

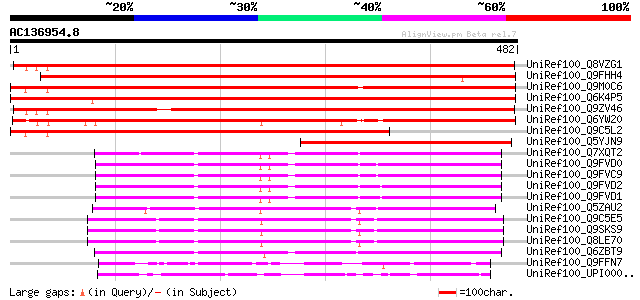
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.8 + phase: 0
(482 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VZG1 At2g18730/MSF3.11 [Arabidopsis thaliana] 656 0.0
UniRef100_Q9FHH4 Diacylglycerol kinase-like protein [Arabidopsis... 639 0.0
UniRef100_Q9M0C6 Hypothetical protein AT4g30340 [Arabidopsis tha... 616 e-175
UniRef100_Q6K4P5 Diacylglycerol kinase-like [Oryza sativa] 604 e-171
UniRef100_Q9ZV46 Putative diacylglycerol kinase [Arabidopsis tha... 585 e-165
UniRef100_Q6YW20 Putative diacylglycerol kinase variant A [Oryza... 499 e-140
UniRef100_Q9C5L2 Hypothetical protein At4g30340 [Arabidopsis tha... 469 e-131
UniRef100_Q5YJN9 Diacylglycerol kinase [Hyacinthus orientalis] 314 3e-84
UniRef100_Q7XQT2 OSJNBa0043L09.15 protein [Oryza sativa] 258 3e-67
UniRef100_Q9FVD0 Diacylglycerol kinase variant A [Lycopersicon e... 249 9e-65
UniRef100_Q9FVC9 Diacylglycerol kinase variant B [Lycopersicon e... 249 9e-65
UniRef100_Q9FVD2 Calmodulin-binding diacylglycerol kinase [Lycop... 247 6e-64
UniRef100_Q9FVD1 Diacylglycerol kinase [Lycopersicon esculentum] 247 6e-64
UniRef100_Q5ZAU2 Putative diacylglycerol kinase [Oryza sativa] 246 8e-64
UniRef100_Q9C5E5 Putative diacylglycerol kinase [Arabidopsis tha... 246 1e-63
UniRef100_Q9SKS9 Putative diacylglycerol kinase [Arabidopsis tha... 246 1e-63
UniRef100_Q8LE70 Putative diacylglycerol kinase [Arabidopsis tha... 243 7e-63
UniRef100_Q6ZBT9 Putative diacylglycerol kinase [Oryza sativa] 237 5e-61
UniRef100_Q9FFN7 Diacylglycerol kinase 2 [Arabidopsis thaliana] 150 8e-35
UniRef100_UPI000017F5C2 UPI000017F5C2 UniRef100 entry 149 2e-34
>UniRef100_Q8VZG1 At2g18730/MSF3.11 [Arabidopsis thaliana]
Length = 488
Score = 656 bits (1693), Expect = 0.0
Identities = 313/484 (64%), Positives = 385/484 (78%), Gaps = 7/484 (1%)
Query: 4 SPSSTTEDSKK--IQVRSSLVES--IRGCGLSGMR---IDKEDLKKQLTLPQYLRFAMRD 56
SP S T+ SK+ + R S +S +RGCGL+ + +DK +L+++L +P+YLR AMRD
Sbjct: 3 SPVSKTDASKEKFVASRPSTADSKTMRGCGLANLAWVGVDKVELRQRLMMPEYLRLAMRD 62
Query: 57 SIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVF 116
I+ +D SA AP +PMVVFIN SGGRHGP LKERLQQLMSEEQVF
Sbjct: 63 CIKRKDSSAIPDHLLLPGGAVADMAPHAPMVVFINPNSGGRHGPVLKERLQQLMSEEQVF 122
Query: 117 DLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQL 176
DL +VKPHEFV YGL CLE +A GD CAKE R +LR+MVAGGDGTVGWVLGCL EL +
Sbjct: 123 DLTEVKPHEFVRYGLGCLEKVAAEGDECAKECRARLRIMVAGGDGTVGWVLGCLGELNKD 182
Query: 177 GREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSIS 236
G+ +PPVG++PLGTGNDLSRSF WGGSFPFAW+SA+KRTL +AS+G V RLDSW++ +S
Sbjct: 183 GKSQIPPVGVIPLGTGNDLSRSFGWGGSFPFAWRSAVKRTLHRASMGPVARLDSWKILVS 242
Query: 237 MPESTTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHR 296
MP V PPY LK AEE LDQG++ + P K+YEGV+YNY SIGMDAQVAYGFH
Sbjct: 243 MPSGEVVDPPYSLKPAEENELDQGLDAGIDAPPLAKAYEGVFYNYLSIGMDAQVAYGFHH 302
Query: 297 LRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQ 356
LR+ KPYLA GPI+NKIIYS + C+QGWF TPC +DPGLRGLRNI+++HIK+V+ S+WE+
Sbjct: 303 LRNTKPYLAQGPISNKIIYSSFGCSQGWFCTPCVNDPGLRGLRNIMKIHIKKVNCSQWEE 362
Query: 357 VAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHAS 416
+A+PK+VR+IVALNLHSYGSG +PWG KP+YLEK+GFVEA DG +EIFG KQGWHAS
Sbjct: 363 IAVPKNVRSIVALNLHSYGSGSHPWGNLKPDYLEKRGFVEAHCDDGLIEIFGFKQGWHAS 422
Query: 417 FVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQS 476
FVM +LI+AKHIAQAAA+R ELRGG W++A+LQMDGEPWKQP+S ++STFVEIK+ P+QS
Sbjct: 423 FVMAELISAKHIAQAAAVRFELRGGDWRDAFLQMDGEPWKQPMSTEYSTFVEIKKVPYQS 482
Query: 477 LVVD 480
L+++
Sbjct: 483 LMIN 486
>UniRef100_Q9FHH4 Diacylglycerol kinase-like protein [Arabidopsis thaliana]
Length = 498
Score = 639 bits (1647), Expect = 0.0
Identities = 304/463 (65%), Positives = 365/463 (78%), Gaps = 11/463 (2%)
Query: 30 LSGMRIDKEDLKKQLTLPQYLRFAMRDSIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVF 89
L+ +R+ K +L++++ LPQYLR A+RD I +D S + ++ P P++VF
Sbjct: 35 LASIRVSKAELRQRVMLPQYLRIAIRDCILRKDDSFDASSSVAPPLENNALTPEVPLMVF 94
Query: 90 INARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETR 149
+N +SGGR GP +KERLQ L+SEEQV+DL +VKP+EF+ YGL CLE A GD CAKE R
Sbjct: 95 VNPKSGGRQGPLIKERLQNLISEEQVYDLTEVKPNEFIRYGLGCLEAFASRGDECAKEIR 154
Query: 150 EKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAW 209
EK+R++VAGGDGTVGWVLGCL EL R PVPPV I+PLGTGNDLSRSF WGGSFPFAW
Sbjct: 155 EKMRIVVAGGDGTVGWVLGCLGELNLQNRLPVPPVSIMPLGTGNDLSRSFGWGGSFPFAW 214
Query: 210 KSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFTLDQGIEIEGELPD 269
KSAIKRTL +ASV + RLDSW + I+MP V PPY LK +E +DQ +EIEGE+P
Sbjct: 215 KSAIKRTLHRASVAPISRLDSWNILITMPSGEIVDPPYSLKATQECYIDQNLEIEGEIPP 274
Query: 270 KVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPC 329
YEGV+YNYFSIGMDAQVAYGFH LR+EKPYLA+GPIANKIIYSGY C+QGWF T C
Sbjct: 275 STNGYEGVFYNYFSIGMDAQVAYGFHHLRNEKPYLANGPIANKIIYSGYGCSQGWFLTHC 334
Query: 330 TSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYL 389
+DPGLRGL+NI+ +HIK++ SSEWE+V +PKSVRA+VALNLHSYGSGRNPWG K +YL
Sbjct: 335 INDPGLRGLKNIMTLHIKKLDSSEWEKVPVPKSVRAVVALNLHSYGSGRNPWGNLKQDYL 394
Query: 390 EKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIA-----------QAAAIRLEL 438
EK+GFVEA DG LEIFGLKQGWHASFVMV+LI+AKHIA QAAAIRLE+
Sbjct: 395 EKRGFVEAQADDGLLEIFGLKQGWHASFVMVELISAKHIAQLMCKNLGFNEQAAAIRLEI 454
Query: 439 RGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDG 481
RGG WK+A++QMDGEPWKQP+++D+STFV+IKR P QSLVV G
Sbjct: 455 RGGDWKDAFMQMDGEPWKQPMTRDYSTFVDIKRVPHQSLVVKG 497
>UniRef100_Q9M0C6 Hypothetical protein AT4g30340 [Arabidopsis thaliana]
Length = 490
Score = 616 bits (1588), Expect = e-175
Identities = 301/493 (61%), Positives = 376/493 (76%), Gaps = 16/493 (3%)
Query: 1 MESSPSSTTEDSK------KIQVRSSLVESIRGCGLSGMR---IDKEDLKKQLTLPQYLR 51
ME +P S E S + ++ ++RGCG + + IDKE+L+ +L +P+YLR
Sbjct: 1 MEETPRSVGEASTTNFVAARPSAKTDDAVTMRGCGFANLALVGIDKEELRGRLAMPEYLR 60
Query: 52 FAMRDSIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMS 111
AMRD I+ +D + AP +PMVVFIN +SGGRHGP LKERLQQLM+
Sbjct: 61 IAMRDCIKRKDSTEISDHLLLPGGAAADMAPHAPMVVFINPKSGGRHGPVLKERLQQLMT 120
Query: 112 EEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLT 171
EEQVFDL +VKPHEFV YGL CL+ LA GD CA+E REK+R+MVAGGDGTVGWVLGCL
Sbjct: 121 EEQVFDLTEVKPHEFVRYGLGCLDTLAAKGDECARECREKIRIMVAGGDGTVGWVLGCLG 180
Query: 172 ELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSW 231
EL + G+ +PPVG++PLGTGNDLSRSF+WGGSFPFAW+SA+KRTL +A++GS+ RLDSW
Sbjct: 181 ELHKDGKSHIPPVGVIPLGTGNDLSRSFSWGGSFPFAWRSAMKRTLHRATLGSIARLDSW 240
Query: 232 RLSISMPESTTVKPPYCLKQA-EEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQV 290
++ +SMP V PPY LK EE LDQ ++ +G++P K KSYEGV+YNYFSIGMDAQV
Sbjct: 241 KIVVSMPSGEVVDPPYSLKPTIEETALDQALDADGDVPPKAKSYEGVFYNYFSIGMDAQV 300
Query: 291 AYGFHRLRDEKPYLASGPIANKI-IYSG-YSCTQGWFFTPCTSDPGLRGLRNILRMHIKR 348
AYGFH LR+EKPYLA GP+ NK+ IY +SC F S RGLRNI+++HIK+
Sbjct: 301 AYGFHHLRNEKPYLAQGPVTNKVAIYQNLHSCLNYLFELQLYS----RGLRNIMKIHIKK 356
Query: 349 VSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFG 408
+ SEWE++ +PKSVR+IV LNL++YGSGR+PWG +P+YLEK+GFVEA DG +EIFG
Sbjct: 357 ANCSEWEEIHVPKSVRSIVVLNLYNYGSGRHPWGNLRPKYLEKRGFVEAHCDDGLIEIFG 416
Query: 409 LKQGWHASFVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVE 468
LKQGWHASFVM ++I+AKHIAQAAAIR ELRGG WKNA+LQMDGEPWKQP+ D+STFVE
Sbjct: 417 LKQGWHASFVMAEIISAKHIAQAAAIRFELRGGDWKNAFLQMDGEPWKQPMKSDYSTFVE 476
Query: 469 IKREPFQSLVVDG 481
IK+ PFQSL+++G
Sbjct: 477 IKKVPFQSLMING 489
>UniRef100_Q6K4P5 Diacylglycerol kinase-like [Oryza sativa]
Length = 488
Score = 604 bits (1558), Expect = e-171
Identities = 290/486 (59%), Positives = 367/486 (74%), Gaps = 5/486 (1%)
Query: 1 MESSPSSTTEDSKKIQVRSSLVESIRGCGLSGMRIDKEDLKKQLTLPQYLRFAMRDSIRL 60
ME + + S R S+ ES+R CG+ G +DK +L++Q+ +P Y R A+ +++
Sbjct: 1 MERAEETPRAPSTAPSARVSIWESVRACGVWGKEVDKAELRRQVVMPLYARRAVAAAVKA 60
Query: 61 QDPSAGETLYRNRAE-GE---DSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVF 116
+D + G R E GE + A +P+VVF+N+RSGGRHGP LK RL +L+SEEQVF
Sbjct: 61 KDEAVGVAAAAERGEEGEVEVEVEAAVTPVVVFVNSRSGGRHGPELKVRLHELISEEQVF 120
Query: 117 DLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQL 176
DL+ VKP +F+ YGL CLE LA GD+CA+ R+KLR+MVAGGDGTVGWVLGCLT+L +L
Sbjct: 121 DLSVVKPSDFINYGLGCLEKLAEQGDNCAETIRKKLRIMVAGGDGTVGWVLGCLTDLYRL 180
Query: 177 GREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSIS 236
REPVPP GI+PLGTGNDL+RSF WGGSFPF W+SA+KR L KA+ LDSW+ +
Sbjct: 181 KREPVPPTGIIPLGTGNDLARSFGWGGSFPFGWRSAVKRYLSKAATAPTCSLDSWQAVVM 240
Query: 237 MPESTTVKPPYCLKQAEEFTLDQGIEIEG-ELPDKVKSYEGVYYNYFSIGMDAQVAYGFH 295
MP+ + PY LK+ E + + G ELP+K Y+GV+YNY SIGMDAQVAYGFH
Sbjct: 241 MPDGEIKELPYALKKTEPADCLELCQENGTELPEKASCYKGVFYNYLSIGMDAQVAYGFH 300
Query: 296 RLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWE 355
LRDEKPYLA GP+ANK+IY+GYSCTQGWF TPCT+ P LRGL+NILR++IK+V+ SEWE
Sbjct: 301 HLRDEKPYLAQGPVANKLIYAGYSCTQGWFCTPCTASPQLRGLKNILRLYIKKVNCSEWE 360
Query: 356 QVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHA 415
QV +P SVR++V LNL++YGSGR+PWG KP+YLEKKGFVEA DG LEIFGLK+GWHA
Sbjct: 361 QVTMPSSVRSLVVLNLYNYGSGRHPWGDLKPDYLEKKGFVEAHSDDGLLEIFGLKEGWHA 420
Query: 416 SFVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQ 475
SFVM +LI AKHIAQAAAI+ E+RGG W AY+QMDGEPWKQPL ++ ST +EI + P+
Sbjct: 421 SFVMAELIKAKHIAQAAAIKFEMRGGQWNRAYVQMDGEPWKQPLLQEQSTIIEINKVPYP 480
Query: 476 SLVVDG 481
SL+++G
Sbjct: 481 SLMING 486
>UniRef100_Q9ZV46 Putative diacylglycerol kinase [Arabidopsis thaliana]
Length = 475
Score = 585 bits (1507), Expect = e-165
Identities = 290/484 (59%), Positives = 361/484 (73%), Gaps = 20/484 (4%)
Query: 4 SPSSTTEDSKK--IQVRSSLVES--IRGCGLSGMR---IDKEDLKKQLTLPQYLRFAMRD 56
SP S T+ SK+ + R S +S +RGCGL+ + +DK +L+++L +P+YLR AMRD
Sbjct: 3 SPVSKTDASKEKFVASRPSTADSKTMRGCGLANLAWVGVDKVELRQRLMMPEYLRLAMRD 62
Query: 57 SIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVF 116
I+ +D SA AP +PMVVFIN SGGRHGP LKERLQQLMSEEQV
Sbjct: 63 CIKRKDSSAIPDHLLLPGGAVADMAPHAPMVVFINPNSGGRHGPVLKERLQQLMSEEQVL 122
Query: 117 DLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQL 176
++ F L E L + VAGGDGTVGWVLGCL EL +
Sbjct: 123 LSLNLYFKSFSLGFWGNFESNLYL-------------LCVAGGDGTVGWVLGCLGELNKD 169
Query: 177 GREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSIS 236
G+ +PPVG++PLGTGNDLSRSF WGGSFPFAW+SA+KRTL +AS+G V RLDSW++ +S
Sbjct: 170 GKSQIPPVGVIPLGTGNDLSRSFGWGGSFPFAWRSAVKRTLHRASMGPVARLDSWKILVS 229
Query: 237 MPESTTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHR 296
MP V PPY LK AEE LDQG++ + P K+YEGV+YNY SIGMDAQVAYGFH
Sbjct: 230 MPSGEVVDPPYSLKPAEENELDQGLDAGIDAPPLAKAYEGVFYNYLSIGMDAQVAYGFHH 289
Query: 297 LRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQ 356
LR+ KPYLA GPI+NKIIYS + C+QGWF TPC +DPGLRGLRNI+++HIK+V+ S+WE+
Sbjct: 290 LRNTKPYLAQGPISNKIIYSSFGCSQGWFCTPCVNDPGLRGLRNIMKIHIKKVNCSQWEE 349
Query: 357 VAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHAS 416
+A+PK+VR+IVALNLHSYGSG +PWG KP+YLEK+GFVEA DG +EIFG KQGWHAS
Sbjct: 350 IAVPKNVRSIVALNLHSYGSGSHPWGNLKPDYLEKRGFVEAHCDDGLIEIFGFKQGWHAS 409
Query: 417 FVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQS 476
FVM +LI+AKHIAQAAA+R ELRGG W++A+LQMDGEPWKQP+S ++STFVEIK+ P+QS
Sbjct: 410 FVMAELISAKHIAQAAAVRFELRGGDWRDAFLQMDGEPWKQPMSTEYSTFVEIKKVPYQS 469
Query: 477 LVVD 480
L+++
Sbjct: 470 LMIN 473
>UniRef100_Q6YW20 Putative diacylglycerol kinase variant A [Oryza sativa]
Length = 527
Score = 499 bits (1286), Expect = e-140
Identities = 276/516 (53%), Positives = 334/516 (64%), Gaps = 47/516 (9%)
Query: 3 SSPSSTTEDSKKIQVRSSLVESI--RGCGLSGMRI---DKEDLKKQLTLPQYLRFAMRDS 57
S P S+T + + L+ES+ R CG + +KEDL+ + LPQ LR A+ +
Sbjct: 19 SPPRSSTPPPQPLV--GGLIESLSFRSCGFGRAAVSAFEKEDLRARAALPQRLRAAVHAA 76
Query: 58 IRLQDPSAGETLY-------RNRAEGEDSA------------------APTSPMVVFINA 92
+R +DPSAG Y R G S AP SP+V F+N
Sbjct: 77 LRAKDPSAGAFAYVAGNAAARGGRGGGGSGGGREVSAANPWFEVAHDDAPESPLVAFVNP 136
Query: 93 RSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKL 152
RSGGR GP LK RLQ+L+ E+QVFDL VKP +FV Y L CLE LA GD AK R L
Sbjct: 137 RSGGRLGPVLKTRLQELIGEDQVFDLTVVKPSDFVQYVLGCLEQLADAGDHSAKSIRHNL 196
Query: 153 RVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSA 212
RVMVAGGDGTVGWVLGCL +L REP+PPV ++PLGTGNDLSRSF WG SFPF WK+A
Sbjct: 197 RVMVAGGDGTVGWVLGCLGDLYVQNREPIPPVAVIPLGTGNDLSRSFGWGASFPFGWKAA 256
Query: 213 IKRTLQKASVGSVHRLDSWRLSISMP----ESTTVKPPYCLKQAEEFTLDQGIEIEGELP 268
KR+L KA GSV LDSW + +SMP E + P+ L+ E T EGELP
Sbjct: 257 AKRSLYKAIFGSVSCLDSWHIVVSMPERGDEEEELDFPHSLRNLGECTFYDDGTAEGELP 316
Query: 269 DKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKII---YSGYSCTQGWF 325
+ V ++GV+YNYFSIGMDAQVAYGFH LRDEKP+LASGP++NK G C F
Sbjct: 317 ETVSCFDGVFYNYFSIGMDAQVAYGFHHLRDEKPFLASGPLSNKGFNWDVIGTWCAASDF 376
Query: 326 FTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPK 385
P GL +I + R S + + VRAIVALNLH+Y SGRNPWG K
Sbjct: 377 HMPAV---GL-AAHDITFFNTYRNS----QAINFDLIVRAIVALNLHNYASGRNPWGNLK 428
Query: 386 PEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRLELRGGGWKN 445
PEYLEK+GFVEA DG LEIFGLKQGWHAS VMV+LI+AKHIAQAAAIRLE++GG W++
Sbjct: 429 PEYLEKRGFVEAQSDDGLLEIFGLKQGWHASLVMVELISAKHIAQAAAIRLEIKGGQWRD 488
Query: 446 AYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDG 481
AY+QMDGEPWKQPL ++STFV+IK+ P+ SL+++G
Sbjct: 489 AYMQMDGEPWKQPLDHEYSTFVDIKKVPYPSLIING 524
>UniRef100_Q9C5L2 Hypothetical protein At4g30340 [Arabidopsis thaliana]
Length = 374
Score = 469 bits (1207), Expect = e-131
Identities = 223/371 (60%), Positives = 280/371 (75%), Gaps = 10/371 (2%)
Query: 1 MESSPSSTTEDSK------KIQVRSSLVESIRGCGLSGMR---IDKEDLKKQLTLPQYLR 51
ME +P S E S + ++ ++RGCG + + IDKE+L+ +L +P+YLR
Sbjct: 1 MEETPRSVGEASTTNFVAARPSAKTDDAVTMRGCGFANLALVGIDKEELRGRLAMPEYLR 60
Query: 52 FAMRDSIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMS 111
AMRD I+ +D + AP +PMVVFIN +SGGRHGP LKERLQQLM+
Sbjct: 61 IAMRDCIKRKDSTEISDHLLLPGGAAADMAPHAPMVVFINPKSGGRHGPVLKERLQQLMT 120
Query: 112 EEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLT 171
EEQVFDL +VKPHEFV YGL CL+ LA GD CA+E REK+R+MVAGGDGTVGWVLGCL
Sbjct: 121 EEQVFDLTEVKPHEFVRYGLGCLDTLAAKGDECARECREKIRIMVAGGDGTVGWVLGCLG 180
Query: 172 ELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSW 231
EL + G+ +PPVG++PLGTGNDLSRSF+WGGSFPFAW+SA+KRTL +A++GS+ RLDSW
Sbjct: 181 ELHKDGKSHIPPVGVIPLGTGNDLSRSFSWGGSFPFAWRSAMKRTLHRATLGSIARLDSW 240
Query: 232 RLSISMPESTTVKPPYCLKQA-EEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQV 290
++ +SMP V PPY LK EE LDQ ++ +G++P K KSYEGV+YNYFSIGMDAQV
Sbjct: 241 KIVVSMPSGEVVDPPYSLKPTIEETALDQALDADGDVPPKAKSYEGVFYNYFSIGMDAQV 300
Query: 291 AYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVS 350
AYGFH LR+EKPYLA GP+ NKIIYS YSCTQGWF TPC ++P LRGLRNI+++HIK+ +
Sbjct: 301 AYGFHHLRNEKPYLAQGPVTNKIIYSSYSCTQGWFCTPCVNNPALRGLRNIMKIHIKKAN 360
Query: 351 SSEWEQVAIPK 361
SEWE++ +PK
Sbjct: 361 CSEWEEIHVPK 371
>UniRef100_Q5YJN9 Diacylglycerol kinase [Hyacinthus orientalis]
Length = 202
Score = 314 bits (805), Expect = 3e-84
Identities = 146/201 (72%), Positives = 164/201 (80%)
Query: 277 VYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLR 336
V+YNY SIGMDAQVAYGFH LRD KPYLA GPI NK+IYSGYSCTQGWF TPC + PGLR
Sbjct: 1 VFYNYLSIGMDAQVAYGFHHLRDTKPYLAQGPITNKMIYSGYSCTQGWFCTPCVATPGLR 60
Query: 337 GLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVE 396
GL+NI +K EWEQV +P VR+IVALNLH+YGSGRNPWG PKPEYLEK+GFVE
Sbjct: 61 GLKNIQGFILKESIVQEWEQVPVPSDVRSIVALNLHNYGSGRNPWGHPKPEYLEKRGFVE 120
Query: 397 ADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWK 456
+ G LEIFGLKQGWHASFVMV+LI AKHIAQAAAIR E+RGG WK AY+QMDGEPWK
Sbjct: 121 SHADHGLLEIFGLKQGWHASFVMVELIDAKHIAQAAAIRFEIRGGRWKKAYMQMDGEPWK 180
Query: 457 QPLSKDFSTFVEIKREPFQSL 477
Q ++ ++ST VEI+ P SL
Sbjct: 181 QAINTEYSTLVEIRTGPLTSL 201
>UniRef100_Q7XQT2 OSJNBa0043L09.15 protein [Oryza sativa]
Length = 499
Score = 258 bits (658), Expect = 3e-67
Identities = 144/396 (36%), Positives = 215/396 (53%), Gaps = 19/396 (4%)
Query: 81 APTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLAC-LEMLAG 139
AP P+VVFIN+RSGG+ G +L + ++L+++ QVFDL++ P E VL+ L C E L
Sbjct: 41 APCCPVVVFINSRSGGQLGSSLIKTYRELLNKAQVFDLSEEAP-EKVLHRLYCNFEKLKS 99
Query: 140 LGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSF 199
GD A + + LR++VAGGDGT W+LG +++L+ PP+ VPLGTGN+L SF
Sbjct: 100 NGDPIAFQIQSNLRLIVAGGDGTASWLLGVVSDLKL---SHPPPIATVPLGTGNNLPFSF 156
Query: 200 NWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP------PYCLKQ 251
WG P + A+K L + +DSW + + M P+ +P P+ L
Sbjct: 157 GWGKKNPTTDQEAVKSFLGQVKKAREMNIDSWHIIMRMRAPQEGPCEPIAPLELPHSLHA 216
Query: 252 AEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIAN 311
+ + +EG +Y G ++NYFS+GMDAQV+Y FH R P + N
Sbjct: 217 FHRVSGSDSLNMEG-----YHTYRGGFWNYFSMGMDAQVSYEFHSERKRNPEKFKNQLTN 271
Query: 312 KIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNL 371
+ Y+ QGWF T P R + + ++ I + +WE++ IP+S+R+IV LNL
Sbjct: 272 QSTYAKLGLKQGWFAASLTH-PSSRNIAQLAKVRIMKRPGGQWEELKIPRSIRSIVCLNL 330
Query: 372 HSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQA 431
S+ G NPWG P ++ + V DG +E+ G + WH ++ +AQA
Sbjct: 331 PSFSGGLNPWGTPGTRKVQDRDLTAPFVDDGLIEVVGFRDAWHGLVLLAPNGHGTRLAQA 390
Query: 432 AAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E G ++ ++++DGEPWKQPL KD T V
Sbjct: 391 HRIRFEFHKGAAEHTFMRIDGEPWKQPLPKDDDTVV 426
>UniRef100_Q9FVD0 Diacylglycerol kinase variant A [Lycopersicon esculentum]
Length = 489
Score = 249 bits (637), Expect = 9e-65
Identities = 137/394 (34%), Positives = 209/394 (52%), Gaps = 18/394 (4%)
Query: 82 PTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
PT P++VF+N++SGG+ G L + L+++ QVFDL D P + +E L G G
Sbjct: 37 PTCPVLVFVNSKSGGQLGGELLRTFRHLLNKYQVFDLGDEAPDSVLRRLYLNIERLKGNG 96
Query: 142 DSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNW 201
D A E E+++++VAGGDGT GW+LG +++L+ PP+ VPLGTGN+L +F W
Sbjct: 97 DHFAAEIEERMKIIVAGGDGTAGWLLGVVSDLKL---SQPPPIATVPLGTGNNLPFAFGW 153
Query: 202 GGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP------PYCLKQAE 253
G P +++ L++ ++DSW + + M P+ + P P+ L
Sbjct: 154 GKKNPGTDLNSVISFLKQVMNAKEMKMDSWHILMRMRAPKVGSCDPVAPLELPHSLHAFH 213
Query: 254 EFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKI 313
+ + +EG ++ G ++NYFS+GMDAQV+Y FH R P + N+
Sbjct: 214 RVSPSDELNVEG-----FHTFRGGFWNYFSMGMDAQVSYAFHSERKMNPDKFKNQLVNQS 268
Query: 314 IYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHS 373
Y CTQGWFF P P + + + ++ I + EW+ + IP SVR+IV LNL S
Sbjct: 269 SYLKLGCTQGWFFAPLIR-PSSKNIAQLTKVKIMK-KQGEWQDLHIPPSVRSIVCLNLPS 326
Query: 374 YGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAA 433
+ G NPWG P + V DG LE+ G + WH ++ +AQA
Sbjct: 327 FSGGLNPWGTPNSNKRRYRDLTPPFVDDGLLEVVGFRDAWHGLVLLAPKGHGTRLAQAHG 386
Query: 434 IRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E + G + ++++DGEPWKQPL ++ T V
Sbjct: 387 IRFEFQKGAADHTFMRIDGEPWKQPLPENDDTVV 420
>UniRef100_Q9FVC9 Diacylglycerol kinase variant B [Lycopersicon esculentum]
Length = 511
Score = 249 bits (637), Expect = 9e-65
Identities = 137/394 (34%), Positives = 209/394 (52%), Gaps = 18/394 (4%)
Query: 82 PTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
PT P++VF+N++SGG+ G L + L+++ QVFDL D P + +E L G G
Sbjct: 37 PTCPVLVFVNSKSGGQLGGELLRTFRHLLNKYQVFDLGDEAPDSVLRRLYLNIERLKGNG 96
Query: 142 DSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNW 201
D A E E+++++VAGGDGT GW+LG +++L+ PP+ VPLGTGN+L +F W
Sbjct: 97 DHFAAEIEERMKIIVAGGDGTAGWLLGVVSDLKL---SQPPPIATVPLGTGNNLPFAFGW 153
Query: 202 GGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP------PYCLKQAE 253
G P +++ L++ ++DSW + + M P+ + P P+ L
Sbjct: 154 GKKNPGTDLNSVISFLKQVMNAKEMKMDSWHILMRMRAPKVGSCDPVAPLELPHSLHAFH 213
Query: 254 EFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKI 313
+ + +EG ++ G ++NYFS+GMDAQV+Y FH R P + N+
Sbjct: 214 RVSPSDELNVEG-----FHTFRGGFWNYFSMGMDAQVSYAFHSERKMNPDKFKNQLVNQS 268
Query: 314 IYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHS 373
Y CTQGWFF P P + + + ++ I + EW+ + IP SVR+IV LNL S
Sbjct: 269 SYLKLGCTQGWFFAPLIR-PSSKNIAQLTKVKIMK-KQGEWQDLHIPPSVRSIVCLNLPS 326
Query: 374 YGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAA 433
+ G NPWG P + V DG LE+ G + WH ++ +AQA
Sbjct: 327 FSGGLNPWGTPNSNKRRYRDLTPPFVDDGLLEVVGFRDAWHGLVLLAPKGHGTRLAQAHG 386
Query: 434 IRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E + G + ++++DGEPWKQPL ++ T V
Sbjct: 387 IRFEFQKGAADHTFMRIDGEPWKQPLPENDDTVV 420
>UniRef100_Q9FVD2 Calmodulin-binding diacylglycerol kinase [Lycopersicon esculentum]
Length = 511
Score = 247 bits (630), Expect = 6e-64
Identities = 136/394 (34%), Positives = 208/394 (52%), Gaps = 18/394 (4%)
Query: 82 PTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
PT P++VF+N++SGG+ G L + L+++ QVFDL D P + +E L G G
Sbjct: 37 PTCPVLVFVNSKSGGQLGGELLRTFRHLLNKYQVFDLGDEAPDSVLRRLYLNIERLKGNG 96
Query: 142 DSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNW 201
D A E E+++++VAGGDGT GW+LG +++L+ PP+ VPLGTGN+L +F W
Sbjct: 97 DHFAAEIEERMKIIVAGGDGTAGWLLGVVSDLKL---SQPPPIATVPLGTGNNLPFAFGW 153
Query: 202 GGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP------PYCLKQAE 253
G P +++ L++ ++DSW + + M P+ + P P+ L
Sbjct: 154 GKKNPGTDLNSVISFLKQVMNAKEMKMDSWHILMRMRAPKVGSCDPVAPLELPHSLHAFH 213
Query: 254 EFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKI 313
+ + +EG ++ G ++NYFS+GMDAQV+Y FH R P + N+
Sbjct: 214 RVSPSDELNVEG-----FHTFRGGFWNYFSMGMDAQVSYAFHSERKMNPDKFKNQLVNQS 268
Query: 314 IYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHS 373
Y CTQGWFF P P + + + ++ I + W+ + IP SVR+IV LNL S
Sbjct: 269 SYLKLGCTQGWFFAPLIH-PSSKNIAQLTKVKIMKKQGG-WQDLHIPPSVRSIVCLNLPS 326
Query: 374 YGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAA 433
+ G NPWG P + V DG LE+ G + WH ++ +AQA
Sbjct: 327 FSGGLNPWGTPNSNKRRYRDLTPPFVNDGLLEVVGFRDAWHGLVLLAPKGHGTRLAQAHG 386
Query: 434 IRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E + G + ++++DGEPWKQPL ++ T V
Sbjct: 387 IRFEFQKGAADHTFMRIDGEPWKQPLPENDDTVV 420
>UniRef100_Q9FVD1 Diacylglycerol kinase [Lycopersicon esculentum]
Length = 489
Score = 247 bits (630), Expect = 6e-64
Identities = 136/394 (34%), Positives = 208/394 (52%), Gaps = 18/394 (4%)
Query: 82 PTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
PT P++VF+N++SGG+ G L + L+++ QVFDL D P + +E L G G
Sbjct: 37 PTCPVLVFVNSKSGGQLGGELLRTFRHLLNKYQVFDLGDEAPDSVLRRLYLNIERLKGNG 96
Query: 142 DSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNW 201
D A E E+++++VAGGDGT GW+LG +++L+ PP+ VPLGTGN+L +F W
Sbjct: 97 DHFAAEIEERMKIIVAGGDGTAGWLLGVVSDLKL---SQPPPIATVPLGTGNNLPFAFGW 153
Query: 202 GGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP------PYCLKQAE 253
G P +++ L++ ++DSW + + M P+ + P P+ L
Sbjct: 154 GKKNPGTDLNSVISFLKQVMNAKEMKMDSWHILMRMRAPKVGSCDPVAPLELPHSLHAFH 213
Query: 254 EFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKI 313
+ + +EG ++ G ++NYFS+GMDAQV+Y FH R P + N+
Sbjct: 214 RVSPSDELNVEG-----FHTFRGGFWNYFSMGMDAQVSYAFHSERKMNPDKFKNQLVNQS 268
Query: 314 IYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHS 373
Y CTQGWFF P P + + + ++ I + W+ + IP SVR+IV LNL S
Sbjct: 269 SYLKLGCTQGWFFAPLIH-PSSKNIAQLTKVKIMKKQGG-WQDLHIPPSVRSIVCLNLPS 326
Query: 374 YGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAA 433
+ G NPWG P + V DG LE+ G + WH ++ +AQA
Sbjct: 327 FSGGLNPWGTPNSNKRRYRDLTPPFVDDGLLEVVGFRDAWHGLVLLAPKGHGTRLAQAHG 386
Query: 434 IRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E + G + ++++DGEPWKQPL ++ T V
Sbjct: 387 IRFEFQKGAADHTFMRIDGEPWKQPLPENDDTVV 420
>UniRef100_Q5ZAU2 Putative diacylglycerol kinase [Oryza sativa]
Length = 487
Score = 246 bits (629), Expect = 8e-64
Identities = 145/391 (37%), Positives = 214/391 (54%), Gaps = 16/391 (4%)
Query: 79 SAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFV--LYGLACLEM 136
S P+ P++VFIN +SGG+ G L ++L++ QVFDL + P + + LYG +E
Sbjct: 31 SQIPSCPVIVFINTKSGGQLGHDLIVTYRKLLNNSQVFDLLEEAPDKVLHKLYGN--MER 88
Query: 137 LAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLS 196
L GD+ A E +LR++VAGGDGT GW+LG +++L+ + PPV VPLGTGN+L
Sbjct: 89 LMRDGDTVAAEIHRRLRLIVAGGDGTAGWLLGVVSDLKLV---HPPPVATVPLGTGNNLP 145
Query: 197 RSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKPPYCLKQAEE 254
SF WG P + ++ LQ ++DSW + + M P+S+T P L
Sbjct: 146 YSFGWGKRNPGTDEKSVLSFLQSVRQAKEMKIDSWHIVMKMESPKSSTCDPIAPLDLPHS 205
Query: 255 FTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKII 314
+ + + ++ G ++NYFS+GMDAQV+Y FH R P ++N+
Sbjct: 206 LHAFHRVPNNPQDKEYSCTFRGGFWNYFSMGMDAQVSYAFHSERKLHPEKFKNQLSNQKT 265
Query: 315 YSGYSCTQGWFFTPCTS--DPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLH 372
Y +CTQGWF C S P R + ++ ++ I + S +WE + IP+S+R+IV LNL
Sbjct: 266 YLKLACTQGWF---CASLCHPMSRNIAHLSKVKIMK-KSGKWETLEIPQSIRSIVCLNLP 321
Query: 373 SYGSGRNPWGKPKPEYLEKKGFVEAD-VADGRLEIFGLKQGWHASFVMVDLITAKHIAQA 431
S+ G NPWG P K+ V V DG LEI G K WH ++ +AQA
Sbjct: 322 SFSGGLNPWGTPSERKQRKRDLVMPPLVDDGLLEIVGFKDAWHGLVLLSPKGHGTRLAQA 381
Query: 432 AAIRLELRGGGWKNAYLQMDGEPWKQPLSKD 462
++ + G +AY+++DGEPW QPL KD
Sbjct: 382 HRVQFKFHKGATDHAYMRLDGEPWNQPLPKD 412
>UniRef100_Q9C5E5 Putative diacylglycerol kinase [Arabidopsis thaliana]
Length = 509
Score = 246 bits (627), Expect = 1e-63
Identities = 143/402 (35%), Positives = 211/402 (51%), Gaps = 16/402 (3%)
Query: 75 EGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACL 134
E E P SP++VFIN++SGG+ G L + L++ QVFDL P + + L
Sbjct: 30 EEESRPTPASPVLVFINSKSGGQLGGELILTYRSLLNHNQVFDLDQETPDKVLRRIYLNL 89
Query: 135 EMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGND 194
E L D A++ REKL+++VAGGDGT GW+LG + +L+ PP+ VPLGTGN+
Sbjct: 90 ERLKD--DDFARQIREKLKIIVAGGDGTAGWLLGVVCDLKL---SHPPPIATVPLGTGNN 144
Query: 195 LSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMP---ESTTVKPPYCLKQ 251
L +F WG P ++A++ L++ V ++D+W + + M E + P L+
Sbjct: 145 LPFAFGWGKKNPGTDRTAVESFLEQVLKAKVMKIDNWHILMRMKTPKEGGSCDPVAPLEL 204
Query: 252 AEEFTLDQGIEIEGEL-PDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIA 310
+ EL + ++ G ++NYFS+GMDAQ++Y FH R P +
Sbjct: 205 PHSLHAFHRVSPTDELNKEGCHTFRGGFWNYFSLGMDAQISYAFHSERKLHPEKFKNQLV 264
Query: 311 NKIIYSGYSCTQGWFFTPCTS--DPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVA 368
N+ Y CTQGWF C S P R + + ++ I + +W+ + IP S+R+IV
Sbjct: 265 NQSTYVKLGCTQGWF---CASLFHPASRNIAQLAKVKIA-TRNGQWQDLHIPHSIRSIVC 320
Query: 369 LNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHI 428
LNL S+ G NPWG P P +G V DG +E+ G + WH ++ +
Sbjct: 321 LNLPSFSGGLNPWGTPNPRKQRDRGLTPPFVDDGLIEVVGFRNAWHGLVLLAPNGHGTRL 380
Query: 429 AQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFST-FVEI 469
AQA IR E G + +++MDGEPWKQPL D T VEI
Sbjct: 381 AQANRIRFEFHKGATDHTFMRMDGEPWKQPLPLDDETVMVEI 422
>UniRef100_Q9SKS9 Putative diacylglycerol kinase [Arabidopsis thaliana]
Length = 491
Score = 246 bits (627), Expect = 1e-63
Identities = 143/402 (35%), Positives = 211/402 (51%), Gaps = 16/402 (3%)
Query: 75 EGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACL 134
E E P SP++VFIN++SGG+ G L + L++ QVFDL P + + L
Sbjct: 30 EEESRPTPASPVLVFINSKSGGQLGGELILTYRSLLNHNQVFDLDQETPDKVLRRIYLNL 89
Query: 135 EMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGND 194
E L D A++ REKL+++VAGGDGT GW+LG + +L+ PP+ VPLGTGN+
Sbjct: 90 ERLKD--DDFARQIREKLKIIVAGGDGTAGWLLGVVCDLKL---SHPPPIATVPLGTGNN 144
Query: 195 LSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMP---ESTTVKPPYCLKQ 251
L +F WG P ++A++ L++ V ++D+W + + M E + P L+
Sbjct: 145 LPFAFGWGKKNPGTDRTAVESFLEQVLKAKVMKIDNWHILMRMKTPKEGGSCDPVAPLEL 204
Query: 252 AEEFTLDQGIEIEGEL-PDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIA 310
+ EL + ++ G ++NYFS+GMDAQ++Y FH R P +
Sbjct: 205 PHSLHAFHRVSPTDELNKEGCHTFRGGFWNYFSLGMDAQISYAFHSERKLHPEKFKNQLV 264
Query: 311 NKIIYSGYSCTQGWFFTPCTS--DPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVA 368
N+ Y CTQGWF C S P R + + ++ I + +W+ + IP S+R+IV
Sbjct: 265 NQSTYVKLGCTQGWF---CASLFHPASRNIAQLAKVKIA-TRNGQWQDLHIPHSIRSIVC 320
Query: 369 LNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHI 428
LNL S+ G NPWG P P +G V DG +E+ G + WH ++ +
Sbjct: 321 LNLPSFSGGLNPWGTPNPRKQRDRGLTPPFVDDGLIEVVGFRNAWHGLVLLAPNGHGTRL 380
Query: 429 AQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFST-FVEI 469
AQA IR E G + +++MDGEPWKQPL D T VEI
Sbjct: 381 AQANRIRFEFHKGATDHTFMRMDGEPWKQPLPLDDETVMVEI 422
>UniRef100_Q8LE70 Putative diacylglycerol kinase [Arabidopsis thaliana]
Length = 491
Score = 243 bits (621), Expect = 7e-63
Identities = 142/402 (35%), Positives = 210/402 (51%), Gaps = 16/402 (3%)
Query: 75 EGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACL 134
E E P SP++VFIN++SGG+ G L + L++ QVFDL P + + L
Sbjct: 30 EQESRPTPASPVLVFINSKSGGQLGGELILTYRSLLNHNQVFDLDQETPDKVLRRIYLNL 89
Query: 135 EMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGND 194
E L D A++ EKL+++VAGGDGT GW+LG + +L+ PP+ VPLGTGN+
Sbjct: 90 ERLKD--DDSARQIGEKLKIIVAGGDGTAGWLLGVVCDLKL---SHPPPIATVPLGTGNN 144
Query: 195 LSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMP---ESTTVKPPYCLKQ 251
L +F WG P ++A++ L++ V ++D+W + + M E + P L+
Sbjct: 145 LPFAFGWGKKNPGTDRTAVESFLEQVLKAKVMKIDNWHILMRMKTPKEGGSCDPVAPLEL 204
Query: 252 AEEFTLDQGIEIEGEL-PDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIA 310
+ EL + ++ G ++NYFS+GMDAQ++Y FH R P +
Sbjct: 205 PHSLHAFHRVSPTDELNKEGCHTFRGGFWNYFSLGMDAQISYAFHSERKLHPEKFKNQLV 264
Query: 311 NKIIYSGYSCTQGWFFTPCTS--DPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVA 368
N+ Y CTQGWF C S P R + + ++ I + +W+ + IP S+R+IV
Sbjct: 265 NQSTYVKLGCTQGWF---CASLFHPASRNIAQLAKVKIA-TRNGQWQDLHIPHSIRSIVC 320
Query: 369 LNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHI 428
LNL S+ G NPWG P P +G V DG +E+ G + WH ++ +
Sbjct: 321 LNLPSFSGGLNPWGTPNPRKQRDRGLTPPFVDDGLIEVVGFRNAWHGLVLLAPNGHGTRL 380
Query: 429 AQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFST-FVEI 469
AQA IR E G + +++MDGEPWKQPL D T VEI
Sbjct: 381 AQANRIRFEFHKGATDHTFMRMDGEPWKQPLPLDDETVMVEI 422
>UniRef100_Q6ZBT9 Putative diacylglycerol kinase [Oryza sativa]
Length = 498
Score = 237 bits (605), Expect = 5e-61
Identities = 132/395 (33%), Positives = 208/395 (52%), Gaps = 17/395 (4%)
Query: 81 APTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGL 140
AP P++VFIN+ SGG+ G +L + ++L+ E QVFD+++ P + + LE L
Sbjct: 37 APPCPVLVFINSGSGGQLGSSLIKTYRELLGEAQVFDVSEEAPDKVLHRLNVNLEKLKMD 96
Query: 141 GDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFN 200
GD A + LR++VAGGDGT W+LG +++L+ PP+ VPLGTGN+L SF
Sbjct: 97 GDILAVQIWRTLRIIVAGGDGTASWLLGVVSDLKL---SHPPPIATVPLGTGNNLPFSFG 153
Query: 201 WGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPES--------TTVKPPYCLKQA 252
WG P + ++K L ++DSW + + M + ++ P+ L
Sbjct: 154 WGKKNPCTDQESVKSFLGLVRHAKEMKIDSWHIMLRMRATKEGPCDPIAPLELPHSLHAF 213
Query: 253 EEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANK 312
+ + +EG ++ G ++NYFS+GMDA+++YGFH R + P + N+
Sbjct: 214 HRVSSSDSLNMEG-----YHTFRGGFWNYFSMGMDAEISYGFHSERKKNPEKFKNQLTNQ 268
Query: 313 IIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLH 372
Y+ QGWF S P R + + + I + + S W+++ I S+R+IV LNL
Sbjct: 269 GTYAKVGLKQGWFCASL-SHPSSRNIAQLASVKIMKRAGSHWQELNIHHSIRSIVCLNLP 327
Query: 373 SYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAA 432
S+ G NPWG P +E++ V D +E+ G + WH ++ +AQA
Sbjct: 328 SFSGGLNPWGTPGTRKVEERELTAPFVDDRLIEVVGFRDAWHGLVLLAPNGHGTRLAQAQ 387
Query: 433 AIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFV 467
IR E G ++ ++++DGEPWKQPL KD T V
Sbjct: 388 RIRFEFHKGAAEHTFMRIDGEPWKQPLPKDDDTVV 422
>UniRef100_Q9FFN7 Diacylglycerol kinase 2 [Arabidopsis thaliana]
Length = 712
Score = 150 bits (379), Expect = 8e-35
Identities = 121/379 (31%), Positives = 173/379 (44%), Gaps = 75/379 (19%)
Query: 85 PMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSC 144
P++VFINA+SGG+ GP L RL L++ QVF+L +C AGL D C
Sbjct: 342 PLLVFINAKSGGQLGPFLHRRLNMLLNPVQVFELG------------SCQGPDAGL-DLC 388
Query: 145 AKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWG-G 203
+K + RV+V GGDGTV WVL + E R E PPV I+PLGTGNDLSR WG G
Sbjct: 389 SKV--KYFRVLVCGGDGTVAWVLDAI-EKRNF--ESPPPVAILPLGTGNDLSRVLQWGRG 443
Query: 204 SFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFTLDQGIEI 263
+ +++ LQ +V LD W S+ + E +T K P ++ +F +
Sbjct: 444 ISVVDGQGSLRTFLQDIDHAAVTMLDRW--SVKIVEESTEKFP--AREGHKFMM------ 493
Query: 264 EGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQG 323
NY IG DA+VAY FH +R EKP NK+ Y+
Sbjct: 494 ----------------NYLGIGCDAKVAYEFHMMRQEKPEKFCSQFVNKLRYA------- 530
Query: 324 WFFTPCTSDPGLRGLRNILRMHIKRVSSSEW-----EQVAIPKSVRAIVALNLHSYGSGR 378
G R+I+ + W + + IPK ++ LN+ SY G
Sbjct: 531 -----------KEGARDIMDRACADLPWQVWLEVDGKDIEIPKDSEGLIVLNIGSYMGGV 579
Query: 379 NPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRLEL 438
+ W + +Y F + D LE+ ++ WH + V L A+ +AQ IR+ +
Sbjct: 580 DLW---QNDYEHDDNFSIQCMHDKTLEVVCVRGAWHLGKLQVGLSQARRLAQGKVIRIHV 636
Query: 439 RGGGWKNAYLQMDGEPWKQ 457
+Q+DGEP+ Q
Sbjct: 637 S----SPFPVQIDGEPFIQ 651
>UniRef100_UPI000017F5C2 UPI000017F5C2 UniRef100 entry
Length = 567
Score = 149 bits (375), Expect = 2e-34
Identities = 110/375 (29%), Positives = 176/375 (46%), Gaps = 64/375 (17%)
Query: 84 SPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDS 143
+P+++ N+RSG G L + L++ QVFD+ P + L++
Sbjct: 216 TPLIILANSRSGTNMGEGLLGEFKMLLNPVQVFDVTKTPP-------IKALQL------- 261
Query: 144 CAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREP-VPPVGIVPLGTGNDLSRSFNWG 202
C +RV+V GGDGTVGWVL + E++ G+E +P V ++PLGTGNDLS + WG
Sbjct: 262 CTLLPYYSVRVLVCGGDGTVGWVLDAIDEMKIKGQEKYIPEVAVLPLGTGNDLSNTLGWG 321
Query: 203 GSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFTLDQGIE 262
+ A + + + L+ +LD W++ + T K Y L++ +EFT++
Sbjct: 322 TGY--AGEIPVAQVLRNVMEADGIKLDRWKVQV------TNKGYYSLRKPKEFTMN---- 369
Query: 263 IEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQ 322
NYFSIG DA +A FH R++ P L S I NK +Y Y T+
Sbjct: 370 -----------------NYFSIGPDALMALNFHAHREKAPSLFSSRILNKAVYLFYG-TR 411
Query: 323 GWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWG 382
C + L + + + + E+V +P ++ ++ LN+ +G G W
Sbjct: 412 DCLVQEC------KDLNKKIELEL------DGERVELP-NLEGVIVLNIGYWGGGCRLW- 457
Query: 383 KPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRLELRGGG 442
E + + + A DG LE+ G+ +H + + V L I QA +RL L+
Sbjct: 458 ----EGVGDETYPLARHDDGLLEVVGVYGSFHCAQIQVKLANPFRIGQAHTVRLTLKCSR 513
Query: 443 WKNAYLQMDGEPWKQ 457
+Q+DGEPW Q
Sbjct: 514 MPMP-MQVDGEPWAQ 527
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 833,014,113
Number of Sequences: 2790947
Number of extensions: 35823336
Number of successful extensions: 73713
Number of sequences better than 10.0: 331
Number of HSP's better than 10.0 without gapping: 225
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 72487
Number of HSP's gapped (non-prelim): 467
length of query: 482
length of database: 848,049,833
effective HSP length: 131
effective length of query: 351
effective length of database: 482,435,776
effective search space: 169334957376
effective search space used: 169334957376
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC136954.8


