

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.8 + phase: 0
(518 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
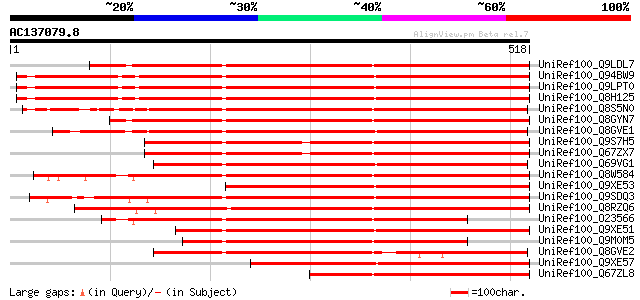
UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thal... 554 e-156
UniRef100_Q94BW9 F17J6.12/F17J6.12 [Arabidopsis thaliana] 553 e-156
UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thal... 552 e-156
UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana] 552 e-156
UniRef100_Q8S5N0 Putative SCARECROW gene regulator-like [Oryza s... 538 e-151
UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis ... 533 e-150
UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein... 511 e-143
UniRef100_Q9S7H5 Putative SCARECROW gene regulator [Arabidopsis ... 480 e-134
UniRef100_Q67ZX7 Putative SCARECROW gene regulator [Arabidopsis ... 479 e-134
UniRef100_Q69VG1 Putative chitin-inducible gibberellin-responsiv... 450 e-125
UniRef100_Q8W584 AT4g17230/dl4650c [Arabidopsis thaliana] 444 e-123
UniRef100_Q9XE53 Scarecrow-like 5 [Arabidopsis thaliana] 426 e-118
UniRef100_Q9SDQ3 Scarecrow-like 1 [Arabidopsis thaliana] 392 e-107
UniRef100_Q8RZQ6 Scarecrow-like protein [Oryza sativa] 385 e-105
UniRef100_O23566 SCARECROW like protein [Arabidopsis thaliana] 381 e-104
UniRef100_Q9XE51 Scarecrow-like 1 [Arabidopsis thaliana] 362 2e-98
UniRef100_Q9M0M5 Scarecrow-like 13 [Arabidopsis thaliana] 351 3e-95
UniRef100_Q8GVE2 Chitin-inducible gibberellin-responsive protein... 344 3e-93
UniRef100_Q9XE57 Scarecrow-like 13 [Arabidopsis thaliana] 309 1e-82
UniRef100_Q67ZL8 Putative SCARECROW gene regulator [Arabidopsis ... 280 9e-74
>UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thaliana]
Length = 490
Score = 554 bits (1428), Expect = e-156
Identities = 276/440 (62%), Positives = 343/440 (77%), Gaps = 10/440 (2%)
Query: 80 SCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMI 139
SC+T +D HKIRE+E M+G D+ D+L D+ E W +E I
Sbjct: 60 SCVTDELNDFKHKIREIETVMMGPDSLDLLVDCTDSF-----DSTASQEINGWRSTLEAI 114
Query: 140 SRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASS 199
SR DL+ L +CAKA+SEND+ A +M +L +MVSVSG PIQRLGAY+LE LVA++ASS
Sbjct: 115 SRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASS 174
Query: 200 GSIIYKSL-KCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHII 258
GS IYK+L +C EP S ELLS+MH+LYE+CPY KFGYMSANG IAEA+K+E+ +HII
Sbjct: 175 GSSIYKALNRCPEP---ASTELLSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHII 231
Query: 259 DFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYN 318
DFQI QG QW++LIQA A +PGGPP+IRITG DD TSAYARGGGL IVG RL+KLA+ +N
Sbjct: 232 DFQIGQGSQWVTLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFN 291
Query: 319 VAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKC 378
V FEF+++ VS SEV+ ++L +R GEA+AVNFA +LHH+PDE V +NHRDRL+R+ K
Sbjct: 292 VPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESV-STENHRDRLLRMVKS 350
Query: 379 LSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLARE 438
LSPKVVTLVEQESNTN FF RF+ETMNYY A+FESIDV LPR+H++RINVEQHCLAR+
Sbjct: 351 LSPKVVTLVEQESNTNTAAFFPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARD 410
Query: 439 VVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEK 498
VVN++ACEGA+RVERHE+L KWRS F MAGFTPYPLS +N +I++LL NY Y L+E+
Sbjct: 411 VVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEER 470
Query: 499 DGALYLGWMNQPLITSSAWR 518
DGALYLGWM++ L+ S AW+
Sbjct: 471 DGALYLGWMHRDLVASCAWK 490
>UniRef100_Q94BW9 F17J6.12/F17J6.12 [Arabidopsis thaliana]
Length = 526
Score = 553 bits (1424), Expect = e-156
Identities = 292/514 (56%), Positives = 366/514 (70%), Gaps = 19/514 (3%)
Query: 7 IRNLDSYCFLQNENLENYSSSDNGSHSTYPSFQALEQNLE-SSNNSPVSKLQSKSYTFTS 65
++ DSYC L+ SSS SH + SSN SP+S+ + + + +
Sbjct: 30 VQTFDSYCTLE-------SSSGTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFN 82
Query: 66 QNSLEIINDS-LENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDP 124
+S E N+S L S N+ +L +++LE AM+ D + YN+ Q+
Sbjct: 83 NHSPEENNNSPLSGSSATNTNETELSLMLKDLETAMMEPDVDNS---YNNQGGFGQQHGV 139
Query: 125 LLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRL 184
+ + + +EMISRGDLK +L+ CAKA+ D+E +WL+S+L +MVSVSG P+QRL
Sbjct: 140 V---SSAMYRSMEMISRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRL 196
Query: 185 GAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGV 244
GAYMLE LVAR+ASSGS IYK+L+CK+P T ELL++MH+LYE CPY KFGY SANG
Sbjct: 197 GAYMLEGLVARLASSGSSIYKALRCKDP---TGPELLTYMHILYEACPYFKFGYESANGA 253
Query: 245 IAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLG 304
IAEA+K+ES +HIIDFQI+QG QW+SLI+AL +PGGPP +RITG DD S++AR GGL
Sbjct: 254 IAEAVKNESFVHIIDFQISQGGQWVSLIRALGARPGGPPNVRITGIDDPRSSFARQGGLE 313
Query: 305 IVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHG 364
+VG+RL KLAE V FEFH + +EV +E L +R GEA+AVNF ++LHH+PDE V
Sbjct: 314 LVGQRLGKLAEMCGVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTV 373
Query: 365 GKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREH 424
+NHRDRL+RL K LSP VVTLVEQE+NTN PF RFVETMN+Y AVFESIDV L R+H
Sbjct: 374 -ENHRDRLLRLVKHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDH 432
Query: 425 RERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQN 484
+ERINVEQHCLAREVVNL+ACEG ER ERHE L KWRS F MAGF PYPLSSY+N +I+
Sbjct: 433 KERINVEQHCLAREVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIEG 492
Query: 485 LLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
LLE+Y YTL+E+DGALYLGW NQPLITS AWR
Sbjct: 493 LLESYSEKYTLEERDGALYLGWKNQPLITSCAWR 526
>UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thaliana]
Length = 526
Score = 552 bits (1423), Expect = e-156
Identities = 292/514 (56%), Positives = 366/514 (70%), Gaps = 19/514 (3%)
Query: 7 IRNLDSYCFLQNENLENYSSSDNGSHSTYPSFQALEQNLE-SSNNSPVSKLQSKSYTFTS 65
++ DSYC L+ SSS SH + SSN SP+S+ + + + +
Sbjct: 30 VQTFDSYCTLE-------SSSGTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFN 82
Query: 66 QNSLEIINDS-LENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDP 124
+S E N+S L S N+ +L +++LE AM+ D + YN+ Q+
Sbjct: 83 NHSPEENNNSPLSGSSATNTNETELSLMLKDLETAMMEPDVDNS---YNNQGGFGQQHGV 139
Query: 125 LLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRL 184
+ + + +EMISRGDLK +L+ CAKA+ D+E +WL+S+L +MVSVSG P+QRL
Sbjct: 140 V---SSAMYRSMEMISRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRL 196
Query: 185 GAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGV 244
GAYMLE LVAR+ASSGS IYK+L+CK+P T ELL++MH+LYE CPY KFGY SANG
Sbjct: 197 GAYMLEGLVARLASSGSSIYKALRCKDP---TGPELLTYMHILYEACPYFKFGYESANGA 253
Query: 245 IAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLG 304
IAEA+K+ES +HIIDFQI+QG QW+SLI+AL +PGGPP +RITG DD S++AR GGL
Sbjct: 254 IAEAVKNESFVHIIDFQISQGGQWVSLIRALGARPGGPPNVRITGIDDPRSSFARQGGLE 313
Query: 305 IVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHG 364
+VG+RL KLAE V FEFH + +EV +E L +R GEA+AVNF ++LHH+PDE V
Sbjct: 314 LVGQRLGKLAEMCGVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTV 373
Query: 365 GKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREH 424
+NHRDRL+RL K LSP VVTLVEQE+NTN PF RFVETMN+Y AVFESIDV L R+H
Sbjct: 374 -ENHRDRLLRLVKHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDH 432
Query: 425 RERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQN 484
+ERINVEQHCLAREVVNL+ACEG ER ERHE L KWRS F MAGF PYPLSSY+N +I+
Sbjct: 433 KERINVEQHCLAREVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKG 492
Query: 485 LLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
LLE+Y YTL+E+DGALYLGW NQPLITS AWR
Sbjct: 493 LLESYSEKYTLEERDGALYLGWKNQPLITSCAWR 526
>UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana]
Length = 597
Score = 552 bits (1423), Expect = e-156
Identities = 292/514 (56%), Positives = 366/514 (70%), Gaps = 19/514 (3%)
Query: 7 IRNLDSYCFLQNENLENYSSSDNGSHSTYPSFQALEQNLE-SSNNSPVSKLQSKSYTFTS 65
++ DSYC L+ SSS SH + SSN SP+S+ + + + +
Sbjct: 101 VQTFDSYCTLE-------SSSGTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFN 153
Query: 66 QNSLEIINDS-LENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDP 124
+S E N+S L S N+ +L +++LE AM+ D + YN+ Q+
Sbjct: 154 NHSPEENNNSPLSGSSATNTNETELSLMLKDLETAMMEPDVDNS---YNNQGGFGQQHGV 210
Query: 125 LLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRL 184
+ + + +EMISRGDLK +L+ CAKA+ D+E +WL+S+L +MVSVSG P+QRL
Sbjct: 211 V---SSAMYRSMEMISRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRL 267
Query: 185 GAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGV 244
GAYMLE LVAR+ASSGS IYK+L+CK+P T ELL++MH+LYE CPY KFGY SANG
Sbjct: 268 GAYMLEGLVARLASSGSSIYKALRCKDP---TGPELLTYMHILYEACPYFKFGYESANGA 324
Query: 245 IAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLG 304
IAEA+K+ES +HIIDFQI+QG QW+SLI+AL +PGGPP +RITG DD S++AR GGL
Sbjct: 325 IAEAVKNESFVHIIDFQISQGGQWVSLIRALGARPGGPPNVRITGIDDPRSSFARQGGLE 384
Query: 305 IVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHG 364
+VG+RL KLAE V FEFH + +EV +E L +R GEA+AVNF ++LHH+PDE V
Sbjct: 385 LVGQRLGKLAEMCGVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTV 444
Query: 365 GKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREH 424
+NHRDRL+RL K LSP VVTLVEQE+NTN PF RFVETMN+Y AVFESIDV L R+H
Sbjct: 445 -ENHRDRLLRLVKHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDH 503
Query: 425 RERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQN 484
+ERINVEQHCLAREVVNL+ACEG ER ERHE L KWRS F MAGF PYPLSSY+N +I+
Sbjct: 504 KERINVEQHCLAREVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKG 563
Query: 485 LLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
LLE+Y YTL+E+DGALYLGW NQPLITS AWR
Sbjct: 564 LLESYSEKYTLEERDGALYLGWKNQPLITSCAWR 597
>UniRef100_Q8S5N0 Putative SCARECROW gene regulator-like [Oryza sativa]
Length = 524
Score = 538 bits (1386), Expect = e-151
Identities = 285/506 (56%), Positives = 366/506 (72%), Gaps = 31/506 (6%)
Query: 13 YCFLQNENLENYSSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKS-YTFTSQNSLEI 71
YC L+ SSS NG+H + S A ++ + SP+S S S +T+ S S
Sbjct: 48 YCTLE-------SSSANGAHPAHSS--ASSHSISPISGSPLSHHDSHSDHTYNSPPSA-- 96
Query: 72 INDSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEK 131
SC+T D L K+RELENA+LG + LDI D+ + + ++ E
Sbjct: 97 --------SCVTEITD-LQIKLRELENAILGPE----LDIAYDSPESALQPN-IMATPEN 142
Query: 132 WNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEA 191
W +++ I+ GDLK+++ C KA++END+ E L+SEL +MVSVSG+P+QRLGAYMLE
Sbjct: 143 WRQLLG-INTGDLKQVIIACGKAVAENDVRLTELLISELGQMVSVSGDPLQRLGAYMLEG 201
Query: 192 LVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKD 251
LVAR++SSGS IYKSLKCKEP TS EL+S+MH+LYEICP+ KFGYMSANG IAEA+K
Sbjct: 202 LVARLSSSGSKIYKSLKCKEP---TSSELMSYMHLLYEICPFFKFGYMSANGAIAEAIKG 258
Query: 252 ESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLS 311
E+ +HIIDFQI QG QWM+LIQALA +PGGPP +RITG DDS SAYARGGGL IVG RL
Sbjct: 259 ENFVHIIDFQIAQGSQWMTLIQALAARPGGPPFLRITGIDDSNSAYARGGGLDIVGMRLY 318
Query: 312 KLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDR 371
K+A+S+ + FEF+A+ + EV LE L++R GE I VNFA LHH PDE V +NHRDR
Sbjct: 319 KVAQSFGLPFEFNAVPAASHEVYLEHLDIRVGEVIVVNFAYQLHHTPDESV-STENHRDR 377
Query: 372 LVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVE 431
++R+ K LSP++VTLVEQESNTN PFF R++ET++YY A+FESIDVALPR+ + R++ E
Sbjct: 378 ILRMVKSLSPRLVTLVEQESNTNTRPFFPRYLETLDYYTAMFESIDVALPRDDKRRMSAE 437
Query: 432 QHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQG 491
QHC+AR++VNL+ACEGAERVERHEV KW++ TMAGF PYPLSS +N +I+ LL Y
Sbjct: 438 QHCVARDIVNLIACEGAERVERHEVFGKWKARLTMAGFRPYPLSSVVNSTIKTLLHTYNS 497
Query: 492 HYTLQEKDGALYLGWMNQPLITSSAW 517
Y L+E+DG LYLGW N+ L+ SSAW
Sbjct: 498 FYRLEERDGVLYLGWKNRVLVVSSAW 523
>UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 411
Score = 533 bits (1374), Expect = e-150
Identities = 266/420 (63%), Positives = 330/420 (78%), Gaps = 10/420 (2%)
Query: 100 MLGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMISRGDLKEILFTCAKAISEND 159
M+G D+ D+L D+ E W +E ISR DL+ L +CAKA+SEND
Sbjct: 1 MMGPDSLDLLVDCTDSF-----DSTASQEINGWRSTLEAISRRDLRADLVSCAKAMSEND 55
Query: 160 METAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSL-KCKEPITATSK 218
+ A +M +L +MVSVSG PIQRLGAY+LE LVA++ASSGS IYK+L +C EP S
Sbjct: 56 LMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASSGSSIYKALNRCPEP---AST 112
Query: 219 ELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGK 278
ELLS+MH+LYE+CPY KFGYMSANG IAEA+K+E+ +HIIDFQI QG QW++LIQA A +
Sbjct: 113 ELLSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAAR 172
Query: 279 PGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDL 338
PGGPP+IRITG DD TSAYARGGGL IVG RL+KLA+ +NV FEF+++ VS SEV+ ++L
Sbjct: 173 PGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNL 232
Query: 339 ELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPF 398
+R GEA+AVNFA +LHH+PDE V +NHRDRL+R+ K LSPKVVTLVEQESNTN F
Sbjct: 233 GVRPGEALAVNFAFVLHHMPDESV-STENHRDRLLRMVKSLSPKVVTLVEQESNTNTAAF 291
Query: 399 FARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLK 458
F RF+ETMNYY A+FESIDV LPR+H++RINVEQHCLAR+VVN++ACEGA+RVERHE+L
Sbjct: 292 FPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIACEGADRVERHELLG 351
Query: 459 KWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
KWRS F MAGFTPYPLS +N +I++LL NY Y L+E+DGALYLGWM++ L+ S AW+
Sbjct: 352 KWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEERDGALYLGWMHRDLVASCAWK 411
>UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein [Oryza sativa]
Length = 544
Score = 511 bits (1317), Expect = e-143
Identities = 263/475 (55%), Positives = 338/475 (70%), Gaps = 20/475 (4%)
Query: 43 QNLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLENESCLTHNQDDLWHKIRELENAMLG 102
Q+ + + SP+S+ S S + + S SC+T + +DL K+++LE MLG
Sbjct: 89 QSFTTRSGSPLSQEDSHS---------DSTDGSPVGASCVTEDPNDLKQKLKDLEAVMLG 139
Query: 103 QDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMET 162
D+ + + N ++ L LE EKW +M+ I RG+LKE+L CA+A+ E +
Sbjct: 140 PDSEIVNSLENSV------ANQLSLEPEKWVRMMG-IPRGNLKELLIACARAVEEKNSFA 192
Query: 163 AEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLS 222
+ ++ EL K+VSVSG P++RLGAYM+E LVAR+ASSG IYK+LKCKEP S +LLS
Sbjct: 193 IDMMIPELRKIVSVSGEPLERLGAYMVEGLVARLASSGISIYKALKCKEP---KSSDLLS 249
Query: 223 HMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGP 282
+MH LYE CPY KFGYMSANG IAEA+K E IHIIDF I+QG QW+SL+QALA +PGGP
Sbjct: 250 YMHFLYEACPYFKFGYMSANGAIAEAVKGEDRIHIIDFHISQGAQWISLLQALAARPGGP 309
Query: 283 PKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRR 342
P +RITG DDS SAYARGGGL +VG RLS +A V FEFH + +S S+V L +
Sbjct: 310 PTVRITGIDDSVSAYARGGGLELVGRRLSHIASLCKVPFEFHPLAISGSKVEAAHLGVIP 369
Query: 343 GEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARF 402
GEA+AVNF + LHH+PDE V NHRDRL+R+ K LSPKV+TLVE ESNTN PF RF
Sbjct: 370 GEALAVNFTLELHHIPDESVSTA-NHRDRLLRMVKSLSPKVLTLVEMESNTNTAPFPQRF 428
Query: 403 VETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRS 462
ET++YY A+FESID+ LPR+ RERIN+EQHCLARE+VNL+ACEG ER ER+E KW++
Sbjct: 429 AETLDYYTAIFESIDLTLPRDDRERINMEQHCLAREIVNLIACEGEERAERYEPFGKWKA 488
Query: 463 CFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAW 517
TMAGF P PLSS +N +I+ LL++Y +Y L E+DGALYLGW ++PL+ SSAW
Sbjct: 489 RLTMAGFRPSPLSSLVNATIRTLLQSYSDNYKLAERDGALYLGWKSRPLVVSSAW 543
>UniRef100_Q9S7H5 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 413
Score = 480 bits (1236), Expect = e-134
Identities = 238/384 (61%), Positives = 299/384 (76%), Gaps = 11/384 (2%)
Query: 135 MIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVA 194
++E ISRGDLK +L CAKA+SEN++ A W M EL MVS+SG PIQRLGAYMLE LVA
Sbjct: 41 IVEAISRGDLKLVLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVA 100
Query: 195 RIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESE 254
R+A+SGS IYKSL+ +EP S E LS+++VL+E+CPY KFGYMSANG IAEA+KDE
Sbjct: 101 RLAASGSSIYKSLQSREP---ESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEER 157
Query: 255 IHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLA 314
IHIIDFQI QG QW++LIQA A +PGG P IRITG D G L V +RL KLA
Sbjct: 158 IHIIDFQIGQGSQWIALIQAFAARPGGAPNIRITGVGD-------GSVLVTVKKRLEKLA 210
Query: 315 ESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVR 374
+ ++V F F+A+ EV +E+L++R GEA+ VNFA MLHH+PDE V +NHRDRL+R
Sbjct: 211 KKFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESV-SMENHRDRLLR 269
Query: 375 LAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHC 434
+ K LSPKVVTLVEQE NTN PF RF+ET++YY A+FESIDV LPR H+ERIN+EQHC
Sbjct: 270 MVKSLSPKVVTLVEQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHC 329
Query: 435 LAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYT 494
+AR+VVN++ACEGAER+ERHE+L KW+S F+MAGF PYPLSS I+ +I+ LL +Y Y
Sbjct: 330 MARDVVNIIACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYA 389
Query: 495 LQEKDGALYLGWMNQPLITSSAWR 518
++E+DGALYLGWM++ L++S AW+
Sbjct: 390 IEERDGALYLGWMDRILVSSCAWK 413
>UniRef100_Q67ZX7 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 413
Score = 479 bits (1234), Expect = e-134
Identities = 238/384 (61%), Positives = 299/384 (76%), Gaps = 11/384 (2%)
Query: 135 MIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVA 194
++E ISRGDLK +L CAKA+SEN++ A W M EL MVS+SG PIQRLGAYMLE LVA
Sbjct: 41 IVEAISRGDLKLVLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVA 100
Query: 195 RIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESE 254
R+A+SGS IYKSL+ +EP S E LS+++VL+E+CPY KFGYMSANG IAEA+KDE
Sbjct: 101 RLAASGSSIYKSLQSREP---ESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEER 157
Query: 255 IHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLA 314
IHIIDFQI QG QW++LIQA A +PGG P IRITG D G L V +RL KLA
Sbjct: 158 IHIIDFQIGQGSQWIALIQAFAARPGGAPNIRITGVGD-------GSVLVTVKKRLEKLA 210
Query: 315 ESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVR 374
+ ++V F F+A+ EV +E+L++R GEA+ VNFA MLHH+PDE V +NHRDRL+R
Sbjct: 211 KKFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESV-SMENHRDRLLR 269
Query: 375 LAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHC 434
+ K LSPKVVTLVEQE NTN PF RF+ET++YY A+FESIDV LPR H+ERIN+EQHC
Sbjct: 270 MVKSLSPKVVTLVEQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHC 329
Query: 435 LAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYT 494
+AR+VVN++ACEGAER+ERHE+L KW+S F+MAGF PYPLSS I+ +I+ LL +Y Y
Sbjct: 330 MARDVVNIMACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYA 389
Query: 495 LQEKDGALYLGWMNQPLITSSAWR 518
++E+DGALYLGWM++ L++S AW+
Sbjct: 390 IEERDGALYLGWMDRILVSSCAWK 413
>UniRef100_Q69VG1 Putative chitin-inducible gibberellin-responsive protein [Oryza
sativa]
Length = 571
Score = 450 bits (1157), Expect = e-125
Identities = 222/374 (59%), Positives = 283/374 (75%), Gaps = 4/374 (1%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
+K++L CA+A+SE+ E L+ E +VS++G PIQRLGAY+LE LVAR +SG+ I
Sbjct: 201 VKQLLTRCAEALSEDRTEEFHKLVQEARGVVSINGEPIQRLGAYLLEGLVARHGNSGTNI 260
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
Y++LKC+EP SKELLS+M +LY ICPY KFGYM+ANG IAEAL+ E+ IHIIDFQI
Sbjct: 261 YRALKCREP---ESKELLSYMRILYNICPYFKFGYMAANGAIAEALRTENNIHIIDFQIA 317
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
QG QW++LIQALA +PGGPP++RITG DD S YARG GL IVG+ L ++E + + EF
Sbjct: 318 QGTQWITLIQALAARPGGPPRVRITGIDDPVSEYARGEGLDIVGKMLKSMSEEFKIPLEF 377
Query: 324 HAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKV 383
+ V ++V E LE+R GEA++VNF + LHH PDE V N RD L+R+ K LSPKV
Sbjct: 378 TPLSVYATQVTKEMLEIRPGEALSVNFTLQLHHTPDESVDVN-NPRDGLLRMVKGLSPKV 436
Query: 384 VTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLV 443
TLVEQES+TN PF RF ETM YY A+FESID LPR+++ERI+VEQHCLA+++VN++
Sbjct: 437 TTLVEQESHTNTTPFLMRFGETMEYYSAMFESIDANLPRDNKERISVEQHCLAKDIVNII 496
Query: 444 ACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALY 503
ACEG +RVERHE+L KW+S TMAGF PYPLSSY+N I+ LL Y YTL EKDGA+
Sbjct: 497 ACEGKDRVERHELLGKWKSRLTMAGFRPYPLSSYVNSVIRKLLACYSDKYTLDEKDGAML 556
Query: 504 LGWMNQPLITSSAW 517
LGW ++ LI++SAW
Sbjct: 557 LGWRSRKLISASAW 570
>UniRef100_Q8W584 AT4g17230/dl4650c [Arabidopsis thaliana]
Length = 529
Score = 444 bits (1141), Expect = e-123
Identities = 247/515 (47%), Positives = 329/515 (62%), Gaps = 35/515 (6%)
Query: 24 YSSSDNGSHSTYPS---FQALEQNLES----SNNSPVSKLQSKSYTFTSQNSLEIIND-- 74
+ + DN S PS F LE + S S +SP + S F+ Q S I+D
Sbjct: 26 FQAKDNKGFSDIPSKENFFTLESSTASGSLPSYDSPSVSITSGQSPFSPQGSQSCISDLH 85
Query: 75 ---------SLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQES--D 123
L S L +++ + KIRELE ++L D +++ S
Sbjct: 86 HSPDNVYGSPLSGVSSLAYDEAGVKSKIRELEVSLLSGDTK-----------VEEFSGFS 134
Query: 124 PLLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQR 183
P ++ W++++ + + DLKE+L A+A+++ D TA + L +MVSVSG+PIQR
Sbjct: 135 PAAGKSWNWDELLALTPQLDLKEVLVEAARAVADGDFATAYGFLDVLEQMVSVSGSPIQR 194
Query: 184 LGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANG 243
LG YM E L AR+ SGS IYKSLKC EP T +EL+S+M VLYEICPY KF Y +AN
Sbjct: 195 LGTYMAEGLRARLEGSGSNIYKSLKCNEP---TGRELMSYMSVLYEICPYWKFAYTTANV 251
Query: 244 VIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGL 303
I EA+ E+ +HIIDFQI QG Q+M LIQ LA +PGGPP +R+TG DDS S YARGGGL
Sbjct: 252 EILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKRPGGPPLLRVTGVDDSQSTYARGGGL 311
Query: 304 GIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVH 363
+VGERL+ LA+S V FEFH +S +V+ E L L G A+ VNF +LHH+PDE V
Sbjct: 312 SLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLGLEPGFAVVVNFPYVLHHMPDESV- 370
Query: 364 GGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPRE 423
+NHRDRL+ L K LSPK+VTLVEQESNTN PF +RFVET++YY A+FESID A PR+
Sbjct: 371 SVENHRDRLLHLIKSLSPKLVTLVEQESNTNTSPFLSRFVETLDYYTAMFESIDAARPRD 430
Query: 424 HRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQ 483
++RI+ EQHC+AR++VN++ACE +ERVERHEVL KWR MAGFT +P+S+ ++
Sbjct: 431 DKQRISAEQHCVARDIVNMIACEESERVERHEVLGKWRVRMMMAGFTGWPVSTSAAFAAS 490
Query: 484 NLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+L+ Y +Y L +GALYL W +P+ T S W+
Sbjct: 491 EMLKAYDKNYKLGGHEGALYLFWKRRPMATCSVWK 525
>UniRef100_Q9XE53 Scarecrow-like 5 [Arabidopsis thaliana]
Length = 306
Score = 426 bits (1095), Expect = e-118
Identities = 208/303 (68%), Positives = 244/303 (79%), Gaps = 1/303 (0%)
Query: 216 TSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQAL 275
T ELL++MH+LYE CPY KFGY SANG IAEA+K+ES +HIIDFQI+QG QW+SLI+AL
Sbjct: 5 TGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWVSLIRAL 64
Query: 276 AGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRL 335
+PGGPP +RITG DD S++AR GGL +VG+RL KLAE V FEFH + +EV +
Sbjct: 65 GARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALCCTEVEI 124
Query: 336 EDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNE 395
E L +R GEA+AVNF ++LHH+PDE V +NHRDRL+RL K LSP VVTLVEQE+NTN
Sbjct: 125 EKLGVRNGEALAVNFPLVLHHMPDESVTV-ENHRDRLLRLVKHLSPNVVTLVEQEANTNT 183
Query: 396 LPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHE 455
PF RFVETMN+Y AVFESIDV L R+H+ERINVEQHCLAREVVNL+ACEG ER ERHE
Sbjct: 184 APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEGVEREERHE 243
Query: 456 VLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSS 515
L KWRS F MAGF PYPLSSY+N +I+ LLE+Y YTL+E+DGALYLGW NQPLITS
Sbjct: 244 PLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDGALYLGWKNQPLITSC 303
Query: 516 AWR 518
AWR
Sbjct: 304 AWR 306
>UniRef100_Q9SDQ3 Scarecrow-like 1 [Arabidopsis thaliana]
Length = 593
Score = 392 bits (1007), Expect = e-107
Identities = 219/519 (42%), Positives = 321/519 (61%), Gaps = 37/519 (7%)
Query: 20 NLENYSSSDNGSHSTYP------SFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIIN 73
++ ++ S D+ + + P FQ + +S+ + Q+ SY+ S++++
Sbjct: 92 SVSSFGSLDSFPYQSRPVLGCSMEFQLPLDSTSTSSTRLLGDYQAVSYS----PSMDVVE 147
Query: 74 DSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVII----------QQESD 123
+ + + + KI+ELE A+LG + M+ I N I Q +
Sbjct: 148 E---------FDDEQMRSKIQELERALLGDEDDKMVGIDNLMEIDSEWSYQNESEQHQDS 198
Query: 124 PLLLEAEKWNKMI---EMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNP 180
P + N + E++S+ K+IL +CA+A+SE +E A +++EL ++VS+ G+P
Sbjct: 199 PKESSSADSNSHVSSKEVVSQATPKQILISCARALSEGKLEEALSMVNELRQIVSIQGDP 258
Query: 181 IQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMS 240
QR+ AYM+E L AR+A+SG IY++LKCKEP S E L+ M VL+E+CP KFG+++
Sbjct: 259 SQRIAAYMVEGLAARMAASGKFIYRALKCKEP---PSDERLAAMQVLFEVCPCFKFGFLA 315
Query: 241 ANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARG 300
ANG I EA+K E E+HIIDF INQG Q+M+LI+++A PG P++R+TG DD S
Sbjct: 316 ANGAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIAELPGKRPRLRLTGIDDPESVQRSI 375
Query: 301 GGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDE 360
GGL I+G RL +LAE V+F+F A+ S V L + GE + VNFA LHH+PDE
Sbjct: 376 GGLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPSTLGCKPGETLIVNFAFQLHHMPDE 435
Query: 361 DVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVAL 420
V N RD L+ + K L+PK+VT+VEQ+ NTN PFF RF+E YY AVFES+D+ L
Sbjct: 436 SVTT-VNQRDELLHMVKSLNPKLVTVVEQDVNTNTSPFFPRFIEAYEYYSAVFESLDMTL 494
Query: 421 PREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINY 480
PRE +ER+NVE+ CLAR++VN+VACEG ER+ER+E KWR+ MAGF P P+S+ +
Sbjct: 495 PRESQERMNVERQCLARDIVNIVACEGEERIERYEAAGKWRARMMMAGFNPKPMSAKVTN 554
Query: 481 SIQNLL-ENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+IQNL+ + Y Y L+E+ G L+ W + LI +SAWR
Sbjct: 555 NIQNLIKQQYCNKYKLKEEMGELHFCWEEKSLIVASAWR 593
>UniRef100_Q8RZQ6 Scarecrow-like protein [Oryza sativa]
Length = 553
Score = 385 bits (988), Expect = e-105
Identities = 206/464 (44%), Positives = 296/464 (63%), Gaps = 14/464 (3%)
Query: 65 SQNSLEIINDSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDP 124
SQ + + I+D+ +E + ++DD+ K++ELE+A+L + +I I + +DP
Sbjct: 94 SQQNSQSISDNQSSELEVEFDEDDIRMKLQELEHALLDDSDDILYEISQAGSINDEWADP 153
Query: 125 L---LLEAEKWNKMIEMISRGDL-------KEILFTCAKAISENDMETAEWLMSELSKMV 174
+ +L + G K++LF CA A+S+ +++ A+ ++++L +MV
Sbjct: 154 MKNVILPNSPKESESSISCAGSNNGEPRTPKQLLFDCAMALSDYNVDEAQAIITDLRQMV 213
Query: 175 SVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYL 234
S+ G+P QR+ AY++E L ARI +SG IYK+L CKEP T LS M +L+EICP
Sbjct: 214 SIQGDPSQRIAAYLVEGLAARIVASGKGIYKALSCKEPPTLYQ---LSAMQILFEICPCF 270
Query: 235 KFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDST 294
+FG+M+AN I EA K E +HIIDF INQG Q+++LIQ L P +RITG DD
Sbjct: 271 RFGFMAANYAILEACKGEDRVHIIDFDINQGSQYITLIQFLKNNANKPRHLRITGVDDPE 330
Query: 295 SAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMML 354
+ GGL ++G+RL KLAE ++FEF A+G + +V L+ GEA+ VNFA L
Sbjct: 331 TVQRTVGGLKVIGQRLEKLAEDCGISFEFRAVGANIGDVTPAMLDCCPGEALVVNFAFQL 390
Query: 355 HHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFE 414
HH+PDE V N RD+L+R+ K L PK+VTLVEQ++NTN PF RF E +YY A+F+
Sbjct: 391 HHLPDESV-SIMNERDQLLRMVKGLQPKLVTLVEQDANTNTAPFQTRFREVYDYYAALFD 449
Query: 415 SIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPL 474
S+D LPRE +R+NVE+ CLARE+VN++ACEG +RVER+EV KWR+ TMAGFTP P
Sbjct: 450 SLDATLPRESPDRMNVERQCLAREIVNILACEGPDRVERYEVAGKWRARMTMAGFTPCPF 509
Query: 475 SSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
SS + I++LL++Y Y +E G L+ GW + LI SSAW+
Sbjct: 510 SSNVISGIRSLLKSYCDRYKFEEDHGGLHFGWGEKTLIVSSAWQ 553
>UniRef100_O23566 SCARECROW like protein [Arabidopsis thaliana]
Length = 375
Score = 381 bits (978), Expect = e-104
Identities = 202/368 (54%), Positives = 260/368 (69%), Gaps = 17/368 (4%)
Query: 92 KIRELENAMLGQDAADMLDIYNDTVIIQQES--DPLLLEAEKWNKMIEMISRGDLKEILF 149
KIRELE ++L D +++ S P ++ W++++ + + DLKE+L
Sbjct: 17 KIRELEVSLLSGDTK-----------VEEFSGFSPAAGKSWNWDELLALTPQLDLKEVLV 65
Query: 150 TCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKC 209
A+A+++ D TA + L +MVSVSG+PIQRLG YM E L AR+ SGS IYKSLKC
Sbjct: 66 EAARAVADGDFATAYGFLDVLEQMVSVSGSPIQRLGTYMAEGLRARLEGSGSNIYKSLKC 125
Query: 210 KEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWM 269
EP T +EL+S+M VLYEICPY KF Y +AN I EA+ E+ +HIIDFQI QG Q+M
Sbjct: 126 NEP---TGRELMSYMSVLYEICPYWKFAYTTANVEILEAIAGETRVHIIDFQIAQGSQYM 182
Query: 270 SLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVS 329
LIQ LA PGGPP +R+TG DDS S YARGGGL +VGERL+ LA+S V FEFH +S
Sbjct: 183 FLIQELAKHPGGPPLLRVTGVDDSQSTYARGGGLSLVGERLATLAQSCGVPFEFHDAIMS 242
Query: 330 PSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQ 389
+V+ E L L G A+ VNF +LHH+PDE V +NHRDRL+ L K LSPK+VTLVEQ
Sbjct: 243 GCKVQREHLGLEPGFAVVVNFPYVLHHMPDESV-SVENHRDRLLHLIKSLSPKLVTLVEQ 301
Query: 390 ESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAE 449
ESNTN PF +RFVET++YY A+FESID A PR+ ++RI+ EQHC+AR++VN++ACE +E
Sbjct: 302 ESNTNTSPFLSRFVETLDYYTAMFESIDAARPRDDKQRISAEQHCVARDIVNMIACEESE 361
Query: 450 RVERHEVL 457
RVERHEVL
Sbjct: 362 RVERHEVL 369
>UniRef100_Q9XE51 Scarecrow-like 1 [Arabidopsis thaliana]
Length = 352
Score = 362 bits (928), Expect = 2e-98
Identities = 182/354 (51%), Positives = 248/354 (69%), Gaps = 5/354 (1%)
Query: 166 LMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMH 225
+++EL ++VS+ G+P QR+ AYM+E L AR+A+SG IY++LKCKEP S E L+ M
Sbjct: 3 MVNELRQIVSIQGDPSQRIAAYMVEGLAARMAASGKFIYRALKCKEP---PSDERLAAMQ 59
Query: 226 VLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKI 285
VL+E+CP KFG+++ANG I EA+K E E+HIIDF INQG Q+M+LI+++A PG P++
Sbjct: 60 VLFEVCPCFKFGFLAANGAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIAELPGKRPRL 119
Query: 286 RITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEA 345
R+TG DD S GGL I+G RL +LAE V+F+F A+ S V L + GE
Sbjct: 120 RLTGIDDPESVQRSIGGLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPSTLGCKPGET 179
Query: 346 IAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVET 405
+ VNFA LHH+PDE V N RD L+ + K L+PK+VT+VEQ+ NTN PFF RF+E
Sbjct: 180 LIVNFAFQLHHMPDESVTT-VNQRDELLHMVKSLNPKLVTVVEQDVNTNTSPFFPRFIEA 238
Query: 406 MNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFT 465
YY AVFES+D+ LPRE +ER+NVE+ CLAR++VN+VACEG ER+ER+E KWR+
Sbjct: 239 YEYYSAVFESLDMTLPRESQERMNVERQCLARDIVNIVACEGEERIERYEAAGKWRARMM 298
Query: 466 MAGFTPYPLSSYINYSIQNLL-ENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
MAGF P P+S+ + +IQNL+ + Y Y L+E+ G L+ W + LI +SAWR
Sbjct: 299 MAGFNPKPMSAKVTNNIQNLIKQQYCNKYKLKEEMGELHFCWEEKSLIVASAWR 352
>UniRef100_Q9M0M5 Scarecrow-like 13 [Arabidopsis thaliana]
Length = 287
Score = 351 bits (901), Expect = 3e-95
Identities = 180/285 (63%), Positives = 218/285 (76%), Gaps = 4/285 (1%)
Query: 173 MVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICP 232
MVSVSG+PIQRLG YM E L AR+ SGS IYKSLKC EP T +EL+S+M VLYEICP
Sbjct: 1 MVSVSGSPIQRLGTYMAEGLRARLEGSGSNIYKSLKCNEP---TGRELMSYMSVLYEICP 57
Query: 233 YLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDD 292
Y KF Y +AN I EA+ E+ +HIIDFQI QG Q+M LIQ LA PGGPP +R+TG DD
Sbjct: 58 YWKFAYTTANVEILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKHPGGPPLLRVTGVDD 117
Query: 293 STSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAM 352
S S YARGGGL +VGERL+ LA+S V FEFH +S +V+ E L L G A+ VNF
Sbjct: 118 SQSTYARGGGLSLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLGLEPGFAVVVNFPY 177
Query: 353 MLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAV 412
+LHH+PDE V +NHRDRL+ L K LSPK+VTLVEQESNTN PF +RFVET++YY A+
Sbjct: 178 VLHHMPDESV-SVENHRDRLLHLIKSLSPKLVTLVEQESNTNTSPFLSRFVETLDYYTAM 236
Query: 413 FESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVL 457
FESID A PR+ ++RI+ EQHC+AR++VN++ACE +ERVERHEVL
Sbjct: 237 FESIDAARPRDDKQRISAEQHCVARDIVNMIACEESERVERHEVL 281
>UniRef100_Q8GVE2 Chitin-inducible gibberellin-responsive protein [Oryza sativa]
Length = 572
Score = 344 bits (883), Expect = 3e-93
Identities = 189/388 (48%), Positives = 246/388 (62%), Gaps = 31/388 (7%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
+K++L CA+A+SE+ E L+ E +VS++G PIQRLGAY+LE LVAR +SG+ I
Sbjct: 201 VKQLLTRCAEALSEDRTEEFHKLVQEARGVVSINGEPIQRLGAYLLEGLVARHGNSGTNI 260
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
Y++LKC+EP SKELLS+M +LY ICPY KFGYM+ANG IAEAL+ E+ IHIIDFQI
Sbjct: 261 YRALKCREP---ESKELLSYMRILYNICPYFKFGYMAANGAIAEALRTENNIHIIDFQIA 317
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
QG QW++LIQALA +PGGPP++RITG DD S YARG GL IVG+ L ++E + + EF
Sbjct: 318 QGTQWITLIQALAARPGGPPRVRITGIDDPVSEYARGEGLDIVGKMLKSMSEEFKIPLEF 377
Query: 324 HAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKV 383
+ V ++V E LE+R GEA++VNF + LHH PDE V N RD L
Sbjct: 378 TPLSVYATQVTKEMLEIRPGEALSVNFTLQLHHTPDESV-DVNNPRDGL----------- 425
Query: 384 VTLVEQESNTNELPFFARFVETMN---YYFAVFESIDVALPREHRERINV---------- 430
L + E E +F R T + V+ V L +R+
Sbjct: 426 --LPDGERAVPEGDYFGRAGVTHQHNAFLDEVWGDHGVLLRHVRVDRLPTCRGTTRRGSA 483
Query: 431 -EQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENY 489
R +VN++ACEG +RVERHE+L KW+S TMAGF PYPLSSY+N I+ LL Y
Sbjct: 484 WSSTASPRHIVNIIACEGKDRVERHELLGKWKSRLTMAGFRPYPLSSYVNSVIRKLLACY 543
Query: 490 QGHYTLQEKDGALYLGWMNQPLITSSAW 517
YTL EKDGA+ LGW ++ LI++SAW
Sbjct: 544 SDKYTLDEKDGAMLLGWRSRKLISASAW 571
>UniRef100_Q9XE57 Scarecrow-like 13 [Arabidopsis thaliana]
Length = 284
Score = 309 bits (792), Expect = 1e-82
Identities = 156/278 (56%), Positives = 201/278 (72%), Gaps = 1/278 (0%)
Query: 241 ANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARG 300
AN I EA+ E+ +HIIDFQI QG Q+M LIQ LA +PGGPP +R+TG DDS S YARG
Sbjct: 1 ANVEILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKRPGGPPLLRVTGVDDSQSTYARG 60
Query: 301 GGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDE 360
GGL +VGERL+ LA+S V FEFH +S +V+ E L L G A+ VNF +LHH+PDE
Sbjct: 61 GGLSLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLGLEPGFAVVVNFPYVLHHMPDE 120
Query: 361 DVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVAL 420
V K +RDRL+ L K LSPK+VTLVEQESNTN P +RFVET++YY A+FESID A
Sbjct: 121 SVSVEK-YRDRLLHLIKSLSPKLVTLVEQESNTNTSPLVSRFVETLDYYTAMFESIDAAR 179
Query: 421 PREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINY 480
PR+ ++RI+ EQHC+AR++VN++ACE +ERVERHEVL KWR MAGFT +P+S+ +
Sbjct: 180 PRDDKQRISAEQHCVARDIVNMIACEESERVERHEVLGKWRVRMMMAGFTGWPVSTSAAF 239
Query: 481 SIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+ +L+ Y +Y L +GALYL W +P+ T S W+
Sbjct: 240 AASEMLKAYDKNYKLGGHEGALYLFWKRRPMATCSVWK 277
>UniRef100_Q67ZL8 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 220
Score = 280 bits (715), Expect = 9e-74
Identities = 133/219 (60%), Positives = 174/219 (78%), Gaps = 1/219 (0%)
Query: 300 GGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPD 359
G L V +RL KLA+ ++V F F+A+ EV +E+L++R GEA+ VNFA MLHH+PD
Sbjct: 3 GSVLVTVKKRLEKLAKKFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPD 62
Query: 360 EDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVA 419
E V +NHRDRL+R+ K LSPKVVTLVEQE NTN PF RF+ET++YY A+FESIDV
Sbjct: 63 ESV-SMENHRDRLLRMVKSLSPKVVTLVEQECNTNTSPFLPRFLETLSYYTAMFESIDVM 121
Query: 420 LPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYIN 479
LPR H+ERIN+EQHC+AR+VVN++ACEGAER+ERHE+L KW+S F+MAGF PYPLSS I+
Sbjct: 122 LPRNHKERINIEQHCMARDVVNIIACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIIS 181
Query: 480 YSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+I+ LL +Y Y ++E+DGALYLGWM++ L++S AW+
Sbjct: 182 ATIRALLRDYSNGYAIEERDGALYLGWMDRILVSSCAWK 220
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 865,405,351
Number of Sequences: 2790947
Number of extensions: 36116649
Number of successful extensions: 111164
Number of sequences better than 10.0: 241
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 110345
Number of HSP's gapped (non-prelim): 310
length of query: 518
length of database: 848,049,833
effective HSP length: 132
effective length of query: 386
effective length of database: 479,644,829
effective search space: 185142903994
effective search space used: 185142903994
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC137079.8


