

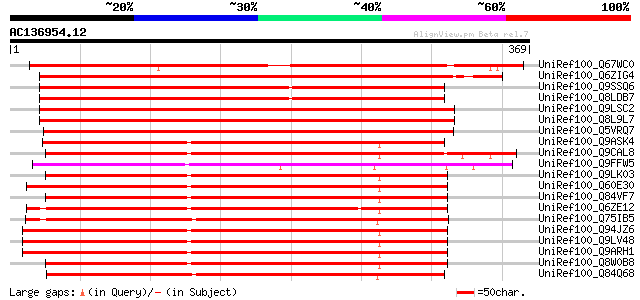
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.12 + phase: 0
(369 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q67WC0 Receptor protein kinase PERK1-like [Oryza sativa] 465 e-130
UniRef100_Q6ZIG4 Receptor protein kinase PERK1-like protein [Ory... 395 e-108
UniRef100_Q9SSQ6 F6D8.24 protein [Arabidopsis thaliana] 335 1e-90
UniRef100_Q8LDB7 Somatic embryogenesis receptor-like kinase, put... 335 1e-90
UniRef100_Q9LSC2 Protein kinase-like protein [Arabidopsis thaliana] 322 1e-86
UniRef100_Q8L9L7 Protein kinase, putative [Arabidopsis thaliana] 320 3e-86
UniRef100_Q5VRQ7 Putative receptor protein kinase PERK1 [Oryza s... 313 4e-84
UniRef100_Q9ASK4 Putative LRR receptor-like protein kinase [Oryz... 298 1e-79
UniRef100_Q9CAL8 Hypothetical protein F24J13.3 [Arabidopsis thal... 293 4e-78
UniRef100_Q9FFW5 Similarity to protein kinase [Arabidopsis thali... 283 4e-75
UniRef100_Q9LK03 Somatic embryogenesis receptor kinase-like prot... 281 3e-74
UniRef100_Q60E30 Hypothetical protein OSJNBb0012L23.7 [Oryza sat... 280 3e-74
UniRef100_Q84VF7 Putative receptor protein kinase [Oryza sativa] 280 5e-74
UniRef100_Q6ZE12 Hypothetical protein P0495H05.37 [Oryza sativa] 280 6e-74
UniRef100_Q75IB5 Hypothetical protein OJ1785_A05.27 [Oryza sativa] 278 2e-73
UniRef100_Q94JZ6 Protein kinase-like protein [Arabidopsis thaliana] 277 3e-73
UniRef100_Q9LV48 Protein kinase-like protein [Arabidopsis thaliana] 277 3e-73
UniRef100_Q9ARH1 Receptor protein kinase PERK1 [Brassica napus] 276 7e-73
UniRef100_Q8W0B8 Putative receptor protein kinase PERK1 [Oryza s... 275 1e-72
UniRef100_Q84Q68 Protein kinase [Oryza sativa] 275 2e-72
>UniRef100_Q67WC0 Receptor protein kinase PERK1-like [Oryza sativa]
Length = 392
Score = 465 bits (1197), Expect = e-130
Identities = 238/379 (62%), Positives = 286/379 (74%), Gaps = 47/379 (12%)
Query: 15 SYGVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMN-S 73
S +++N+WRIF+YKELH ATNGFS++ KLGEGGFGSVYWG+TSDGLQIAVKKLKA N S
Sbjct: 21 SASMSSNTWRIFSYKELHAATNGFSEENKLGEGGFGSVYWGKTSDGLQIAVKKLKATNTS 80
Query: 74 KAEMEFAVEVEVLGRVRHKNLLGLRGYCVGD---DQRLIVYDYMPNLSLLSHLHGQYAGE 130
KAEMEFAVEVEVL RVRHKNLLGLRGYC G DQR+IVYDYMPNLSLLSHLHGQ+A +
Sbjct: 81 KAEMEFAVEVEVLARVRHKNLLGLRGYCAGGAAGDQRMIVYDYMPNLSLLSHLHGQFAAD 140
Query: 131 VQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP 190
V+L+W +RM++A+G+AEG+++LHHE TPHIIHRDIKASNVLLDS F PLVADFG
Sbjct: 141 VRLDWARRMAVAVGAAEGLVHLHHEATPHIIHRDIKASNVLLDSGFAPLVADFG------ 194
Query: 191 EGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRT 250
VKGTLGYLAPEYAMWGKVS +CDVYSFGILLLELV+GRKPIE+LP G KRT
Sbjct: 195 ---------VKGTLGYLAPEYAMWGKVSGACDVYSFGILLLELVSGRKPIERLPSGAKRT 245
Query: 251 ITEWAEPLITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
+TEWAEPLI +GR D+VDP+LRG FD Q+ + V AALCVQ+EPE+RP+M+ VV +L+
Sbjct: 246 VTEWAEPLIARGRLADLVDPRLRGAFDAAQLARAVEAAALCVQAEPERRPDMRAVVRILR 305
Query: 311 GQEPDQGKVTKMRIDSVKYNDELLALDQPS-----------DDDYD-------------G 346
G +R+ S+KY D L+ +D+ S ++D D
Sbjct: 306 G----DADAKPVRMKSIKYADHLMEMDKSSVYYGEDGGGDGEEDMDDEEVEEYSLMEDKS 361
Query: 347 NSSYGVFSAIDVQKMKDPY 365
+ ++GVF A+ VQ M DPY
Sbjct: 362 SVNFGVFGAMPVQTMHDPY 380
>UniRef100_Q6ZIG4 Receptor protein kinase PERK1-like protein [Oryza sativa]
Length = 377
Score = 395 bits (1015), Expect = e-108
Identities = 199/329 (60%), Positives = 247/329 (74%), Gaps = 8/329 (2%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
+WRIF+ KEL +ATN F+ D KLGEGGFGSVYWG+ DG QIAVK+LK+ ++KAE EFA+
Sbjct: 25 TWRIFSLKELQSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKSWSNKAETEFAI 84
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVEVL VRHK+LL LRGYC +RLIVYDYMPNLSL SHLHGQ+A E L W++RM I
Sbjct: 85 EVEVLATVRHKSLLSLRGYCAEGQERLIVYDYMPNLSLHSHLHGQHAAECHLGWERRMKI 144
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVK 201
AI SAEGI YLHH+ TPHIIHRDIK+SNVLLD +F VADFGFAKLIP+G +H+TT+VK
Sbjct: 145 AIDSAEGIAYLHHQATPHIIHRDIKSSNVLLDKNFQARVADFGFAKLIPDGATHVTTKVK 204
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK 261
GTLGYLAPEYAM GK SESCDV+SFG+LLLEL +G++P+EKL K TITEWA PL
Sbjct: 205 GTLGYLAPEYAMLGKASESCDVFSFGVLLLELASGKRPVEKLNPTTKLTITEWALPLARD 264
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQGKVTK 321
+F+++ DPKL+ F E ++K+ V V C Q++ E+RP M +VV LLKG+ + K++
Sbjct: 265 KKFKEIADPKLKDVFVEAELKRMVLVGLACSQNKQEQRPIMSEVVELLKGESAE--KLSN 322
Query: 322 MRIDSVKYNDELLALDQPSDDDYDGNSSY 350
+ NDE+ D S +SS+
Sbjct: 323 LE------NDEMFKPDLTSSFQDSSHSSH 345
>UniRef100_Q9SSQ6 F6D8.24 protein [Arabidopsis thaliana]
Length = 347
Score = 335 bits (859), Expect = 1e-90
Identities = 163/288 (56%), Positives = 215/288 (74%), Gaps = 1/288 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWRIF+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LKA +S+ E++FAV
Sbjct: 21 SWRIFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKAWSSREEIDFAV 80
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RLIVYDYMPNLSL+SHLHGQ++ E L+W +RM+I
Sbjct: 81 EVEILARIRHKNLLSVRGYCAEGQERLIVYDYMPNLSLVSHLHGQHSSESLLDWTRRMNI 140
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVK 201
A+ SA+ I YLHH TP I+H D++ASNVLLDS+F V DFG+ KL+P+ ++ +T+
Sbjct: 141 AVSSAQAIAYLHHFATPRIVHGDVRASNVLLDSEFEARVTDFGYDKLMPDDGANKSTK-G 199
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK 261
+GYL+PE GK S+ DVYSFG+LLLELVTG++P E++ KR ITEW PL+ +
Sbjct: 200 NNIGYLSPECIESGKESDMGDVYSFGVLLLELVTGKRPTERVNLTTKRGITEWVLPLVYE 259
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
+F ++VD +L G + E ++K+ V V +C Q E EKRP M +VV +L
Sbjct: 260 RKFGEIVDQRLNGKYVEEELKRIVLVGLMCAQRESEKRPTMSEVVEML 307
>UniRef100_Q8LDB7 Somatic embryogenesis receptor-like kinase, putative [Arabidopsis
thaliana]
Length = 350
Score = 335 bits (859), Expect = 1e-90
Identities = 163/288 (56%), Positives = 215/288 (74%), Gaps = 1/288 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWRIF+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LKA +S+ E++FAV
Sbjct: 24 SWRIFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKAWSSREEIDFAV 83
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RLIVYDYMPNLSL+SHLHGQ++ E L+W +RM+I
Sbjct: 84 EVEILARIRHKNLLSVRGYCAEGQERLIVYDYMPNLSLVSHLHGQHSSESLLDWTRRMNI 143
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVK 201
A+ SA+ I YLHH TP I+H D++ASNVLLDS+F V DFG+ KL+P+ ++ +T+
Sbjct: 144 AVSSAQAIAYLHHFATPRIVHGDVRASNVLLDSEFEARVTDFGYDKLMPDDGANKSTK-G 202
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK 261
+GYL+PE GK S+ DVYSFG+LLLELVTG++P E++ KR ITEW PL+ +
Sbjct: 203 NNIGYLSPECIESGKESDMGDVYSFGVLLLELVTGKRPTERVNLTTKRGITEWVLPLVYE 262
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
+F ++VD +L G + E ++K+ V V +C Q E EKRP M +VV +L
Sbjct: 263 RKFGEIVDQRLNGKYVEEELKRIVLVGLMCAQRESEKRPTMSEVVEML 310
>UniRef100_Q9LSC2 Protein kinase-like protein [Arabidopsis thaliana]
Length = 361
Score = 322 bits (824), Expect = 1e-86
Identities = 155/296 (52%), Positives = 210/296 (70%), Gaps = 1/296 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWR+F+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LK +++ E++FAV
Sbjct: 23 SWRVFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKEWSNREEIDFAV 82
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RL+VY+YM NLSL+SHLHGQ++ E L+W KRM I
Sbjct: 83 EVEILARIRHKNLLSVRGYCAEGQERLLVYEYMQNLSLVSHLHGQHSAECLLDWTKRMKI 142
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSHMTTRV 200
AI SA+ I YLH TPHI+H D++ASNVLLDS+F V DFG+ KL+P + T+
Sbjct: 143 AISSAQAIAYLHDHATPHIVHGDVRASNVLLDSEFEARVTDFGYGKLMPDDDTGDGATKA 202
Query: 201 KGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLIT 260
K GY++PE GK SE+ DVYSFGILL+ LV+G++P+E+L R ITEW PL+
Sbjct: 203 KSNNGYISPECDASGKESETSDVYSFGILLMVLVSGKRPLERLNPTTTRCITEWVLPLVY 262
Query: 261 KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQ 316
+ F ++VD +L ++K+ V V +C Q++P+KRP M +VV +L + ++
Sbjct: 263 ERNFGEIVDKRLSEEHVAEKLKKVVLVGLMCAQTDPDKRPTMSEVVEMLVNESKEK 318
>UniRef100_Q8L9L7 Protein kinase, putative [Arabidopsis thaliana]
Length = 361
Score = 320 bits (821), Expect = 3e-86
Identities = 154/296 (52%), Positives = 210/296 (70%), Gaps = 1/296 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWR+F+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LK +++ E++FAV
Sbjct: 23 SWRVFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKEWSNREEIDFAV 82
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RL+VY+YM NLSL+SHLHGQ++ E L+W KRM I
Sbjct: 83 EVEILARIRHKNLLSVRGYCAEGQERLLVYEYMQNLSLVSHLHGQHSAECLLDWTKRMKI 142
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSHMTTRV 200
A+ SA+ I YLH TPHI+H D++ASNVLLDS+F V DFG+ KL+P + T+
Sbjct: 143 AMSSAQAIAYLHDHATPHIVHGDVRASNVLLDSEFEARVTDFGYGKLMPDDDTGDGATKA 202
Query: 201 KGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLIT 260
K GY++PE GK SE+ DVYSFGILL+ LV+G++P+E+L R ITEW PL+
Sbjct: 203 KSNNGYISPECDASGKESETSDVYSFGILLMVLVSGKRPLERLNPTTTRCITEWVLPLVY 262
Query: 261 KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQ 316
+ F ++VD +L ++K+ V V +C Q++P+KRP M +VV +L + ++
Sbjct: 263 ERNFGEIVDKRLSEEHVAEKLKKVVLVGLMCAQTDPDKRPTMSEVVEMLVNESKEK 318
>UniRef100_Q5VRQ7 Putative receptor protein kinase PERK1 [Oryza sativa]
Length = 367
Score = 313 bits (803), Expect = 4e-84
Identities = 150/291 (51%), Positives = 199/291 (67%)
Query: 25 IFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVE 84
+F+ +EL +ATN F+ D K+GEG FGSVYWG+ DG QIAVKKLK + E EFA +VE
Sbjct: 35 VFSLRELRSATNSFNYDNKIGEGPFGSVYWGQVWDGSQIAVKKLKCAKNGTETEFASDVE 94
Query: 85 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIG 144
+LGRVRHKNLL RGYC +R++VYD+MPN SL +HLHG ++ E L+W++R IAIG
Sbjct: 95 ILGRVRHKNLLSFRGYCADGPERVLVYDFMPNSSLYAHLHGTHSTECLLDWRRRTFIAIG 154
Query: 145 SAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTL 204
+A + YLHH TP IIH +KA+NVLLDS+F + DFG + IP+GV H +
Sbjct: 155 AARALAYLHHHATPQIIHGSVKATNVLLDSNFQAHLGDFGLIRFIPDGVDHDKIISENQR 214
Query: 205 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKGRF 264
GYLAPEY M+GK + CDVYSFGI+LLEL +GR+P+E+ + W PL GR+
Sbjct: 215 GYLAPEYIMFGKPTIGCDVYSFGIILLELSSGRRPVERSGSAKMCGVRNWVLPLAKDGRY 274
Query: 265 RDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPD 315
++VD KL + E+++K+ V V C EPEKRP M +VVS+LKG+ +
Sbjct: 275 DEIVDSKLNDKYSESELKRVVLVGLACTHREPEKRPTMLEVVSMLKGESKE 325
>UniRef100_Q9ASK4 Putative LRR receptor-like protein kinase [Oryza sativa]
Length = 698
Score = 298 bits (764), Expect = 1e-79
Identities = 148/290 (51%), Positives = 200/290 (68%), Gaps = 6/290 (2%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 83
R FTY+ELH TNGF+ LGEGGFGSVY G +DG ++AVKKLK + E EF EV
Sbjct: 346 RFFTYEELHQITNGFAAKNLLGEGGFGSVYKGCLADGREVAVKKLKGGGGQGEREFQAEV 405
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAI 143
E++ RV H++L+ L GYC+ DQRL+VYD++PN +L HLHG+ G L W R+ IA
Sbjct: 406 EIISRVHHRHLVSLVGYCISGDQRLLVYDFVPNDTLHHHLHGR--GMPVLEWSARVKIAA 463
Query: 144 GSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGT 203
GSA GI YLH + P IIHRDIK+SN+LLD++F VADFG A+L + V+H+TTRV GT
Sbjct: 464 GSARGIAYLHEDCHPRIIHRDIKSSNILLDNNFEAQVADFGLARLAMDAVTHVTTRVMGT 523
Query: 204 LGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK-- 261
GYLAPEYA GK++E DV+SFG++LLEL+TGRKP++ ++ EWA PL+T+
Sbjct: 524 FGYLAPEYASSGKLTERSDVFSFGVVLLELITGRKPVDASKPLGDESLVEWARPLLTEAI 583
Query: 262 --GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
G +++D +L NF+E ++ + + AA C++ +RP M QVV +L
Sbjct: 584 ETGNVGELIDSRLDKNFNEAEMFRMIEAAAACIRHSASRRPRMSQVVRVL 633
>UniRef100_Q9CAL8 Hypothetical protein F24J13.3 [Arabidopsis thaliana]
Length = 710
Score = 293 bits (751), Expect = 4e-78
Identities = 156/349 (44%), Positives = 222/349 (62%), Gaps = 17/349 (4%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY+EL T GFS LGEGGFG VY G+ +DG +AVK+LK + + + EF EVE+
Sbjct: 341 FTYEELTDITEGFSKHNILGEGGFGCVYKGKLNDGKLVAVKQLKVGSGQGDREFKAEVEI 400
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ D +RL++Y+Y+PN +L HLHG+ G L W +R+ IAIGS
Sbjct: 401 ISRVHHRHLVSLVGYCIADSERLLIYEYVPNQTLEHHLHGK--GRPVLEWARRVRIAIGS 458
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIK++N+LLD +F VADFG AKL +H++TRV GT G
Sbjct: 459 AKGLAYLHEDCHPKIIHRDIKSANILLDDEFEAQVADFGLAKLNDSTQTHVSTRVMGTFG 518
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK+++ DV+SFG++LLEL+TGRKP+++ + ++ EWA PL+ K
Sbjct: 519 YLAPEYAQSGKLTDRSDVFSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAIET 578
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQGKVT- 320
G F ++VD +L ++ EN+V + + AA CV+ KRP M QVV L E D G ++
Sbjct: 579 GDFSELVDRRLEKHYVENEVFRMIETAAACVRHSGPKRPRMVQVVRAL-DSEGDMGDISN 637
Query: 321 ------KMRIDSVKYNDELLALDQPS---DDDYDGNSSYGVFSAIDVQK 360
DS +YN++ + + + DD D G +S D +K
Sbjct: 638 GNKVGQSSAYDSGQYNNDTMKFRKMAFGFDDSSDSGMYSGDYSVQDSRK 686
>UniRef100_Q9FFW5 Similarity to protein kinase [Arabidopsis thaliana]
Length = 681
Score = 283 bits (725), Expect = 4e-75
Identities = 157/360 (43%), Positives = 214/360 (58%), Gaps = 21/360 (5%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAE 76
G+ +N F+Y EL T+GFS+ LGEGGFG VY G SDG ++AVK+LK S+ E
Sbjct: 318 GMVSNQRSWFSYDELSQVTSGFSEKNLLGEGGFGCVYKGVLSDGREVAVKQLKIGGSQGE 377
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF EVE++ RV H++L+ L GYC+ + RL+VYDY+PN +L HLH G + W+
Sbjct: 378 REFKAEVEIISRVHHRHLVTLVGYCISEQHRLLVYDYVPNNTLHYHLHA--PGRPVMTWE 435
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPE--GVS 194
R+ +A G+A GI YLH + P IIHRDIK+SN+LLD+ F LVADFG AK+ E +
Sbjct: 436 TRVRVAAGAARGIAYLHEDCHPRIIHRDIKSSNILLDNSFEALVADFGLAKIAQELDLNT 495
Query: 195 HMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEW 254
H++TRV GT GY+APEYA GK+SE DVYS+G++LLEL+TGRKP++ ++ EW
Sbjct: 496 HVSTRVMGTFGYMAPEYATSGKLSEKADVYSYGVILLELITGRKPVDTSQPLGDESLVEW 555
Query: 255 AEPL----ITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL- 309
A PL I F ++VDP+L NF ++ + V AA CV+ KRP M QVV L
Sbjct: 556 ARPLLGQAIENEEFDELVDPRLGKNFIPGEMFRMVEAAAACVRHSAAKRPKMSQVVRALD 615
Query: 310 ---------KGQEPDQGKVTKMRIDSVK---YNDELLALDQPSDDDYDGNSSYGVFSAID 357
G P Q +V R S + + S D +D + S+ + + D
Sbjct: 616 TLEEATDITNGMRPGQSQVFDSRQQSAQIRMFQRMAFGSQDYSSDFFDRSQSHSSWGSRD 675
>UniRef100_Q9LK03 Somatic embryogenesis receptor kinase-like protein [Arabidopsis
thaliana]
Length = 458
Score = 281 bits (718), Expect = 3e-74
Identities = 139/290 (47%), Positives = 196/290 (66%), Gaps = 6/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
F Y+EL ATNGFS+ LG+GGFG V+ G +G ++AVK+LK +S+ E EF EV +
Sbjct: 83 FNYEELSRATNGFSEANLLGQGGFGYVFKGMLRNGKEVAVKQLKEGSSQGEREFQAEVGI 142
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ D QRL+VY+++PN +L HLHG+ G + W R+ IA+GS
Sbjct: 143 ISRVHHRHLVALVGYCIADAQRLLVYEFVPNNTLEFHLHGK--GRPTMEWSSRLKIAVGS 200
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH P IIHRDIKASN+L+D F VADFG AK+ + +H++TRV GT G
Sbjct: 201 AKGLSYLHENCNPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFG 260
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK++E DV+SFG++LLEL+TGR+PI+ ++ +WA PL+ +
Sbjct: 261 YLAPEYASSGKLTEKSDVFSFGVVLLELITGRRPIDVNNVHADNSLVDWARPLLNQVSEL 320
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
G F +VD KL +D+ ++ + V AA CV+S +RP M QV +L+G
Sbjct: 321 GNFEVVVDKKLNNEYDKEEMARMVACAAACVRSTAPRRPRMDQVARVLEG 370
>UniRef100_Q60E30 Hypothetical protein OSJNBb0012L23.7 [Oryza sativa]
Length = 471
Score = 280 bits (717), Expect = 3e-74
Identities = 142/304 (46%), Positives = 203/304 (66%), Gaps = 7/304 (2%)
Query: 13 PTSYGVANN-SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAM 71
P S G A S FTY+EL AT+GFSD LG+GGFG V+ G G +IAVK+LK
Sbjct: 71 PPSPGAALGFSKSTFTYEELLRATDGFSDANLLGQGGFGYVHRGVLPTGKEIAVKQLKVG 130
Query: 72 NSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEV 131
+ + E EF EVE++ RV HK+L+ L GYC+ +RL+VY+++PN +L HLHG+ G
Sbjct: 131 SGQGEREFQAEVEIISRVHHKHLVSLVGYCISGGKRLLVYEFVPNNTLEFHLHGK--GRP 188
Query: 132 QLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPE 191
+ W R+ IA+G+A+G+ YLH + P IIHRDIKASN+LLD F VADFG AK +
Sbjct: 189 TMEWPTRLKIALGAAKGLAYLHEDCHPKIIHRDIKASNILLDFKFESKVADFGLAKFTSD 248
Query: 192 GVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTI 251
+H++TRV GT GYLAPEYA GK++E DV+S+G++LLEL+TGR+P++ + ++
Sbjct: 249 NNTHVSTRVMGTFGYLAPEYASSGKLTEKSDVFSYGVMLLELITGRRPVDTSQTYMDDSL 308
Query: 252 TEWAEPLITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVS 307
+WA PL+ + G + ++VDP+L +F+ N++ + + AA CV+ +RP M QVV
Sbjct: 309 VDWARPLLMQALENGNYEELVDPRLGKDFNPNEMARMIACAAACVRHSARRRPRMSQVVR 368
Query: 308 LLKG 311
L+G
Sbjct: 369 ALEG 372
>UniRef100_Q84VF7 Putative receptor protein kinase [Oryza sativa]
Length = 394
Score = 280 bits (716), Expect = 5e-74
Identities = 137/290 (47%), Positives = 198/290 (68%), Gaps = 6/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY+EL AT+GFSD LG+GGFG V+ G G +IAVK+LK + + E EF EVE+
Sbjct: 8 FTYEELLRATDGFSDANLLGQGGFGYVHRGVLPTGKEIAVKQLKVGSGQGEREFQAEVEI 67
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV HK+L+ L GYC+ +RL+VY+++PN +L HLHG+ G + W R+ IA+G+
Sbjct: 68 ISRVHHKHLVSLVGYCISGGKRLLVYEFVPNNTLEFHLHGK--GRPTMEWPTRLKIALGA 125
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIKASN+LLD F VADFG AK + +H++TRV GT G
Sbjct: 126 AKGLAYLHEDCHPKIIHRDIKASNILLDFKFESKVADFGLAKFTSDNNTHVSTRVMGTFG 185
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK++E DV+S+G++LLEL+TGR+P++ + ++ +WA PL+ +
Sbjct: 186 YLAPEYASSGKLTEKSDVFSYGVMLLELITGRRPVDTSQTYMDDSLVDWARPLLMQALEN 245
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
G + ++VDP+L +F+ N++ + + AA CV+ +RP M QVV L+G
Sbjct: 246 GNYEELVDPRLGKDFNPNEMARMIACAAACVRHSARRRPRMSQVVRALEG 295
>UniRef100_Q6ZE12 Hypothetical protein P0495H05.37 [Oryza sativa]
Length = 517
Score = 280 bits (715), Expect = 6e-74
Identities = 142/306 (46%), Positives = 200/306 (64%), Gaps = 13/306 (4%)
Query: 13 PTSYGVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMN 72
P G+ N+ F Y EL AT GFS+ LG+GGFG VY G DG ++AVK+L A
Sbjct: 132 PVPAGLDENA---FGYDELAAATGGFSEGNMLGQGGFGYVYRGVLGDGKEVAVKQLSAGG 188
Query: 73 SKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQ 132
+ E EF EV+++ RV H++L+ L GYC+ QRL+VYD++PN +L HLH + G
Sbjct: 189 GQGEREFQAEVDMISRVHHRHLVPLVGYCIAGAQRLLVYDFVPNRTLEHHLHEK--GLPV 246
Query: 133 LNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG 192
+ W R+ IA+GSA+G+ YLH E P IIHRDIK++N+LLD++F PLVADFG AKL E
Sbjct: 247 MKWTTRLRIAVGSAKGLAYLHEECNPRIIHRDIKSANILLDNNFEPLVADFGMAKLTSEN 306
Query: 193 VSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTIT 252
V+H++TRV GT GYLAPEYA GK+++ DV+S+G++LLEL+TGR+P ++ G +
Sbjct: 307 VTHVSTRVMGTFGYLAPEYASSGKLTDKSDVFSYGVMLLELLTGRRPADRSSYGAD-CLV 365
Query: 253 EWAEPLITK-------GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQV 305
+WA + + G + D+VDP+LRG +D + + A CV+ +RP M QV
Sbjct: 366 DWARQALPRAMAAGGGGGYDDIVDPRLRGEYDRAEAARVAACAVACVRHAGRRRPKMSQV 425
Query: 306 VSLLKG 311
V +L+G
Sbjct: 426 VKVLEG 431
>UniRef100_Q75IB5 Hypothetical protein OJ1785_A05.27 [Oryza sativa]
Length = 520
Score = 278 bits (711), Expect = 2e-73
Identities = 135/305 (44%), Positives = 201/305 (65%), Gaps = 9/305 (2%)
Query: 12 RPTSYGVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAM 71
RP S + S +Y +L AT GFS D +G+GGFG VY GR DG ++A+KKLK
Sbjct: 180 RPHSISIDGGS---LSYDQLAAATGGFSPDNVIGQGGFGCVYRGRLQDGTEVAIKKLKTE 236
Query: 72 NSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEV 131
+ + + EF E +++ RV H+NL+ L GYC+ + RL+VY+++PN +L +HLHG
Sbjct: 237 SKQGDREFRAEADIITRVHHRNLVSLVGYCISGNDRLLVYEFVPNKTLDTHLHGDKWPP- 295
Query: 132 QLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPE 191
L+WQ+R IA+GSA G+ YLH + +P IIHRD+KASN+LLD F P VADFG AK P
Sbjct: 296 -LDWQQRWKIAVGSARGLAYLHDDCSPKIIHRDVKASNILLDHGFEPKVADFGLAKYQPG 354
Query: 192 GVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTI 251
+H++TR+ GT GY+APE+ GK+++ DV++FG++LLEL+TGR P++ + T+
Sbjct: 355 NHTHVSTRIMGTFGYIAPEFLSSGKLTDKADVFAFGVVLLELITGRLPVQSSESYMDSTL 414
Query: 252 TEWAEPLIT----KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVS 307
WA+PLI+ +G F +VDP + ++DEN++ + + AA V+ RP+M Q++
Sbjct: 415 VGWAKPLISEAMEEGNFDILVDPDIGDDYDENKMMRMMECAAAAVRQSAHLRPSMVQILK 474
Query: 308 LLKGQ 312
L+GQ
Sbjct: 475 HLQGQ 479
>UniRef100_Q94JZ6 Protein kinase-like protein [Arabidopsis thaliana]
Length = 652
Score = 277 bits (709), Expect = 3e-73
Identities = 142/307 (46%), Positives = 201/307 (65%), Gaps = 7/307 (2%)
Query: 10 VCRPTSYG-VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKL 68
V P S G V S FTY+EL ATNGFS+ LG+GGFG V+ G G ++AVK+L
Sbjct: 251 VLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQL 310
Query: 69 KAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYA 128
KA + + E EF EVE++ RV H++L+ L GYC+ QRL+VY+++PN +L HLHG+
Sbjct: 311 KAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGK-- 368
Query: 129 GEVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKL 188
G + W R+ IA+GSA+G+ YLH + P IIHRDIKASN+L+D F VADFG AK+
Sbjct: 369 GRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASNILIDFKFEAKVADFGLAKI 428
Query: 189 IPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLK 248
+ +H++TRV GT GYLAPEYA GK++E DV+SFG++LLEL+TGR+P++ +
Sbjct: 429 ASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVVLLELITGRRPVDANNVYVD 488
Query: 249 RTITEWAEPLITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQ 304
++ +WA PL+ + G F + D K+ +D ++ + V AA CV+ +RP M Q
Sbjct: 489 DSLVDWARPLLNRASEEGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQ 548
Query: 305 VVSLLKG 311
+V L+G
Sbjct: 549 IVRALEG 555
>UniRef100_Q9LV48 Protein kinase-like protein [Arabidopsis thaliana]
Length = 652
Score = 277 bits (709), Expect = 3e-73
Identities = 142/307 (46%), Positives = 201/307 (65%), Gaps = 7/307 (2%)
Query: 10 VCRPTSYG-VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKL 68
V P S G V S FTY+EL ATNGFS+ LG+GGFG V+ G G ++AVK+L
Sbjct: 251 VLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQL 310
Query: 69 KAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYA 128
KA + + E EF EVE++ RV H++L+ L GYC+ QRL+VY+++PN +L HLHG+
Sbjct: 311 KAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGK-- 368
Query: 129 GEVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKL 188
G + W R+ IA+GSA+G+ YLH + P IIHRDIKASN+L+D F VADFG AK+
Sbjct: 369 GRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASNILIDFKFEAKVADFGLAKI 428
Query: 189 IPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLK 248
+ +H++TRV GT GYLAPEYA GK++E DV+SFG++LLEL+TGR+P++ +
Sbjct: 429 ASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVVLLELITGRRPVDANNVYVD 488
Query: 249 RTITEWAEPLITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQ 304
++ +WA PL+ + G F + D K+ +D ++ + V AA CV+ +RP M Q
Sbjct: 489 DSLVDWARPLLNRASEEGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQ 548
Query: 305 VVSLLKG 311
+V L+G
Sbjct: 549 IVRALEG 555
>UniRef100_Q9ARH1 Receptor protein kinase PERK1 [Brassica napus]
Length = 647
Score = 276 bits (706), Expect = 7e-73
Identities = 140/307 (45%), Positives = 200/307 (64%), Gaps = 7/307 (2%)
Query: 10 VCRPTSYG-VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKL 68
V P S G V S FTY+EL ATNGFS+ LG+GGFG V+ G G ++AVK+L
Sbjct: 246 VLPPPSPGLVLGFSKSTFTYEELARATNGFSEANLLGQGGFGYVHKGVLPSGKEVAVKQL 305
Query: 69 KAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYA 128
K + + E EF EVE++ RV H++L+ L GYC+ +RL+VY+++PN +L HLHG+
Sbjct: 306 KVGSGQGEREFQAEVEIISRVHHRHLVSLVGYCIAGAKRLLVYEFVPNNNLELHLHGE-- 363
Query: 129 GEVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKL 188
G + W R+ IA+GSA+G+ YLH + P IIHRDIKASN+L+D F VADFG AK+
Sbjct: 364 GRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASNILIDFKFEAKVADFGLAKI 423
Query: 189 IPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLK 248
+ +H++TRV GT GYLAPEYA GK++E DV+SFG++LLEL+TGR+P++ +
Sbjct: 424 ASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVVLLELITGRRPVDANNVYVD 483
Query: 249 RTITEWAEPLITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQ 304
++ +WA PL+ + G F + D K+ +D ++ + V AA CV+ +RP M Q
Sbjct: 484 DSLVDWARPLLNRASEQGDFEGLADAKMNNGYDREEMARMVACAAACVRHSARRRPRMSQ 543
Query: 305 VVSLLKG 311
+V L+G
Sbjct: 544 IVRALEG 550
>UniRef100_Q8W0B8 Putative receptor protein kinase PERK1 [Oryza sativa]
Length = 597
Score = 275 bits (703), Expect = 1e-72
Identities = 133/290 (45%), Positives = 198/290 (67%), Gaps = 6/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY++L AT+GFSD LG+GGFG V+ G +G ++AVK+L+ + + E EF EVE+
Sbjct: 211 FTYEDLSAATDGFSDANLLGQGGFGYVHKGVLPNGTEVAVKQLRDGSGQGEREFQAEVEI 270
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV HK+L+ L GYC+ +RL+VY+Y+PN +L HLHG+ G + W R+ IA+G+
Sbjct: 271 ISRVHHKHLVTLVGYCISGGKRLLVYEYVPNNTLELHLHGR--GRPTMEWPTRLRIALGA 328
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIK++N+LLD+ F VADFG AKL + +H++TRV GT G
Sbjct: 329 AKGLAYLHEDCHPKIIHRDIKSANILLDARFEAKVADFGLAKLTSDNNTHVSTRVMGTFG 388
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA G+++E DV+SFG++LLEL+TGR+P+ + ++ +WA PL+ +
Sbjct: 389 YLAPEYASSGQLTEKSDVFSFGVMLLELITGRRPVRSNQSQMDDSLVDWARPLMMRASDD 448
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
G + +VDP+L ++ N++ + + AA CV+ +RP M QVV L+G
Sbjct: 449 GNYDALVDPRLGQEYNGNEMARMIACAAACVRHSARRRPRMSQVVRALEG 498
>UniRef100_Q84Q68 Protein kinase [Oryza sativa]
Length = 503
Score = 275 bits (702), Expect = 2e-72
Identities = 130/287 (45%), Positives = 195/287 (67%), Gaps = 6/287 (2%)
Query: 27 TYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVL 86
+Y +L AT+GFS D +G+GGFG VY G DG ++A+KKLK + + + EF EVE++
Sbjct: 216 SYDQLAAATDGFSPDNVIGQGGFGCVYRGTLQDGTEVAIKKLKTESKQGDREFRAEVEII 275
Query: 87 GRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSA 146
RV H+NL+ L G+C+ ++RL+VY+++PN +L +HLHG L+WQ+R IA+GSA
Sbjct: 276 TRVHHRNLVSLVGFCISGNERLLVYEFVPNKTLDTHLHGNKGPP--LDWQQRWKIAVGSA 333
Query: 147 EGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLGY 206
G+ YLH + +P IIHRD+KASN+LLD DF P VADFG AK P +H++TR+ GT GY
Sbjct: 334 RGLAYLHDDCSPKIIHRDVKASNILLDHDFEPKVADFGLAKYQPGNHTHVSTRIMGTFGY 393
Query: 207 LAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLIT----KG 262
+APE+ GK+++ DV++FG++LLEL+TGR P++ + T+ WA+PL++ +G
Sbjct: 394 IAPEFLSSGKLTDKADVFAFGVVLLELITGRLPVQSSESYMDSTLVAWAKPLLSEATEEG 453
Query: 263 RFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
F +VDP + ++DEN + + + AA V+ RP+M QV L+
Sbjct: 454 NFDILVDPDIGDDYDENIMMRMIECAAAAVRQSAHLRPSMVQVRFLI 500
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 643,546,301
Number of Sequences: 2790947
Number of extensions: 27547435
Number of successful extensions: 113811
Number of sequences better than 10.0: 17780
Number of HSP's better than 10.0 without gapping: 7340
Number of HSP's successfully gapped in prelim test: 10468
Number of HSP's that attempted gapping in prelim test: 80058
Number of HSP's gapped (non-prelim): 20035
length of query: 369
length of database: 848,049,833
effective HSP length: 129
effective length of query: 240
effective length of database: 488,017,670
effective search space: 117124240800
effective search space used: 117124240800
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC136954.12


