK+GRIK+ VNI+ +FSM G
Sbjct: 288 EDNYELLEGSWQMIWSSQMYTDSWIENAANGLMGRQIIEKDGRIKFEVNIIPAFRFSMKG 347
Query: 325 IFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFILEILFNDGKVRISRGDNEIIFVHAR 384
F+KS Y++ MDDAAIIGG FGYP++ L+IL+ D K+RISRG + IIFVH R
Sbjct: 348 KFIKSESSTYDLKMDDAAIIGGAFGYPVDITNNIELKILYTDEKMRISRGFDNIIFVHIR 407
>UniRef100_Q69WE1 Plastid-lipid associated protein PAP/fibrillin family-like [Oryza
sativa]
Length = 442
Score = 350 bits (899), Expect = 3e-95
Identities = 190/351 (54%), Positives = 243/351 (69%), Gaps = 32/351 (9%)
Query: 52 SENSLIQALVGIQGRGRSSSPQQLNA-IERAIQVLEHIGGVSD----------------- 93
+E L+ AL G+QGRGR +P+QL A +ER +L+ + GV+D
Sbjct: 66 AETELLDALAGVQGRGRGVAPRQLEASMERTRCLLKCLCGVADISSCYVCAGGGERCSGS 125
Query: 94 ------------PTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDP 141
TNSSLIEG WQLIFTTRPG+ASPIQRTFVGVD F +FQEVYL+T+DP
Sbjct: 126 GSTGRFARSGGCTTNSSLIEGSWQLIFTTRPGSASPIQRTFVGVDSFKIFQEVYLRTDDP 185
Query: 142 RVTNIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEA 201
RV N+V FS++IGELKVEA A+I DGKRILFRFDRAAF+FKFLPFKVPYPVPFKLLGDEA
Sbjct: 186 RVINVVKFSESIGELKVEAEATIEDGKRILFRFDRAAFNFKFLPFKVPYPVPFKLLGDEA 245
Query: 202 KGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNK 261
KGWLDTTYLS +GN+RISRGNKGTTFVLQK + RQ LL+AIS+G GV+EAID L S ++
Sbjct: 246 KGWLDTTYLSQTGNIRISRGNKGTTFVLQKSADQRQLLLSAISAGTGVKEAIDDLTS-SR 304
Query: 262 NSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFS 321
E D GEWQ++W+S+T +SW A+ GL G QI++++G++K +V G+ +
Sbjct: 305 QGIEADLNTLAGEWQLLWSSKTEDESWSFVASAGLKGVQIIKEDGQLKNLVRPFPGVSLN 364
Query: 322 MSGIFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFILEILFNDGKVRIS 372
SG + ++++ AI G +PL+ +F +EI + D K+RIS
Sbjct: 365 ASGNIKNEDGNNFNLSINKGAIQAGGLQFPLDARGEFAMEI-YIDNKIRIS 414
>UniRef100_Q9C686 Hypothetical protein F23H24.16 [Arabidopsis thaliana]
Length = 257
Score = 279 bits (713), Expect = 1e-73
Identities = 141/204 (69%), Positives = 166/204 (81%), Gaps = 6/204 (2%)
Query: 31 NSRFVSAGCSKVEQISIVTEES-----ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVL 85
N R + CS ++ T++S E LI AL+GIQGRG+S+SP+QLN +E A++VL
Sbjct: 48 NCRTLRISCSSSSTVTDQTQQSSFNDAELKLIDALIGIQGRGKSASPKQLNDVESAVKVL 107
Query: 86 EHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQ-TNDPRVT 144
E + G+ +PT+S LIEGRW+L+FTTRPGTASPIQRTF GVD F+VFQ+VYL+ TNDPRV+
Sbjct: 108 EGLEGIQNPTDSDLIEGRWRLMFTTRPGTASPIQRTFTGVDVFTVFQDVYLKATNDPRVS 167
Query: 145 NIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGW 204
NIV FSD IGELKVEA ASI DGKR+LFRFDRAAF KFLPFKVPYPVPF+LLGDEAKGW
Sbjct: 168 NIVKFSDFIGELKVEAVASIKDGKRVLFRFDRAAFDLKFLPFKVPYPVPFRLLGDEAKGW 227
Query: 205 LDTTYLSHSGNLRISRGNKGTTFV 228
LDTTYLS SGNLRISRGNK F+
Sbjct: 228 LDTTYLSPSGNLRISRGNKVNEFL 251
>UniRef100_Q9C687 Hypothetical protein F23H24.17 [Arabidopsis thaliana]
Length = 110
Score = 140 bits (353), Expect = 6e-32
Identities = 66/108 (61%), Positives = 83/108 (76%)
Query: 277 MIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEV 336
MIW+SQ TDSW+ENAANGLMG+QI+EK+GRIK+ VNI+ +FSM G F+KS Y++
Sbjct: 1 MIWSSQMYTDSWIENAANGLMGRQIIEKDGRIKFEVNIIPAFRFSMKGKFIKSESSTYDL 60
Query: 337 TMDDAAIIGGPFGYPLEFGKKFILEILFNDGKVRISRGDNEIIFVHAR 384
MDDAAIIGG FGYP++ L+IL+ D K+RISRG + IIFVH R
Sbjct: 61 KMDDAAIIGGAFGYPVDITNNIELKILYTDEKMRISRGFDNIIFVHIR 108
>UniRef100_Q9WWP2 Fibrillin [Nostoc sp.]
Length = 194
Score = 66.2 bits (160), Expect = 1e-09
Identities = 53/195 (27%), Positives = 90/195 (45%), Gaps = 21/195 (10%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSS-LIEGRWQLIFTTR 111
+++LI + G RG ++ QQ AI AI LE + P ++ L++G W+L++TT
Sbjct: 4 KSTLIDTIAGTN-RGLLANEQQKQAILAAIARLEDLNPTPRPVEATNLLDGNWRLLYTTS 62
Query: 112 PGTASPIQRTFVGVDFFSVFQEVYLQTNDP-RVTNIVSFSDAIGELKVEAAASIGDGKRI 170
+ + F + ++Q + ++T + I G + V A G+R+
Sbjct: 63 KALLNLDRVPFYKLG--QIYQCIRVETTSVYNIAEIYGLPYLEGLISVRAKFEPVSGRRV 120
Query: 171 LFRFDRAAFSFKFL--------------PFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNL 216
+F+R+ K L K + F + D +GWLD TY+ + +L
Sbjct: 121 QVKFERSIIGLKSLIGYTSVENFIQQIETGKKFIAIDFPISSDTQQGWLDITYIDN--DL 178
Query: 217 RISRGNKGTTFVLQK 231
RI RGN+G+ FVL K
Sbjct: 179 RIGRGNEGSVFVLSK 193
>UniRef100_Q8YP81 Fibrillin [Anabaena sp.]
Length = 194
Score = 62.8 bits (151), Expect = 2e-08
Identities = 55/205 (26%), Positives = 93/205 (44%), Gaps = 41/205 (20%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSS-LIEGRWQLIFTTR 111
+ +L+ A+ G RG +S QQ AI AI LE + P ++ L+EG W+L++TT
Sbjct: 4 KTALLDAIAGTN-RGLLASEQQKQAILAAIATLEDLNPTPRPVETANLLEGNWRLLYTT- 61
Query: 112 PGTASPIQRTFVGVDFFSVFQ--EVYLQTNDPRVTNIVSFSDAIGELKVEAAASIG---- 165
+ + +D V++ ++Y Q T++ + ++ G +E S+
Sbjct: 62 -------SKALLNLDRVPVYKLGQIY-QCIRVETTSVYNIAEIYGLPYLEGLVSVAAKFE 113
Query: 166 --DGKRILFRFDRAAFSFKFLPFKVPYPVPFKLL-----------------GDEAKGWLD 206
+R+ +F R+ + L + Y P + D +GWLD
Sbjct: 114 PVSERRVQVKFQRSIVGLQRL---IGYTSPGDFIQQIEAGKKFTALDVLIKSDTQQGWLD 170
Query: 207 TTYLSHSGNLRISRGNKGTTFVLQK 231
TY+ + NLRI RGN+G+ FVL K
Sbjct: 171 ITYIDN--NLRIGRGNEGSVFVLSK 193
>UniRef100_Q75IA6 Putative PAP_fibrillin [Oryza sativa]
Length = 262
Score = 61.6 bits (148), Expect = 4e-08
Identities = 48/174 (27%), Positives = 82/174 (46%), Gaps = 19/174 (10%)
Query: 66 RGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGV 125
RG ++ + +E+ +Q LE + V +P S L+ G+W+L++TT P + F+
Sbjct: 106 RGVDATAEDKERVEKIVQQLEEVNQVKEPLKSDLLNGKWELLYTTSESILQPQRPKFLR- 164
Query: 126 DFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLP 185
F +++Q + T+ R N+ ++ +V A + +R+ RFD F +
Sbjct: 165 PFGTIYQAI--NTDTLRAQNMETWPYF---NQVTANLVPLNSRRVAVRFDYFKI-FNLIS 218
Query: 186 FKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
K P KG L+ TYL LR SRG+KG F+L K +P ++
Sbjct: 219 IKAP---------GSGKGELEITYLDE--ELRASRGDKGNLFIL-KMVDPTYRV 260
>UniRef100_P73219 Fibrillin [Synechocystis sp.]
Length = 202
Score = 58.9 bits (141), Expect = 2e-07
Identities = 54/202 (26%), Positives = 97/202 (47%), Gaps = 35/202 (17%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNS-SLIEGRWQLIFTTR 111
+ +L++A+ G + RG +S + AI A++ LE P +L++G W+L++T+
Sbjct: 12 KTNLLEAIAG-KNRGLLASDRDRVAILSAVEKLEDYNPHPKPLQEKNLLDGNWRLLYTS- 69
Query: 112 PGTASPIQRTFVGVDFFSVFQ--EVYLQTNDPRVTNIVSFSDAIGELKVEAAASIG---- 165
++ +G++ + Q ++Y Q D + +V+ ++ G +E+ S+
Sbjct: 70 -------SQSILGLNRLPLLQLGQIY-QYIDVAGSRVVNLAEIEGIPFLESLVSVVASFI 121
Query: 166 --DGKRILFRFDRAAFSF-KFLPFKVPY-------------PVPFKLLGDEAKGWLDTTY 209
KRI +F+R+ K L ++ P P F L G + WL+ TY
Sbjct: 122 PVSDKRIEVKFERSILGLQKILNYQSPLKFIQQISTGKRFLPADFNLPGRDNAAWLEITY 181
Query: 210 LSHSGNLRISRGNKGTTFVLQK 231
L +LRISRGN+G F+L K
Sbjct: 182 LDE--DLRISRGNEGNVFILAK 201
>UniRef100_Q8W4A9 Hypothetical protein At3g26090; MPE11.24 [Arabidopsis thaliana]
Length = 217
Score = 56.2 bits (134), Expect = 2e-06
Identities = 46/193 (23%), Positives = 92/193 (46%), Gaps = 27/193 (13%)
Query: 50 EESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFT 109
++ + L++A+ ++ RG ++SP I++ + +E + +P S L+ G+W+LI+T
Sbjct: 47 KQLKQELLEAIEPLE-RGATASPDDQLRIDQLARKVEAVNPTKEPLKSDLVNGKWELIYT 105
Query: 110 TRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVS---FSDAIGELKVEAAASIGD 166
T + F+ S+ + + +V N+ + ++ G++K + +
Sbjct: 106 TSASILQAKKPRFLR----SITNYQSINVDTLKVQNMETWPFYNSVTGDIKPLNSKKVA- 160
Query: 167 GKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTT 226
K +F+ F+P K P D A+G L+ TY+ LR+SRG+KG
Sbjct: 161 VKLQVFKI------LGFIPIKAP---------DSARGELEITYVDE--ELRLSRGDKGNL 203
Query: 227 FVLQKQTEPRQKL 239
F+L K +P ++
Sbjct: 204 FIL-KMFDPTYRI 215
>UniRef100_Q9LU85 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MPE11
[Arabidopsis thaliana]
Length = 242
Score = 56.2 bits (134), Expect = 2e-06
Identities = 46/193 (23%), Positives = 92/193 (46%), Gaps = 27/193 (13%)
Query: 50 EESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFT 109
++ + L++A+ ++ RG ++SP I++ + +E + +P S L+ G+W+LI+T
Sbjct: 72 KQLKQELLEAIEPLE-RGATASPDDQLRIDQLARKVEAVNPTKEPLKSDLVNGKWELIYT 130
Query: 110 TRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVS---FSDAIGELKVEAAASIGD 166
T + F+ S+ + + +V N+ + ++ G++K + +
Sbjct: 131 TSASILQAKKPRFLR----SITNYQSINVDTLKVQNMETWPFYNSVTGDIKPLNSKKVA- 185
Query: 167 GKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTT 226
K +F+ F+P K P D A+G L+ TY+ LR+SRG+KG
Sbjct: 186 VKLQVFKI------LGFIPIKAP---------DSARGELEITYVDE--ELRLSRGDKGNL 228
Query: 227 FVLQKQTEPRQKL 239
F+L K +P ++
Sbjct: 229 FIL-KMFDPTYRI 240
>UniRef100_Q94KU8 Plastid-lipid associated protein PAP2 [Brassica campestris]
Length = 319
Score = 53.9 bits (128), Expect = 7e-06
Identities = 59/228 (25%), Positives = 92/228 (39%), Gaps = 45/228 (19%)
Query: 49 TEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSS-LIEGRWQLI 107
TE + SL+ +L G RG S+S + I I LE PT++ L+ G+W L
Sbjct: 92 TEVLKRSLVDSLYGTD-RGLSASSETRAEIGDLITQLESKNPTPAPTDALFLLNGKWILA 150
Query: 108 FTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAAS--IG 165
+T+ G + R V + + + +++ V N V F+ + + A I
Sbjct: 151 YTSFVGLFPLLSRGIVPLVKVDEISQT-IDSDNFTVENSVLFAGPLATTSISTNAKFEIR 209
Query: 166 DGKRILFRFDRAAFS-------------FKFLPFKVPY---------------------- 190
KR+ +F+ +FL K+
Sbjct: 210 SPKRVQIKFEEGVIGTPQLTDSIEIPEYVEFLGQKIDLTPIRGLLTSVQDTATSVARTIS 269
Query: 191 ---PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
P+ F L GD A+ WL TTYL ++RISRG+ G+ FVL K+ P
Sbjct: 270 SQPPLKFSLPGDSAQSWLLTTYLDK--DIRISRGDGGSVFVLIKEGSP 315
>UniRef100_Q94KU6 Plastid-lipid associated protein PAP2 [Brassica campestris]
Length = 319
Score = 53.9 bits (128), Expect = 7e-06
Identities = 59/228 (25%), Positives = 92/228 (39%), Gaps = 45/228 (19%)
Query: 49 TEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSS-LIEGRWQLI 107
TE + SL+ +L G RG S+S + I I LE PT++ L+ G+W L
Sbjct: 92 TEVLKRSLVDSLYGTD-RGLSASSETRAEIGDLITQLESKNPTPAPTDALFLLNGKWILA 150
Query: 108 FTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAAS--IG 165
+T+ G + R V + + + +++ V N V F+ + + A I
Sbjct: 151 YTSFVGLFPLLSRGIVPLVKVDEISQT-IDSDNFTVENSVLFAGPLATTSISTNAKFEIR 209
Query: 166 DGKRILFRFDRAAFS-------------FKFLPFKVPY---------------------- 190
KR+ +F+ +FL K+
Sbjct: 210 SPKRVQIKFEEGVIGTPQLTDSIEIPEYVEFLGQKIDLTPIRGLLTSVQDTATSVARTIS 269
Query: 191 ---PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
P+ F L GD A+ WL TTYL ++RISRG+ G+ FVL K+ P
Sbjct: 270 SQPPLKFSLPGDSAQSWLLTTYLDK--DIRISRGDGGSVFVLIKEGSP 315
>UniRef100_Q7FAT7 OSJNBa0010H02.5 protein [Oryza sativa]
Length = 221
Score = 49.7 bits (117), Expect = 1e-04
Identities = 51/187 (27%), Positives = 84/187 (44%), Gaps = 26/187 (13%)
Query: 53 ENSLIQALVGIQGRG--RSSSPQQLNAIERAIQVLEHIGGVSDP-TNSSLIEGRWQLIFT 109
E + L+ Q RG S P +L I I L SD +++ + G W+L++T
Sbjct: 46 EKDELLRLIADQRRGLDTQSDPSRLADIVSCIDALAAAAPGSDTVSDADKLSGTWRLLWT 105
Query: 110 TRPGTA-----SPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASI 164
T +P RT G D F V + + N+++F + G V + I
Sbjct: 106 TEHEQLFIVRNAPFFRTAAG-DVFQV-----IDVPGGALNNVITFPPS-GAFVVNGSIEI 158
Query: 165 GDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKG 224
+R+ FRF RA + ++VP+P PF KGW DT YL ++R+++ +G
Sbjct: 159 QPPQRVNFRFTRAML--RGSNWEVPFP-PF------GKGWFDTVYL--DDDIRVAKDIRG 207
Query: 225 TTFVLQK 231
V+++
Sbjct: 208 DYLVVER 214
>UniRef100_O24141 Plastid-lipid-Associated Protein [Nicotiana tabacum]
Length = 270
Score = 49.7 bits (117), Expect = 1e-04
Identities = 60/230 (26%), Positives = 89/230 (38%), Gaps = 51/230 (22%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNS-SLIEGRWQLIFTTR 111
+ L+ + G RG S+S + I I LE PT + L+ G+W L +T+
Sbjct: 47 KKQLVDSFYGTN-RGLSASSETRAEIVELITKLESKNPTPAPTEALPLLNGKWILAYTSF 105
Query: 112 PGTASPIQRTFVGVDFFSVFQEVYLQTNDPR---VTNIVSFSDAIGELKVEAAAS--IGD 166
G + R + + V E QT D V N V F+ + + A +
Sbjct: 106 SGLFPLLSRGTLPL----VRVEEISQTIDSEAFTVQNSVVFAGPLATTSITTNAKFEVRS 161
Query: 167 GKRILFRFDRAAF-------------SFKFLPFKV---PY-------------------- 190
KR+ +FD + +FL K+ P+
Sbjct: 162 PKRVQIKFDEGVIGTPQLTDSIELPENIEFLGQKIDLSPFKGLVNSVQDTASSVAKSISS 221
Query: 191 --PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQK 238
P+ F + A+ WL TTYL H LRISRG+ G+ FVL K+ P K
Sbjct: 222 QPPIKFPISNSNAQSWLLTTYLDH--ELRISRGDGGSVFVLIKEGSPLLK 269
>UniRef100_Q7XS47 OSJNBa0035M09.17 protein [Oryza sativa]
Length = 219
Score = 46.6 bits (109), Expect = 0.001
Identities = 36/144 (25%), Positives = 66/144 (45%), Gaps = 13/144 (9%)
Query: 91 VSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFS 150
+ DP S LI G W++++ + P + + RT +G F + + V N VSFS
Sbjct: 87 IDDPVKSPLIFGEWEVVYCSVPTSPGGLYRTPLGRLIFKTDEMAQVVQAPDVVKNKVSFS 146
Query: 151 --DAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTT 208
G + ++ ++ DGK I F+ L F+ G E++ L+ T
Sbjct: 147 VFGFDGAVSLKGKLNVLDGKWIQVIFEPPEVKVGSLGFQ---------YGGESEVKLEIT 197
Query: 209 YLSHSGNLRISRGNKGTTFVLQKQ 232
Y+ +R+ +G++G+ FV ++
Sbjct: 198 YVDE--KIRLGKGSRGSLFVFMRR 219
>UniRef100_UPI00002A2AA1 UPI00002A2AA1 UniRef100 entry
Length = 179
Score = 44.7 bits (104), Expect = 0.005
Identities = 45/160 (28%), Positives = 73/160 (45%), Gaps = 15/160 (9%)
Query: 78 IERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQ 137
+E+ I++ E V P + G W+L +++ + SP+ ++ F + L
Sbjct: 20 LEKLIKLSELESAVDIPNQFERLIGVWELRWSS---SKSPVLNYSPLLNNFQI-----LD 71
Query: 138 TNDPRVTNIVSFSDAIGEL---KVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPF 194
+ R N +S ++G+L + A I D KRI F++A + K V
Sbjct: 72 LDKSRALNFLSPKGSLGKLLSTNILAKLDIIDQKRINVTFEKAGIIGPQVFGK--NMVFL 129
Query: 195 KLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTE 234
+ GWLDTT L+ LR+ RG KGTTF L K+ +
Sbjct: 130 SEIKKAQTGWLDTTVLTDK--LRVCRGYKGTTFALLKRED 167
>UniRef100_UPI00003294C9 UPI00003294C9 UniRef100 entry
Length = 180
Score = 43.1 bits (100), Expect = 0.013
Identities = 22/41 (53%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Query: 195 KLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
K + +E GWL+ TYLS+ LRI RG+KGT FVL+K P
Sbjct: 129 KEINNEQLGWLEITYLSNK--LRICRGDKGTLFVLRKINSP 167
>UniRef100_UPI0000311B17 UPI0000311B17 UniRef100 entry
Length = 176
Score = 43.1 bits (100), Expect = 0.013
Identities = 22/41 (53%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Query: 195 KLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
K + +E GWL+ TYLS+ LRI RG+KGT FVL+K P
Sbjct: 125 KEINNEQLGWLEITYLSNK--LRICRGDKGTLFVLRKINSP 163
>UniRef100_UPI000029F438 UPI000029F438 UniRef100 entry
Length = 128
Score = 43.1 bits (100), Expect = 0.013
Identities = 22/41 (53%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Query: 195 KLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
K + +E GW + TYLS+ NLRI RG+KGT FVL+K P
Sbjct: 77 KEINNEQLGWSEITYLSN--NLRICRGDKGTLFVLRKINSP 115
>UniRef100_UPI000029F0EC UPI000029F0EC UniRef100 entry
Length = 153
Score = 43.1 bits (100), Expect = 0.013
Identities = 22/41 (53%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Query: 195 KLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
K + +E GWL+ TYLS+ LRI RG+KGT FVL+K P
Sbjct: 102 KEINNEQLGWLEITYLSNK--LRICRGDKGTLFVLRKINSP 140
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,473,403
Number of Sequences: 2790947
Number of extensions: 27242675
Number of successful extensions: 67475
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 55
Number of HSP's that attempted gapping in prelim test: 67380
Number of HSP's gapped (non-prelim): 114
length of query: 388
length of database: 848,049,833
effective HSP length: 129
effective length of query: 259
effective length of database: 488,017,670
effective search space: 126396576530
effective search space used: 126396576530
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC137603.10


