

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.8 - phase: 0 /pseudo
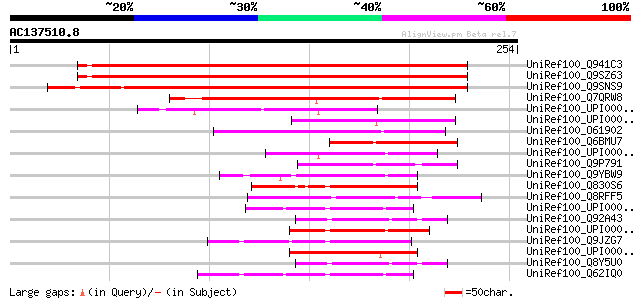
(254 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q941C3 AT4g11980/F16J13_50 [Arabidopsis thaliana] 292 5e-78
UniRef100_Q9SZ63 Hypothetical protein F16J13.50 [Arabidopsis tha... 276 3e-73
UniRef100_Q9SNS9 MutT/nudix protein-like [Oryza sativa] 263 4e-69
UniRef100_Q7QRW8 GLP_549_31342_32085 [Giardia lamblia ATCC 50803] 121 2e-26
UniRef100_UPI0000236600 UPI0000236600 UniRef100 entry 72 2e-11
UniRef100_UPI000031B0E2 UPI000031B0E2 UniRef100 entry 56 1e-06
UniRef100_O61902 Nudix family protein 2 [Caenorhabditis elegans] 53 9e-06
UniRef100_Q6BMU7 Similar to CA5909|CaYSA1 Candida albicans CaYSA... 53 9e-06
UniRef100_UPI000031FA7C UPI000031FA7C UniRef100 entry 51 3e-05
UniRef100_Q9P791 SPBP35G2.12 protein [Schizosaccharomyces pombe] 48 2e-04
UniRef100_Q9YBW9 Hypothetical protein APE1481 [Aeropyrum pernix] 48 2e-04
UniRef100_Q830S6 MutT/nudix family protein [Enterococcus faecalis] 48 3e-04
UniRef100_Q8RFF5 Phosphohydrolase [Fusobacterium nucleatum] 48 3e-04
UniRef100_UPI00003327D2 UPI00003327D2 UniRef100 entry 47 5e-04
UniRef100_Q92A43 Lin2079 protein [Listeria innocua] 47 5e-04
UniRef100_UPI000026B673 UPI000026B673 UniRef100 entry 47 6e-04
UniRef100_Q9JZG7 Hypothetical protein NMB1064 [Neisseria meningi... 47 6e-04
UniRef100_UPI00002D84F6 UPI00002D84F6 UniRef100 entry 46 8e-04
UniRef100_Q8Y5U0 Lmo1965 protein [Listeria monocytogenes] 46 8e-04
UniRef100_Q62IQ0 Pyrophosphatase, MutT/nudix family [Burkholderi... 46 8e-04
>UniRef100_Q941C3 AT4g11980/F16J13_50 [Arabidopsis thaliana]
Length = 309
Score = 292 bits (748), Expect = 5e-78
Identities = 152/195 (77%), Positives = 169/195 (85%), Gaps = 2/195 (1%)
Query: 35 KMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSSLFKQWLHNLQTENGI 94
KMSSS S SLT SITLPS+ +EPV + A G+SSSDF AIDSSLF+ WL NL++E+GI
Sbjct: 35 KMSSS--SSSLTQSITLPSQPNEPVLVSATAGISSSDFRDAIDSSLFRNWLRNLESESGI 92
Query: 95 LANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLES 154
LA+ +MTL+QVLIQGVDMFGKRIGFLKF A+I DKETG KVPGIVFARGPAVA+LILLES
Sbjct: 93 LADGSMTLKQVLIQGVDMFGKRIGFLKFKADIFDKETGQKVPGIVFARGPAVAVLILLES 152
Query: 155 EGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLT 214
+GETYAVLTEQ RVP G+I+LELPAGMLDDDKGD VGTAVREVEEE GIKL EDMVDLT
Sbjct: 153 DGETYAVLTEQVRVPTGKIVLELPAGMLDDDKGDFVGTAVREVEEEIGIKLKKEDMVDLT 212
Query: 215 AFLDSSTGSTVFPSP 229
AFLD STG +FPSP
Sbjct: 213 AFLDPSTGYRIFPSP 227
>UniRef100_Q9SZ63 Hypothetical protein F16J13.50 [Arabidopsis thaliana]
Length = 310
Score = 276 bits (707), Expect = 3e-73
Identities = 147/196 (75%), Positives = 164/196 (83%), Gaps = 3/196 (1%)
Query: 35 KMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSSLFK-QWLHNLQTENG 93
KMSSS S SLT SITLPS+ +EPV + A G+SSSDF +D F WL NL++E+G
Sbjct: 35 KMSSS--SSSLTQSITLPSQPNEPVLVSATAGISSSDFRRVLDIRGFPLNWLRNLESESG 92
Query: 94 ILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLE 153
ILA+ +MTL+QVLIQGVDMFGKRIGFLKF A+I DKETG KVPGIVFARGPAVA+LILLE
Sbjct: 93 ILADGSMTLKQVLIQGVDMFGKRIGFLKFKADIFDKETGQKVPGIVFARGPAVAVLILLE 152
Query: 154 SEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDL 213
S+GETYAVLTEQ RVP G+I+LELPAGMLDDDKGD VGTAVREVEEE GIKL EDMVDL
Sbjct: 153 SDGETYAVLTEQVRVPTGKIVLELPAGMLDDDKGDFVGTAVREVEEEIGIKLKKEDMVDL 212
Query: 214 TAFLDSSTGSTVFPSP 229
TAFLD STG +FPSP
Sbjct: 213 TAFLDPSTGYRIFPSP 228
>UniRef100_Q9SNS9 MutT/nudix protein-like [Oryza sativa]
Length = 325
Score = 263 bits (671), Expect = 4e-69
Identities = 132/210 (62%), Positives = 166/210 (78%), Gaps = 3/210 (1%)
Query: 20 APNLIPKKKKNGFFYKMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSS 79
AP+ P ++ SS +P L+ ++ +P + PV ++AAPG++ +DF SA++SS
Sbjct: 28 APSPAPSSRRGARM--ASSGDHAPQLSTAVAVPGAGA-PVRVVAAPGLTEADFTSAVESS 84
Query: 80 LFKQWLHNLQTENGILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIV 139
LF+QWL NLQ E G+L + LRQ+LIQGVDMFGKR+GF+KF A+IID+ET K+PGIV
Sbjct: 85 LFRQWLKNLQEEKGVLTYGRLNLRQILIQGVDMFGKRVGFVKFKADIIDEETKAKIPGIV 144
Query: 140 FARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEE 199
FARGPAVA+LILLES+G+TYAVLTEQ RVPVG+ ILELPAGMLDD+KGD VGTAVREVEE
Sbjct: 145 FARGPAVAVLILLESKGQTYAVLTEQVRVPVGKFILELPAGMLDDEKGDFVGTAVREVEE 204
Query: 200 ETGIKLNVEDMVDLTAFLDSSTGSTVFPSP 229
ETGIKLN+EDM+DLTA L+ TG + PSP
Sbjct: 205 ETGIKLNLEDMIDLTALLNPDTGCRMLPSP 234
>UniRef100_Q7QRW8 GLP_549_31342_32085 [Giardia lamblia ATCC 50803]
Length = 247
Score = 121 bits (303), Expect = 2e-26
Identities = 70/146 (47%), Positives = 93/146 (62%), Gaps = 12/146 (8%)
Query: 81 FKQWLHNLQTENGILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVF 140
F++W + ++ + +R + +Q VD FG RIGFLKF AE K G +VPGIVF
Sbjct: 36 FREWCEEI--------DENLDVRGLTVQSVDYFGARIGFLKFSAEAYSKIHGQRVPGIVF 87
Query: 141 ARGPAVAMLILL--ESEGETYAVLTEQARVPVGRI-ILELPAGMLDDDKGDIVGTAVREV 197
RG +V +L +L E E Y VLTEQARVPVG+ LE+PAGMLD++ GD+VG AV+E+
Sbjct: 88 MRGGSVGVLPVLIDEKTAEKYIVLTEQARVPVGKASFLEIPAGMLDEN-GDVVGVAVQEM 146
Query: 198 EEETGIKLNVEDMVDLTAFLDSSTGS 223
EETGI L D+ L ++ S GS
Sbjct: 147 AEETGISLKRSDLCSLGSYYTSPGGS 172
>UniRef100_UPI0000236600 UPI0000236600 UniRef100 entry
Length = 332
Score = 71.6 bits (174), Expect = 2e-11
Identities = 52/136 (38%), Positives = 69/136 (50%), Gaps = 20/136 (14%)
Query: 65 PGVSSSDFWSAIDSSLFKQWLHNLQTE-------NGILANDTMTLRQVLIQGVDMF-GKR 116
PG+S D F+ W LQ + D LR++ +Q VD F G R
Sbjct: 160 PGLSKEDLSRF---PAFRVWFATLQRSLSRQKDPSHEFHKDPYLLRKIEVQAVDFFQGGR 216
Query: 117 IGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLE-------SEGETYAVLTEQARVP 169
+GF+K AEI G +PG VF RG +V ML+LL+ E + +AVLT Q R+P
Sbjct: 217 LGFVKLRAEI-SNAGGESLPGSVFLRGGSVGMLLLLQPHDVPSTEEDDKWAVLTVQPRIP 275
Query: 170 VGRIIL-ELPAGMLDD 184
G + E+PAGMLDD
Sbjct: 276 AGSLAFSEIPAGMLDD 291
>UniRef100_UPI000031B0E2 UPI000031B0E2 UniRef100 entry
Length = 161
Score = 55.8 bits (133), Expect = 1e-06
Identities = 33/88 (37%), Positives = 50/88 (56%), Gaps = 6/88 (6%)
Query: 142 RGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGML------DDDKGDIVGTAVR 195
R AVA+++ +GE +A+L Q R P G+ +LE AGML +DD +VG AV
Sbjct: 2 RADAVAVILTEVVDGEEWALLVNQPRAPCGKWMLECVAGMLQKMEGDEDDNFTVVGKAVD 61
Query: 196 EVEEETGIKLNVEDMVDLTAFLDSSTGS 223
E++EE G+ L +V L + S+ G+
Sbjct: 62 ELKEEAGLDLAAGKLVKLGTYWPSTGGT 89
>UniRef100_O61902 Nudix family protein 2 [Caenorhabditis elegans]
Length = 223
Score = 52.8 bits (125), Expect = 9e-06
Identities = 33/116 (28%), Positives = 62/116 (53%), Gaps = 1/116 (0%)
Query: 103 RQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVL 162
++V+ G + +++GF ++ ++ ++ V A V+++ + +G+ Y VL
Sbjct: 34 QEVVWNGKWIQTRQVGFKTHTGQVGVWQSVHRNTKPVEASADGVSIIARVRKQGKLYIVL 93
Query: 163 TEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLD 218
+Q R+P G++ LELPAG++D + A+RE++EETG M FLD
Sbjct: 94 VKQYRIPCGKLCLELPAGLIDAGE-TAQQAAIRELKEETGYVSGKVVMESKLCFLD 148
>UniRef100_Q6BMU7 Similar to CA5909|CaYSA1 Candida albicans CaYSA1 sugar-nucleotide
hydrolase [Debaryomyces hansenii]
Length = 248
Score = 52.8 bits (125), Expect = 9e-06
Identities = 30/64 (46%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query: 161 VLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLDSS 220
VLT+Q R PVG +++ELPAG++ D K + TAVRE+ EETG + D A L S
Sbjct: 113 VLTKQFRPPVGAVVIELPAGLV-DPKESVESTAVRELIEETGYHSTFNHLTDSMADLVSD 171
Query: 221 TGST 224
G T
Sbjct: 172 PGLT 175
>UniRef100_UPI000031FA7C UPI000031FA7C UniRef100 entry
Length = 195
Score = 50.8 bits (120), Expect = 3e-05
Identities = 30/88 (34%), Positives = 49/88 (55%), Gaps = 3/88 (3%)
Query: 129 KETGNKVPGIVFARGPAVAMLILLE--SEGETYAVLTEQARVPVGRIILELPAGMLDDDK 186
K + P + RG V ++ +L+ +G Y +L EQ R+ G + LE PAGM+D D
Sbjct: 32 KNSDISFPRCLILRGDTVIIVPILKCIDDGHIYTILVEQFRIIDGGLSLEFPAGMIDYDI 91
Query: 187 GDIVGTAVREVEEETGIKLNVEDMVDLT 214
G +A+ E +EE I +N +D++ L+
Sbjct: 92 GS-KASAIIECKEELNIDINDKDLIPLS 118
>UniRef100_Q9P791 SPBP35G2.12 protein [Schizosaccharomyces pombe]
Length = 205
Score = 48.1 bits (113), Expect = 2e-04
Identities = 27/80 (33%), Positives = 46/80 (56%), Gaps = 4/80 (5%)
Query: 145 AVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIK 204
AVA+L ++ +G + + +Q R P+G+ +E+PAG++ D K A+RE+ EETG
Sbjct: 55 AVAILAIVPIDGSPHVLCQKQFRPPIGKFCIEIPAGLV-DSKESCEDAAIRELREETGY- 112
Query: 205 LNVEDMVDLTAFLDSSTGST 224
V ++D T + + G T
Sbjct: 113 --VGTVMDSTTVMYNDPGLT 130
>UniRef100_Q9YBW9 Hypothetical protein APE1481 [Aeropyrum pernix]
Length = 188
Score = 48.1 bits (113), Expect = 2e-04
Identities = 37/101 (36%), Positives = 54/101 (52%), Gaps = 8/101 (7%)
Query: 106 LIQGVDMFGKRIGFLKFIAEIIDKETGNK--VPGIVFARGPAVAMLILLESEGETYAVLT 163
L V+ G+R+ KF A + G + V +VF +VA+L L+E +GE + VL
Sbjct: 7 LALNVECRGRRV---KFEARMETLPNGRQILVDRVVFP--DSVAVLPLVEKDGEWHVVLV 61
Query: 164 EQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIK 204
Q R +G+ LE PAG L + + A RE+EEE G+K
Sbjct: 62 RQFRPSIGKWTLEAPAGTLKEGETP-EAAAARELEEEAGLK 101
>UniRef100_Q830S6 MutT/nudix family protein [Enterococcus faecalis]
Length = 198
Score = 47.8 bits (112), Expect = 3e-04
Identities = 34/84 (40%), Positives = 52/84 (61%), Gaps = 5/84 (5%)
Query: 122 FIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGM 181
F+ ++ G +VF G AVAM I L +EG+ VL +Q R P+ ++ILE+PAG
Sbjct: 30 FLDDVALPTGGTAKRELVFHSG-AVAM-IPLTAEGKI--VLVKQFRKPLEQVILEIPAGK 85
Query: 182 LD-DDKGDIVGTAVREVEEETGIK 204
+D ++ + TA+RE+EEETG +
Sbjct: 86 IDPGEENQLETTAMRELEEETGYR 109
>UniRef100_Q8RFF5 Phosphohydrolase [Fusobacterium nucleatum]
Length = 171
Score = 47.8 bits (112), Expect = 3e-04
Identities = 40/117 (34%), Positives = 55/117 (46%), Gaps = 8/117 (6%)
Query: 120 LKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPA 179
LKF+ +D + N + A+A LIL S + V Q R V I E+PA
Sbjct: 9 LKFLKVGVDTDPLNNHNLEYLEKQNAIAALILNHSGDKVLFV--NQYRAGVHNYIYEVPA 66
Query: 180 GMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLDSSTGSTVFPSPSAESVLI 236
G++++D+ IV REV EETG K D DS+TG V P + E + I
Sbjct: 67 GLIENDEKPIVALE-REVREETGYKRE-----DYDILYDSNTGFLVSPGYTTEKIYI 117
>UniRef100_UPI00003327D2 UPI00003327D2 UniRef100 entry
Length = 196
Score = 47.0 bits (110), Expect = 5e-04
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query: 119 FLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELP 178
FLK + + G K + + P M+I L +G ++ Q R P+G+++ E P
Sbjct: 26 FLKLKRDTVRLPDGKKATR-EYVQHPGAVMVIPLFDDGRV--LMESQYRYPIGKVMAEFP 82
Query: 179 AGMLDDDKGDIVGTAVREVEEETG 202
AG LD ++G + AVRE+ EETG
Sbjct: 83 AGKLDPNEG-ALACAVRELREETG 105
>UniRef100_Q92A43 Lin2079 protein [Listeria innocua]
Length = 185
Score = 47.0 bits (110), Expect = 5e-04
Identities = 29/76 (38%), Positives = 45/76 (59%), Gaps = 5/76 (6%)
Query: 144 PAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGI 203
P +I ++G Y L EQ R P+ + I+E+PAG ++ + +V TA RE+EEETG
Sbjct: 43 PGAVAIIPFSADGRMY--LVEQFRKPLEKNIIEIPAGKMEPGEDPLV-TAKRELEEETGF 99
Query: 204 KLNVEDMVDLTAFLDS 219
+ +D+ LT+F S
Sbjct: 100 Q--SDDLTYLTSFYTS 113
>UniRef100_UPI000026B673 UPI000026B673 UniRef100 entry
Length = 165
Score = 46.6 bits (109), Expect = 6e-04
Identities = 28/70 (40%), Positives = 42/70 (60%), Gaps = 3/70 (4%)
Query: 141 ARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEE 200
+R AV ++ L S G VL Q R P G+ +LE PAG++D+ + TA+RE++EE
Sbjct: 20 SRSSAVVIVPRLVSSGSY--VLIRQFRPPTGKYLLEFPAGLIDEGESP-AETALRELKEE 76
Query: 201 TGIKLNVEDM 210
TG N+E +
Sbjct: 77 TGYFGNIESV 86
>UniRef100_Q9JZG7 Hypothetical protein NMB1064 [Neisseria meningitidis]
Length = 178
Score = 46.6 bits (109), Expect = 6e-04
Identities = 35/102 (34%), Positives = 51/102 (49%), Gaps = 5/102 (4%)
Query: 100 MTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETY 159
M LR+V + G ++ GF+ + + GN+ IV R P A ++ + EG+
Sbjct: 1 MDLREVKLGGETIYEG--GFVSISRDKVRLPNGNEGQRIVI-RHPGAACVLAVTDEGKV- 56
Query: 160 AVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEET 201
VL Q R + LELPAG LD D+ A+RE+ EET
Sbjct: 57 -VLVRQWRYAANQATLELPAGKLDVAGEDMAACALRELAEET 97
>UniRef100_UPI00002D84F6 UPI00002D84F6 UniRef100 entry
Length = 139
Score = 46.2 bits (108), Expect = 8e-04
Identities = 26/67 (38%), Positives = 42/67 (61%), Gaps = 3/67 (4%)
Query: 141 ARGPAVAMLILLESEGE-TYAVLTEQARVPVGRIILELPAGMLDD--DKGDIVGTAVREV 197
ARG A+++ ++ + + +L +Q RVP+GR +ELPAG++ D + D A RE+
Sbjct: 33 ARGIHAAVILAIDDADDGPHVLLVDQYRVPLGRRCIELPAGLVGDHEEGEDASIAAAREL 92
Query: 198 EEETGIK 204
EEETG +
Sbjct: 93 EEETGFR 99
>UniRef100_Q8Y5U0 Lmo1965 protein [Listeria monocytogenes]
Length = 185
Score = 46.2 bits (108), Expect = 8e-04
Identities = 29/76 (38%), Positives = 44/76 (57%), Gaps = 5/76 (6%)
Query: 144 PAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGI 203
P +I +G Y L EQ R P+ + I+E+PAG ++ + +V TA RE+EEETG
Sbjct: 43 PGAVAIIPFSEDGGMY--LVEQYRKPLEKTIIEIPAGKMEPGEDPLV-TARRELEEETGF 99
Query: 204 KLNVEDMVDLTAFLDS 219
+ +D+ LT+F S
Sbjct: 100 Q--SDDLTYLTSFYTS 113
>UniRef100_Q62IQ0 Pyrophosphatase, MutT/nudix family [Burkholderia mallei]
Length = 196
Score = 46.2 bits (108), Expect = 8e-04
Identities = 33/108 (30%), Positives = 52/108 (47%), Gaps = 6/108 (5%)
Query: 95 LANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLES 154
L N TL + I+ ++ FLK + + G + P M+I L
Sbjct: 4 LPNHDATLTETCIESESVYDG--AFLKVKRDTVRLPDGKHATR-EYVTHPGAVMVIPLFD 60
Query: 155 EGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETG 202
+G ++ Q R P+G+++ E PAG LD ++G + AVRE+ EETG
Sbjct: 61 DGRV--LMESQYRYPIGKVMAEFPAGKLDPNEG-ALACAVRELREETG 105
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 401,674,040
Number of Sequences: 2790947
Number of extensions: 16353622
Number of successful extensions: 46422
Number of sequences better than 10.0: 292
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 236
Number of HSP's that attempted gapping in prelim test: 46250
Number of HSP's gapped (non-prelim): 299
length of query: 254
length of database: 848,049,833
effective HSP length: 124
effective length of query: 130
effective length of database: 501,972,405
effective search space: 65256412650
effective search space used: 65256412650
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC137510.8


