

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.9 + phase: 0
(354 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
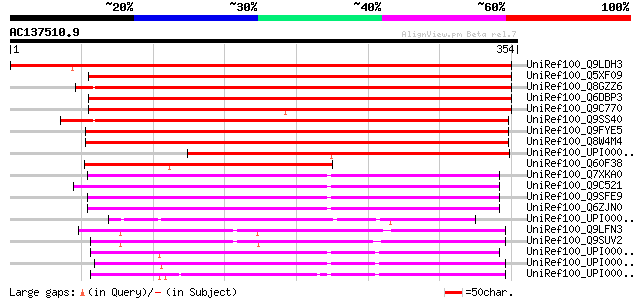
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LDH3 T12C24.5 [Arabidopsis thaliana] 517 e-145
UniRef100_Q5XF09 At3g11320 [Arabidopsis thaliana] 399 e-110
UniRef100_Q8GZZ6 Putative phosphate/phosphoenolpyruvate transloc... 398 e-109
UniRef100_Q6DBP3 At5g05820 [Arabidopsis thaliana] 394 e-108
UniRef100_Q9C770 Integral membrane protein, putative; 85705-8418... 380 e-104
UniRef100_Q9SS40 F14P13.11 protein [Arabidopsis thaliana] 371 e-101
UniRef100_Q9FYE5 Phosphate/phosphoenolpyruvate translocator-like... 370 e-101
UniRef100_Q8W4M4 Phosphate/phosphoenolpyruvate translocator-like... 369 e-101
UniRef100_UPI000032B618 UPI000032B618 UniRef100 entry 227 4e-58
UniRef100_Q60F38 Putative phosphoenolpyruvate translocator [Oryz... 211 2e-53
UniRef100_Q7XKA0 OSJNBb0020J19.10 protein [Oryza sativa] 178 2e-43
UniRef100_Q9C521 Hypothetical protein At1g77610 [Arabidopsis tha... 174 3e-42
UniRef100_Q9SFE9 T26F17.9 [Arabidopsis thaliana] 170 5e-41
UniRef100_Q6ZJN0 Putative glucose-6-phosphate/phosphate transloc... 170 6e-41
UniRef100_UPI000032CC96 UPI000032CC96 UniRef100 entry 150 5e-35
UniRef100_Q9LFN3 Hypothetical protein F2I11_120 [Arabidopsis tha... 134 5e-30
UniRef100_Q9SUV2 Hypothetical protein F8B4.90 [Arabidopsis thali... 129 1e-28
UniRef100_UPI000036626E UPI000036626E UniRef100 entry 128 3e-28
UniRef100_UPI00002BACDF UPI00002BACDF UniRef100 entry 127 6e-28
UniRef100_UPI000024880F UPI000024880F UniRef100 entry 126 8e-28
>UniRef100_Q9LDH3 T12C24.5 [Arabidopsis thaliana]
Length = 361
Score = 517 bits (1332), Expect = e-145
Identities = 267/356 (75%), Positives = 299/356 (83%), Gaps = 6/356 (1%)
Query: 1 MVEAQTWTTRRMSNPRLHTLDTNDQLQLDIPQTPPSDQRNNG------SNINNNNLVTTS 54
MVEAQ+WTTRRMSNPR T +DIP TPP ++ S+ + + T+
Sbjct: 1 MVEAQSWTTRRMSNPRFDAAATAAPTIVDIPGTPPHSSASSPLKPFFLSSPTVSPTILTA 60
Query: 55 LIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIH 114
IIA+W+ SNIGVLLLNKYLL +YG+RYPIFLTM HMLSCAAYS A IN+ VP Q I
Sbjct: 61 AIIAAWFGSNIGVLLLNKYLLFYYGFRYPIFLTMTHMLSCAAYSSAVINIAGIVPRQHIL 120
Query: 115 SKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAE 174
S++QFLKI +LSAIFC SVVCGNTSLRY+PVSFNQAIGATTPFFTA+F+FLITCK E+ E
Sbjct: 121 SRRQFLKILSLSAIFCLSVVCGNTSLRYIPVSFNQAIGATTPFFTAVFSFLITCKTESTE 180
Query: 175 VYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNL 234
VYLALLPVV GIV+++NSEP FHLFGFL+CV STAGRALKSVVQGIILTSE+EKLHSMNL
Sbjct: 181 VYLALLPVVSGIVLASNSEPSFHLFGFLICVASTAGRALKSVVQGIILTSESEKLHSMNL 240
Query: 235 LLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTK 294
LLYMAP+AA ILLP TLYIEGNV + IEKAR+DP I+FLL GNATVAYLVNLTNFLVTK
Sbjct: 241 LLYMAPMAACILLPFTLYIEGNVLRVLIEKARTDPLIIFLLAGNATVAYLVNLTNFLVTK 300
Query: 295 HTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
HTSALTLQVLGN KAAVAA VSVLIFRNPVTVMG+ GFG+TIMGVVLYSEA+KRSK
Sbjct: 301 HTSALTLQVLGNGKAAVAAGVSVLIFRNPVTVMGIAGFGVTIMGVVLYSEARKRSK 356
>UniRef100_Q5XF09 At3g11320 [Arabidopsis thaliana]
Length = 308
Score = 399 bits (1024), Expect = e-110
Identities = 201/295 (68%), Positives = 241/295 (81%)
Query: 56 IIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHS 115
++ASWY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ SY AI ++ VP Q I S
Sbjct: 14 LVASWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLLSYVAIAWMKMVPMQTIRS 73
Query: 116 KKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEV 175
+ QFLKI ALS +FC SVV GN SLR+LPVSFNQAIGATTPFFTA+FA+LIT K+E
Sbjct: 74 RVQFLKIAALSLVFCVSVVFGNISLRFLPVSFNQAIGATTPFFTAVFAYLITFKREAWLT 133
Query: 176 YLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLL 235
Y L+PVV G+V+++ SEP FHLFGF++C+ +TA RALKSV+QGI+L+SE EKL+SMNLL
Sbjct: 134 YFTLVPVVTGVVIASGSEPSFHLFGFIMCIAATAARALKSVLQGILLSSEGEKLNSMNLL 193
Query: 236 LYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKH 295
LYMAP+A + LLP TL +E NV ITI AR D IV+ L+ N+ +AY VNLTNFLVTKH
Sbjct: 194 LYMAPIAVVFLLPATLIMEKNVVGITIALARDDFRIVWYLLFNSALAYFVNLTNFLVTKH 253
Query: 296 TSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
TSALTLQVLGNAK AVA VVS+LIFRNPV+V GM G+ +T+ GV+LYSEAKKRSK
Sbjct: 254 TSALTLQVLGNAKGAVAVVVSILIFRNPVSVTGMLGYSLTVCGVILYSEAKKRSK 308
>UniRef100_Q8GZZ6 Putative phosphate/phosphoenolpyruvate translocator protein [Oryza
sativa]
Length = 322
Score = 398 bits (1022), Expect = e-109
Identities = 197/304 (64%), Positives = 248/304 (80%), Gaps = 1/304 (0%)
Query: 47 NNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQ 106
N T L+ A WY SNIGVLLLNKYLLS YG++YPIFLTM HM +CA SYAAI ++
Sbjct: 19 NGRFFTVGLVTA-WYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMSACALLSYAAIAWLR 77
Query: 107 FVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLI 166
VP Q + S+ Q KI ALS +FC SVV GN SLRYLPVSFNQA+GATTPFFTA+FA+++
Sbjct: 78 VVPMQLVRSRVQLAKIAALSLVFCGSVVSGNVSLRYLPVSFNQAVGATTPFFTAVFAYIM 137
Query: 167 TCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEA 226
T K+E+ YL L+PVV G+++++ EP FHLFGF++C+G+TA RALK+V+QGI+L+SE
Sbjct: 138 TVKRESWVTYLTLVPVVTGVMIASGGEPSFHLFGFIMCIGATAARALKTVLQGILLSSEG 197
Query: 227 EKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVN 286
EKL+SMNLLLYMAP+A ++LLP T+++E NV ITIE A+ D IV+LL+ N+ +AY VN
Sbjct: 198 EKLNSMNLLLYMAPIAVILLLPATIFMEDNVVGITIELAKKDTTIVWLLLFNSCLAYFVN 257
Query: 287 LTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAK 346
LTNFLVTKHTSALTLQVLGNAK AVA VVS+LIFRNPV+V GM G+ +T++GV+LYSE+K
Sbjct: 258 LTNFLVTKHTSALTLQVLGNAKGAVAVVVSILIFRNPVSVTGMLGYTLTVIGVILYSESK 317
Query: 347 KRSK 350
KR+K
Sbjct: 318 KRNK 321
>UniRef100_Q6DBP3 At5g05820 [Arabidopsis thaliana]
Length = 309
Score = 394 bits (1012), Expect = e-108
Identities = 198/295 (67%), Positives = 240/295 (81%)
Query: 56 IIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHS 115
++ASWY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ SY AI ++ VP Q I S
Sbjct: 14 LVASWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLLSYVAIAWLKMVPMQTIRS 73
Query: 116 KKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEV 175
+ QF KI ALS +FC SVV GN SLR+LPVSFNQAIGATTPFFTA+FA+L+T KKE
Sbjct: 74 RVQFFKIAALSLVFCVSVVFGNISLRFLPVSFNQAIGATTPFFTAVFAYLMTRKKEAWLT 133
Query: 176 YLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLL 235
Y L+PVV G+V+++ EP FHLFGFL+C+ +TA RALKSV+QGI+L+SE EKL+SMNLL
Sbjct: 134 YFTLVPVVTGVVIASGGEPSFHLFGFLMCIAATAARALKSVLQGILLSSEGEKLNSMNLL 193
Query: 236 LYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKH 295
LYMAP+A ++LLP TL +E NV ITI AR D IV+ L+ N+ +AYLVNLTNFLVT H
Sbjct: 194 LYMAPIAVVLLLPATLIMEKNVVGITIALARDDFRIVWYLLFNSALAYLVNLTNFLVTNH 253
Query: 296 TSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
TSALTLQVLGNAK AVA VVS+LIF+NPV+V GM G+ +T+ GV+LYSEAKKR+K
Sbjct: 254 TSALTLQVLGNAKGAVAVVVSILIFKNPVSVTGMLGYSLTVCGVILYSEAKKRNK 308
>UniRef100_Q9C770 Integral membrane protein, putative; 85705-84183 [Arabidopsis
thaliana]
Length = 344
Score = 380 bits (977), Expect = e-104
Identities = 201/331 (60%), Positives = 241/331 (72%), Gaps = 36/331 (10%)
Query: 56 IIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHS 115
++ASWY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ SY AI ++ VP Q I S
Sbjct: 14 LVASWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLLSYVAIAWMKMVPMQTIRS 73
Query: 116 KKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEV 175
+ QFLKI ALS +FC SVV GN SLR+LPVSFNQAIGATTPFFTA+FA+LIT K+E
Sbjct: 74 RVQFLKIAALSLVFCVSVVFGNISLRFLPVSFNQAIGATTPFFTAVFAYLITFKREAWLT 133
Query: 176 YLALLPVVLGIVVSTN------------------------------------SEPLFHLF 199
Y L+PVV G+V+++ SEP FHLF
Sbjct: 134 YFTLVPVVTGVVIASGKHPWDLPELDHFMICSLYAAQIRLEVVLLCVHSVNPSEPSFHLF 193
Query: 200 GFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFA 259
GF++C+ +TA RALKSV+QGI+L+SE EKL+SMNLLLYMAP+A + LLP TL +E NV
Sbjct: 194 GFIMCIAATAARALKSVLQGILLSSEGEKLNSMNLLLYMAPIAVVFLLPATLIMEKNVVG 253
Query: 260 ITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLI 319
ITI AR D IV+ L+ N+ +AY VNLTNFLVTKHTSALTLQVLGNAK AVA VVS+LI
Sbjct: 254 ITIALARDDFRIVWYLLFNSALAYFVNLTNFLVTKHTSALTLQVLGNAKGAVAVVVSILI 313
Query: 320 FRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
FRNPV+V GM G+ +T+ GV+LYSEAKKRSK
Sbjct: 314 FRNPVSVTGMLGYSLTVCGVILYSEAKKRSK 344
>UniRef100_Q9SS40 F14P13.11 protein [Arabidopsis thaliana]
Length = 355
Score = 371 bits (952), Expect = e-101
Identities = 182/313 (58%), Positives = 242/313 (77%), Gaps = 1/313 (0%)
Query: 36 SDQRNNGSNINNNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCA 95
SD ++ S+ L +SLII WY SNIGVLLLNK+LLS YG+++PIFLTM HM +CA
Sbjct: 42 SDMSSSSSSPKKQTLFISSLIIL-WYTSNIGVLLLNKFLLSNYGFKFPIFLTMCHMSACA 100
Query: 96 AYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATT 155
SY +I ++ VP Q + S+ QFLK+ LS +FC SVV GN SLRYLPVSFNQA+GATT
Sbjct: 101 ILSYVSIVFLKLVPLQYLKSRSQFLKVATLSIVFCASVVGGNISLRYLPVSFNQAVGATT 160
Query: 156 PFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKS 215
PFFTA+FA+++T K+E Y AL+PVV G+V+++ EP FH FGF++C+ +TA RA KS
Sbjct: 161 PFFTALFAYIMTFKREAWVTYGALVPVVTGVVIASGGEPGFHWFGFIMCISATAARAFKS 220
Query: 216 VVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLL 275
V+QGI+L+SE E+L+SMNL+LYM+P+A + LLPVT+++E +V ++T+ R ++ LL
Sbjct: 221 VLQGILLSSEGERLNSMNLMLYMSPIAVIALLPVTIFMEPDVMSVTLTLGRQHKYMYILL 280
Query: 276 IGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGIT 335
+ N+ +AY NL NFLVTKHTSALTLQVLGNAK AVA V+S+L+FRNPVTVMG+ G+ IT
Sbjct: 281 LVNSVMAYSANLLNFLVTKHTSALTLQVLGNAKGAVAVVISILLFRNPVTVMGIGGYSIT 340
Query: 336 IMGVVLYSEAKKR 348
++GVV Y E K+R
Sbjct: 341 VLGVVAYGETKRR 353
>UniRef100_Q9FYE5 Phosphate/phosphoenolpyruvate translocator-like protein
[Arabidopsis thaliana]
Length = 309
Score = 370 bits (949), Expect = e-101
Identities = 180/295 (61%), Positives = 235/295 (79%)
Query: 54 SLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQI 113
S +I SWY SNIGVLLLNK+LLS YG+++PIFLTM HM +CA SY +I ++ VP Q +
Sbjct: 13 STLIISWYSSNIGVLLLNKFLLSNYGFKFPIFLTMCHMSACAILSYISIVFLKLVPLQHL 72
Query: 114 HSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETA 173
S+ QFLK+ LS +FC SVV GN SLRYLPVSFNQA+GATTPFFTA+FA+L+T K+E
Sbjct: 73 KSRSQFLKVATLSIVFCASVVGGNISLRYLPVSFNQAVGATTPFFTALFAYLMTFKREAW 132
Query: 174 EVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMN 233
Y AL+PVV G+V+++ EP FH FGF++C+ +TA RA KSV+QGI+L+SE EKL+SMN
Sbjct: 133 VTYGALVPVVAGVVIASGGEPGFHWFGFIMCISATAARAFKSVLQGILLSSEGEKLNSMN 192
Query: 234 LLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVT 293
L+LYM+P+A + LLPVTL++E +V ++T+ A+ ++ LL+ N+ +AY NL NFLVT
Sbjct: 193 LMLYMSPIAVIALLPVTLFMEPDVISVTLTLAKQHQYMWILLLVNSVMAYSANLLNFLVT 252
Query: 294 KHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKR 348
KHTSALTLQVLGNAK AVA V+S+LIF+NPVTVMG+ G+ IT++GVV Y E K+R
Sbjct: 253 KHTSALTLQVLGNAKGAVAVVISILIFQNPVTVMGIGGYSITVLGVVAYGETKRR 307
>UniRef100_Q8W4M4 Phosphate/phosphoenolpyruvate translocator-like protein
[Arabidopsis thaliana]
Length = 309
Score = 369 bits (948), Expect = e-101
Identities = 180/295 (61%), Positives = 235/295 (79%)
Query: 54 SLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQI 113
S +I SWY SNIGVLLLNK+LLS YG+++PIFLTM HM +CA SY +I ++ VP Q +
Sbjct: 13 STLIISWYSSNIGVLLLNKFLLSNYGFKFPIFLTMCHMSACAILSYISIVFLKLVPLQHL 72
Query: 114 HSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETA 173
S+ QFLK+ LS +FC SVV GN SLRYLPVSFNQA+GATTPFFTA+FA+L+T K+E
Sbjct: 73 KSRSQFLKVATLSIVFCASVVGGNISLRYLPVSFNQAVGATTPFFTALFAYLMTFKREAW 132
Query: 174 EVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMN 233
Y AL+PVV G+V+++ EP FH FGF++C+ +TA RA KSV+QGI+L+SE EKL+SMN
Sbjct: 133 VTYGALVPVVAGVVIASGGEPGFHWFGFIMCISATAARAFKSVLQGILLSSEGEKLNSMN 192
Query: 234 LLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVT 293
L+LYM+P+A + LLPVTL++E +V ++T+ A+ ++ LL+ N+ +AY NL NFLVT
Sbjct: 193 LMLYMSPVAVIALLPVTLFMEPDVISVTLTLAKQHQYMWILLLVNSVMAYSANLLNFLVT 252
Query: 294 KHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKR 348
KHTSALTLQVLGNAK AVA V+S+LIF+NPVTVMG+ G+ IT++GVV Y E K+R
Sbjct: 253 KHTSALTLQVLGNAKGAVAVVISILIFQNPVTVMGIGGYSITVLGVVAYGETKRR 307
>UniRef100_UPI000032B618 UPI000032B618 UniRef100 entry
Length = 252
Score = 227 bits (578), Expect = 4e-58
Identities = 120/237 (50%), Positives = 156/237 (65%), Gaps = 12/237 (5%)
Query: 125 LSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLPVVL 184
L+ F SV+ GN SLRY+PVSFNQA+GATTPFFTAIFA+ + KKE+ Y++L+PVV
Sbjct: 1 LAVTFALSVLGGNVSLRYIPVSFNQALGATTPFFTAIFAYALLKKKESTATYMSLVPVVA 60
Query: 185 GIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILT------------SEAEKLHSM 232
GI ++T EP F+ GF+ C+ RALKSV+QG +LT S KL SM
Sbjct: 61 GITIATWGEPSFNFIGFVACLVGVCCRALKSVLQGWLLTPVGDKEAEKISNSNENKLDSM 120
Query: 233 NLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLV 292
+LL YMAP+A + L T +E + E +P + +L+ N +AYLVNLTNFLV
Sbjct: 121 SLLYYMAPVAVVTLGVCTFIMEPEAISAFYEATEFNPNFIVILLSNCFIAYLVNLTNFLV 180
Query: 293 TKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRS 349
T H ALTLQVLGNAK V VVS+L+FRNP+T + G+ IT++GV LYS +K+RS
Sbjct: 181 TAHVGALTLQVLGNAKGVVCTVVSILLFRNPITFRSVAGYTITMIGVWLYSSSKRRS 237
>UniRef100_Q60F38 Putative phosphoenolpyruvate translocator [Oryza sativa]
Length = 216
Score = 211 bits (538), Expect = 2e-53
Identities = 101/175 (57%), Positives = 131/175 (74%), Gaps = 2/175 (1%)
Query: 53 TSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPY-- 110
T+ ++ASWY SNIGVLLLNK+LLS YG+RYP+FLT HM +CA SYAA P
Sbjct: 41 TAGLVASWYASNIGVLLLNKFLLSTYGFRYPVFLTACHMSACALLSYAAAAASAAAPRAA 100
Query: 111 QQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKK 170
+ S+ Q ++ L A+FC SVV GN SLRYLPVSFNQA+GATTPFFTA+ A+ + ++
Sbjct: 101 RPRRSRGQLARVALLGAVFCASVVAGNVSLRYLPVSFNQAVGATTPFFTAVLAYAVAARR 160
Query: 171 ETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSE 225
E Y AL+PVV G+V++T EP FHLFGF++C+G+TA RALK+V+QGI+L+SE
Sbjct: 161 EACATYAALIPVVAGVVIATGGEPSFHLFGFIMCIGATAARALKTVLQGILLSSE 215
>UniRef100_Q7XKA0 OSJNBb0020J19.10 protein [Oryza sativa]
Length = 350
Score = 178 bits (452), Expect = 2e-43
Identities = 95/288 (32%), Positives = 165/288 (56%), Gaps = 2/288 (0%)
Query: 55 LIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIH 114
L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y AI+V++ P Q+
Sbjct: 20 LAILQWWGFNVTVIIINKWIFQKLDFKFPLTVSCVHFICSSIGAYIAIHVLKAKPLIQVE 79
Query: 115 SKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAE 174
+ ++ +IF +S +FC ++V GN SLRY+PVSF Q I + TP T I +L+ K
Sbjct: 80 PEDRWRRIFPMSFVFCINIVLGNVSLRYIPVSFMQTIKSFTPATTVILQWLVWSKHFEWR 139
Query: 175 VYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNL 234
++ +L+P+V GI++++ +E F++FGF + + K+++ +L K S+N
Sbjct: 140 IWASLVPIVGGILLTSITELSFNMFGFCAAMVGCLATSTKTILAESLL--HGYKFDSINT 197
Query: 235 LLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTK 294
+ YMAP A MIL + +EG S + ++IG+ +A+ +N + F V
Sbjct: 198 VYYMAPFATMILALPAVLLEGGGVVTWFYTHDSIASALVIIIGSGVLAFCLNFSIFYVIH 257
Query: 295 HTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
T+A+T V GN K AVA +VS LIFRNP++ M G IT++G Y
Sbjct: 258 STTAVTFNVAGNLKVAVAVLVSWLIFRNPISPMNAIGCAITLVGCTFY 305
>UniRef100_Q9C521 Hypothetical protein At1g77610 [Arabidopsis thaliana]
Length = 336
Score = 174 bits (442), Expect = 3e-42
Identities = 92/298 (30%), Positives = 168/298 (55%), Gaps = 2/298 (0%)
Query: 45 INNNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINV 104
+ ++ + L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y I V
Sbjct: 1 MEEGSMFRSLLAILQWWGFNVTVIIMNKWIFQKLDFKFPLSVSCVHFICSSIGAYIVIKV 60
Query: 105 VQFVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAF 164
++ P + + ++ +IF +S +FC ++V GN SLRY+PVSF Q I + TP T + +
Sbjct: 61 LKLKPLIVVDPEDRWRRIFPMSFVFCINIVLGNVSLRYIPVSFMQTIKSFTPATTVVLQW 120
Query: 165 LITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTS 224
L+ K ++ +L+P+V GI++++ +E F++FGF + + K+++ +L
Sbjct: 121 LVWRKYFDWRIWASLVPIVGGILLTSVTELSFNMFGFCAALFGCLATSTKTILAESLL-- 178
Query: 225 EAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYL 284
K S+N + YMAP A MIL L +EG+ E + + +++ + +A+
Sbjct: 179 HGYKFDSINTVYYMAPFATMILGIPALLLEGSGILSWFEAHPAPWSALIIILSSGVLAFC 238
Query: 285 VNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
+N + F V T+A+T V GN K AVA +VS LIFRNP++ M G GIT++G Y
Sbjct: 239 LNFSIFYVIHSTTAVTFNVAGNLKVAVAVMVSWLIFRNPISYMNAVGCGITLVGCTFY 296
>UniRef100_Q9SFE9 T26F17.9 [Arabidopsis thaliana]
Length = 341
Score = 170 bits (431), Expect = 5e-41
Identities = 92/288 (31%), Positives = 161/288 (54%), Gaps = 2/288 (0%)
Query: 55 LIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIH 114
L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y I V++ P +
Sbjct: 17 LSILQWWGFNVTVIIMNKWIFQKLDFKFPLSVSCVHFICSSIGAYIVIKVLKLKPLIVVD 76
Query: 115 SKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAE 174
+ ++ +IF +S +FC ++V GN SLRY+PVSF Q I + TP T + +L+ K
Sbjct: 77 PEDRWRRIFPMSFVFCINIVLGNISLRYIPVSFMQTIKSLTPATTVVLQWLVWRKYFDWR 136
Query: 175 VYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNL 234
++ +L+P+V GI++++ +E F++FGF + + K+++ +L K S+N
Sbjct: 137 IWASLVPIVGGILLTSITELSFNVFGFCAALFGCLATSTKTILAESLL--HGYKFDSINT 194
Query: 235 LLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTK 294
+ YMAP A MIL +E N E S + +L + +A+ +N + F V +
Sbjct: 195 VYYMAPFATMILGLPAFLLERNGILDWFEAHPSPWSALIILFNSGVLAFCLNFSIFYVIQ 254
Query: 295 HTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
T+A+T V GN K AVA VS +IFRNP++ M G GIT++G Y
Sbjct: 255 STTAVTFNVAGNLKVAVAVFVSWMIFRNPISPMNAVGCGITLVGCTFY 302
>UniRef100_Q6ZJN0 Putative glucose-6-phosphate/phosphate translocator [Oryza sativa]
Length = 337
Score = 170 bits (430), Expect = 6e-41
Identities = 90/288 (31%), Positives = 164/288 (56%), Gaps = 2/288 (0%)
Query: 55 LIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIH 114
L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y AI +++ P ++
Sbjct: 16 LAILQWWGFNVTVIIMNKWIFQKLEFKFPLTVSCVHFICSSIGAYIAIKILKMKPLIEVA 75
Query: 115 SKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAE 174
+ ++ +IF +S +FC ++V GN SLRY+PVSF Q I + TP T I +L+ K
Sbjct: 76 PEDRWRRIFPMSFVFCINIVLGNVSLRYIPVSFMQTIKSFTPATTVILQWLVWRKYFEWR 135
Query: 175 VYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNL 234
++ +L+P+V GI++++ +E F++FGF + + K+++ +L K S+N
Sbjct: 136 IWASLVPIVGGIMLTSITELSFNMFGFCAAMVGCLATSTKTILAESLL--HGYKFDSINT 193
Query: 235 LLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTK 294
+ YMAP A MIL + +EG+ + S + ++ + +A+ +N + F V
Sbjct: 194 VYYMAPFATMILSVPAIVLEGSGVINWLYTYDSIVPALIIITTSGVLAFCLNFSIFYVIH 253
Query: 295 HTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
T+A+T V GN K AVA +VS +IFRNP++ M G IT++G Y
Sbjct: 254 STTAVTFNVAGNLKVAVAVLVSWMIFRNPISAMNAVGCAITLVGCTFY 301
>UniRef100_UPI000032CC96 UPI000032CC96 UniRef100 entry
Length = 272
Score = 150 bits (379), Expect = 5e-35
Identities = 97/259 (37%), Positives = 144/259 (55%), Gaps = 9/259 (3%)
Query: 70 LNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIF 129
+NK L+ + + P+FLT LHML + ++ + + I S+ + K+F LS +
Sbjct: 2 MNKVLMGEH-FALPVFLTFLHMLVSFLWCEFSMTM-GWTARGAIKSRAEGWKVFFLSQVM 59
Query: 130 CFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVS 189
SV+ S +Y+ VS QA+ A++P FTA +I K+E +V+L LLPVV G ++S
Sbjct: 60 ALSVLLAVASFKYVDVSLEQALAASSPAFTAAMGVVILKKRERGKVWLTLLPVVGGAMIS 119
Query: 190 TNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPV 249
P FG + + S R KS +Q ++L +A L S+NLL YMA + + LLP
Sbjct: 120 AGGVPEVSWFGVTLVILSNIARGTKSCMQELLLGKDA--LDSINLLRYMAAFSCLTLLPF 177
Query: 250 TLYIEGNVFAITIEK---ARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGN 306
+ IEG AI +E+ D I L+ N T A++VNL F VT++ AL++QVLGN
Sbjct: 178 SFVIEGP--AIIVERLSYVSRDGTIAAALVANCTGAFMVNLFQFQVTENVGALSMQVLGN 235
Query: 307 AKAAVAAVVSVLIFRNPVT 325
K + VSV +FRN VT
Sbjct: 236 LKNVFTSTVSVFVFRNAVT 254
>UniRef100_Q9LFN3 Hypothetical protein F2I11_120 [Arabidopsis thaliana]
Length = 351
Score = 134 bits (336), Expect = 5e-30
Identities = 87/303 (28%), Positives = 156/303 (50%), Gaps = 12/303 (3%)
Query: 49 NLVTTSLIIASWYFSNIGVLLLNKYLLS--FYGYRYPIFLTMLHMLSCAAYSYAAINVVQ 106
N+V + +A W F + V++ NKY+L Y + +PI LTM+HM C+ ++ I V +
Sbjct: 14 NIVLSYSYVAIWIFLSFTVIVYNKYILDKKMYNWPFPISLTMIHMSFCSTLAFLIIKVFK 73
Query: 107 FVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLI 166
FV ++ + + + A++ S+ N++ YL VSF Q + A P A+++ +
Sbjct: 74 FVEPVKMTRETYLRSVVPIGALYALSLWLSNSAYIYLSVSFIQMLKALMP--VAVYSIGV 131
Query: 167 TCKKE--TAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTS 224
KKE ++ + +L + G+ ++ E F ++G ++ +G+ A A + V+ I+L
Sbjct: 132 LFKKEGFKSDTMMNMLSISFGVAIAAYGEARFDVWGVILQLGAVAFEATRLVLIQILLGD 191
Query: 225 EAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIG-NATVAY 283
+ KL+ + L Y+AP L +Y+E V T S + + + G N+ A+
Sbjct: 192 KGIKLNPITSLYYVAPCCLAFLFIPWIYVEFPVLRDT-----SSFHLDYAIFGANSFCAF 246
Query: 284 LVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYS 343
+NL FL+ TSALT+ V G K + S + ++ VT + + G+GI +GV Y+
Sbjct: 247 ALNLAVFLLVGKTSALTMNVAGVVKDWLLIAFSWSVIKDTVTPINLFGYGIAFLGVAYYN 306
Query: 344 EAK 346
AK
Sbjct: 307 HAK 309
>UniRef100_Q9SUV2 Hypothetical protein F8B4.90 [Arabidopsis thaliana]
Length = 350
Score = 129 bits (324), Expect = 1e-28
Identities = 83/295 (28%), Positives = 150/295 (50%), Gaps = 12/295 (4%)
Query: 57 IASWYFSNIGVLLLNKYLLS--FYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIH 114
+A W F + V++ NKY+L Y + +PI LTM+HM C++ + I V + V +
Sbjct: 22 VAIWIFLSFTVIVYNKYILDKKMYNWPFPITLTMIHMAFCSSLAVILIKVFKIVEPVSMS 81
Query: 115 SKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKET-- 172
+ + A++ S+ N++ YL VSF Q + A P A+++ + KKE+
Sbjct: 82 RDTYIRSVVPIGALYSLSLWLSNSAYIYLSVSFIQMLKALMP--VAVYSIGVLLKKESFK 139
Query: 173 AEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSM 232
+E +L + G+ ++ E F +G ++ +G+ A A + V+ I+LTS+ L+ +
Sbjct: 140 SETMTNMLSISFGVAIAAYGEAKFDTWGVMLQLGAVAFEATRLVLIQILLTSKGINLNPI 199
Query: 233 NLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIG-NATVAYLVNLTNFL 291
L Y+AP + L +++E + I + S F++ G N+ A+ +NL FL
Sbjct: 200 TSLYYVAPCCLVFLFFPWIFVE-----LPILRETSSFHFDFVIFGTNSVCAFALNLAVFL 254
Query: 292 VTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAK 346
+ TSALT+ V G K + S + ++ VT + + G+G+ +GV Y+ K
Sbjct: 255 LVGKTSALTMNVAGVVKDWLLIAFSWSVIKDTVTPLNLFGYGLAFLGVAYYNHCK 309
>UniRef100_UPI000036626E UPI000036626E UniRef100 entry
Length = 307
Score = 128 bits (321), Expect = 3e-28
Identities = 87/289 (30%), Positives = 144/289 (49%), Gaps = 7/289 (2%)
Query: 57 IASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAIN--VVQFVPY-QQI 113
+ W + + LNK++ + + +RYP+ L+ LHML+ Y I VV+ + +Q
Sbjct: 22 VVVWLVTGSTISSLNKWIFAVFNFRYPLLLSALHMLTAIVVDYGLIKLRVVRHIGVREQD 81
Query: 114 HSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETA 173
+ K+F LS FC S+ GN L Y+ +SF Q I TTP FT + L+ K+
Sbjct: 82 LTPSAKCKVFMLSLTFCASIAFGNMGLNYVQLSFAQMIYTTTPIFTLAISTLVLGKQHHI 141
Query: 174 EVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMN 233
Y A++P+ LG S E F G L +T R +KS+ Q I+L + EK++S+
Sbjct: 142 LKYTAMMPICLGASFSIMGEVQFDQTGCLFVFAATMLRGVKSIQQSILL--QEEKINSVF 199
Query: 234 LLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVT 293
LL M+ + IL L +E +A+ D + ++ + + L NL + V
Sbjct: 200 LLYLMSIPSFCILAVAALALEN--WALLEWPLHYDRRLWLFILLSCLGSVLYNLASCCVI 257
Query: 294 KHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
TSA+TL +LGN ++S L+F + ++ + G +T+ G+ +Y
Sbjct: 258 SLTSAVTLHILGNLNVVGNLLLSQLLFGSELSALSCAGAVLTLSGMFIY 306
>UniRef100_UPI00002BACDF UPI00002BACDF UniRef100 entry
Length = 344
Score = 127 bits (318), Expect = 6e-28
Identities = 86/290 (29%), Positives = 145/290 (49%), Gaps = 7/290 (2%)
Query: 60 WYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINV--VQFVPY-QQIHSK 116
W + + LNK++ + + +RYP+ L+ LHML+ Y I + ++ V QQ +
Sbjct: 49 WLVTGSTISSLNKWIFAVFNFRYPLLLSALHMLTAMVVDYGLIKLRLIRHVGVRQQDLTP 108
Query: 117 KQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVY 176
K+F LS FC S+ GN L Y+ +SF Q I TTP FT + L+ K+ Y
Sbjct: 109 GAKCKVFMLSLTFCASIAFGNVGLNYVQLSFAQMIYTTTPIFTLAISTLVLGKQHHILKY 168
Query: 177 LALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLL 236
A++P+ LG S E F G +T R +KS+ Q I+L + EK++S+ LL
Sbjct: 169 TAMMPICLGASFSIMGEVQFDQTGCFFVFAATMLRGVKSIQQSILL--QEEKINSVFLLY 226
Query: 237 YMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHT 296
M+ + IL L +E +A+ D + ++ + + L NL + V T
Sbjct: 227 LMSIPSFCILAVAALALEN--WALLEWPLHYDRRLWVFILLSCLGSVLYNLASCCVISLT 284
Query: 297 SALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAK 346
SA+TL +LGN ++S L+F + ++ + G +T+ G+++Y ++
Sbjct: 285 SAVTLHILGNLNVVGNLLLSQLLFGSELSTLSCAGAVLTLSGMLIYQNSE 334
>UniRef100_UPI000024880F UPI000024880F UniRef100 entry
Length = 302
Score = 126 bits (317), Expect = 8e-28
Identities = 89/294 (30%), Positives = 149/294 (50%), Gaps = 10/294 (3%)
Query: 57 IASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAIN--VVQF--VPYQQ 112
++ W + + LNK++ + Y +RYP+ L+ LHML+ Y I VV+ V Q
Sbjct: 10 VSVWLVTGTTISSLNKWIFAVYNFRYPLLLSALHMLTAIVVDYGLIKSRVVRHKGVGEQD 69
Query: 113 IHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKET 172
+ + + K+F LS FC S+ GN L Y+ +SF Q I TTP FT + LI K+
Sbjct: 70 LTTSAK-CKVFLLSLTFCASIAFGNVGLNYVQLSFAQMIYTTTPLFTLAISALILGKQHH 128
Query: 173 AEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSM 232
Y A++P+ LG S E F G L +T R +K+ +Q I+L + EK++S+
Sbjct: 129 FLKYTAMMPICLGASFSIMGEVQFDQTGCLFVFAATMLRGVKT-IQRILL--QEEKINSV 185
Query: 233 NLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLV 292
LL M+ + IL L +E +A + D + ++ + + L NL + V
Sbjct: 186 FLLYLMSIPSFCILAVAALALEN--WAALQSPFQYDHHLWGFILLSCLGSVLYNLASCCV 243
Query: 293 TKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAK 346
TSA+TL +LGN ++S ++F + +T + G +T+ G+++Y ++
Sbjct: 244 ITLTSAVTLHILGNLNVVGNLLLSQVLFGHELTALSCAGAALTLSGMIIYQNSE 297
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 540,960,149
Number of Sequences: 2790947
Number of extensions: 21672831
Number of successful extensions: 118916
Number of sequences better than 10.0: 780
Number of HSP's better than 10.0 without gapping: 300
Number of HSP's successfully gapped in prelim test: 480
Number of HSP's that attempted gapping in prelim test: 118033
Number of HSP's gapped (non-prelim): 895
length of query: 354
length of database: 848,049,833
effective HSP length: 128
effective length of query: 226
effective length of database: 490,808,617
effective search space: 110922747442
effective search space used: 110922747442
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137510.9


