

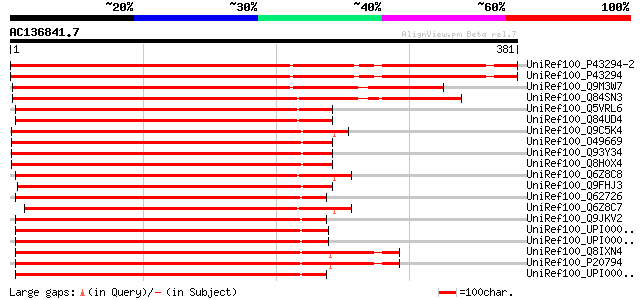
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.7 + phase: 0 /pseudo
(381 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P43294-2 Splice isoform Short of P43294 [Arabidopsis t... 417 e-115
UniRef100_P43294 Serine/threonine-protein kinase MHK [Arabidopsi... 417 e-115
UniRef100_Q9M3W7 Putative Cdc2-related protein kinase CRK2 [Beta... 388 e-106
UniRef100_Q84SN3 Hypothetical protein OSJNBb0043P23.8 [Oryza sat... 355 2e-96
UniRef100_Q5VRL6 Putative GAMYB-binding protein [Oryza sativa] 301 2e-80
UniRef100_Q84UD4 GAMYB-binding protein [Hordeum vulgare var. dis... 296 5e-79
UniRef100_Q9C5K4 Putative kinase [Arabidopsis thaliana] 295 1e-78
UniRef100_O49669 Kinase-like protein [Arabidopsis thaliana] 294 2e-78
UniRef100_Q93Y34 Serine/threonine-protein kinase Mak (Male germ ... 292 9e-78
UniRef100_Q8H0X4 Serine/threonine-protein kinase Mak (Male germ ... 292 9e-78
UniRef100_Q6Z8C8 Putative serine/threonine-protein kinase Mak [O... 290 4e-77
UniRef100_Q9FHJ3 Serine/threonine-protein kinase Mak (Male germ ... 288 2e-76
UniRef100_Q62726 Serine/threonine kinase ICK [Rattus norvegicus] 281 3e-74
UniRef100_Q6Z8C7 Putative serine/threonine-protein kinase Mak [O... 280 5e-74
UniRef100_Q9JKV2 Serine/threonine kinase ICK [Mus musculus] 280 5e-74
UniRef100_UPI00003ABC95 UPI00003ABC95 UniRef100 entry 279 8e-74
UniRef100_UPI00003ABC94 UPI00003ABC94 UniRef100 entry 279 8e-74
UniRef100_Q8IXN4 MAK protein [Homo sapiens] 279 1e-73
UniRef100_P20794 Serine/threonine-protein kinase MAK [Homo sapiens] 279 1e-73
UniRef100_UPI000013ED70 UPI000013ED70 UniRef100 entry 278 2e-73
>UniRef100_P43294-2 Splice isoform Short of P43294 [Arabidopsis thaliana]
Length = 435
Score = 417 bits (1072), Expect = e-115
Identities = 218/382 (57%), Positives = 275/382 (71%), Gaps = 15/382 (3%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMD 60
+ T+E+VAVK++KRKF +WEE NLRE+KALRK+NH +IIKL+E+VRE+NELFFIFE MD
Sbjct: 24 LETYEVVAVKKMKRKFYYWEECVNLREVKALRKLNHPHIIKLKEIVREHNELFFIFECMD 83
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADF 120
NLY ++KERE+PFSE EIR FM QMLQGL+HMHK G+FHRDLKPENLLVTN++LKIADF
Sbjct: 84 HNLYHIMKERERPFSEGEIRSFMSQMLQGLAHMHKNGYFHRDLKPENLLVTNNILKIADF 143
Query: 121 GLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGES 180
GLAREV+SMPPYT+YVSTRWYRAPEVLLQS YTPAVDMWA+GAILAEL+ LTP+FPGES
Sbjct: 144 GLAREVASMPPYTEYVSTRWYRAPEVLLQSSLYTPAVDMWAVGAILAELYALTPLFPGES 203
Query: 181 EIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITA 240
EIDQ+YKI C+LG PD T F + SR++ + H +++D++PNA+ EAIDLI
Sbjct: 204 EIDQLYKICCVLGKPDWTTFPEAKSISRIMS-ISHTEFPQTRIADLLPNAAPEAIDLINR 262
Query: 241 RCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLH 300
C ++ H F + Q + + LEL+L N PNLEL L
Sbjct: 263 LCSWDPLKRPTADEALNHP---FFSMATQASYPI-----HDLELRLDNMAALPNLELNLW 314
Query: 301 DFGPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGENLV*LDAL*KCRICYFARILMTIQ 360
DF +P++CFLGLTLAVKPS L++++N Q M EN + + R L++
Sbjct: 315 DFNREPEECFLGLTLAVKPSAPKLEMLRNVSQDMSENFLFCPGVNNDREPSVFWSLLS-- 372
Query: 361 TNQPDQNGVHSSAETS-LSLSF 381
PD+NG+H+ E+S LSLSF
Sbjct: 373 ---PDENGLHAPVESSPLSLSF 391
>UniRef100_P43294 Serine/threonine-protein kinase MHK [Arabidopsis thaliana]
Length = 443
Score = 417 bits (1072), Expect = e-115
Identities = 218/382 (57%), Positives = 275/382 (71%), Gaps = 15/382 (3%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMD 60
+ T+E+VAVK++KRKF +WEE NLRE+KALRK+NH +IIKL+E+VRE+NELFFIFE MD
Sbjct: 32 LETYEVVAVKKMKRKFYYWEECVNLREVKALRKLNHPHIIKLKEIVREHNELFFIFECMD 91
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADF 120
NLY ++KERE+PFSE EIR FM QMLQGL+HMHK G+FHRDLKPENLLVTN++LKIADF
Sbjct: 92 HNLYHIMKERERPFSEGEIRSFMSQMLQGLAHMHKNGYFHRDLKPENLLVTNNILKIADF 151
Query: 121 GLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGES 180
GLAREV+SMPPYT+YVSTRWYRAPEVLLQS YTPAVDMWA+GAILAEL+ LTP+FPGES
Sbjct: 152 GLAREVASMPPYTEYVSTRWYRAPEVLLQSSLYTPAVDMWAVGAILAELYALTPLFPGES 211
Query: 181 EIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITA 240
EIDQ+YKI C+LG PD T F + SR++ + H +++D++PNA+ EAIDLI
Sbjct: 212 EIDQLYKICCVLGKPDWTTFPEAKSISRIMS-ISHTEFPQTRIADLLPNAAPEAIDLINR 270
Query: 241 RCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLH 300
C ++ H F + Q + + LEL+L N PNLEL L
Sbjct: 271 LCSWDPLKRPTADEALNHP---FFSMATQASYPI-----HDLELRLDNMAALPNLELNLW 322
Query: 301 DFGPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGENLV*LDAL*KCRICYFARILMTIQ 360
DF +P++CFLGLTLAVKPS L++++N Q M EN + + R L++
Sbjct: 323 DFNREPEECFLGLTLAVKPSAPKLEMLRNVSQDMSENFLFCPGVNNDREPSVFWSLLS-- 380
Query: 361 TNQPDQNGVHSSAETS-LSLSF 381
PD+NG+H+ E+S LSLSF
Sbjct: 381 ---PDENGLHAPVESSPLSLSF 399
>UniRef100_Q9M3W7 Putative Cdc2-related protein kinase CRK2 [Beta vulgaris]
Length = 434
Score = 388 bits (997), Expect = e-106
Identities = 196/324 (60%), Positives = 240/324 (73%), Gaps = 8/324 (2%)
Query: 3 TFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCN 62
++EIVAVK++KRKF FWEE NLRE+K+L + +IIKL+EVVRENN+LFFIFEYM N
Sbjct: 26 SYEIVAVKKMKRKFYFWEECVNLREVKSLPSIESSHIIKLKEVVRENNDLFFIFEYMQYN 85
Query: 63 LYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGL 122
LYQ++K+R +PF+EEEIR F+ Q+LQGL+HMH+ G+FHRDLKPENLLVTNDV+KIADFGL
Sbjct: 86 LYQIMKDRHRPFTEEEIRNFLTQVLQGLAHMHRNGYFHRDLKPENLLVTNDVIKIADFGL 145
Query: 123 AREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEI 182
AREVSS+PPYT+YVSTRWYRAPEVLL+S YTPA+DMWA+GA+LAELFT PIFPGESE
Sbjct: 146 AREVSSIPPYTEYVSTRWYRAPEVLLKSSLYTPAIDMWAVGAVLAELFTSCPIFPGESET 205
Query: 183 DQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
DQ+YKI C+LG PD F N S+L + + + P LS+IIPNA+ EAIDLI+ C
Sbjct: 206 DQLYKICCVLGAPDWAVFPEAKNISQLTS-ISYSQMLPANLSEIIPNANWEAIDLISQLC 264
Query: 243 GSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLHDF 302
++ H + V RVPR + DP K + KPNLEL L DF
Sbjct: 265 SWDPLKRPTAEQALHHPFFH-------VALRVPRPIHDPFHSKPDYTKTKPNLELNLWDF 317
Query: 303 GPDPDDCFLGLTLAVKPSVSNLDV 326
DDCFLGLTLAV P VS+L++
Sbjct: 318 DSKADDCFLGLTLAVNPGVSSLEM 341
>UniRef100_Q84SN3 Hypothetical protein OSJNBb0043P23.8 [Oryza sativa]
Length = 433
Score = 355 bits (910), Expect = 2e-96
Identities = 190/337 (56%), Positives = 231/337 (68%), Gaps = 9/337 (2%)
Query: 3 TFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCN 62
T EIVAVK++KRKF WEE +LRE+KAL+K+NH NI+KL+EV EN+ELFFIFE M+CN
Sbjct: 26 TNEIVAVKKMKRKFFQWEECISLREVKALQKLNHPNIVKLKEVTMENHELFFIFENMECN 85
Query: 63 LYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGL 122
LY +I+ER+ FSEEEIR FM Q+LQGL++MH G+FHRDLKPENLLVT+ +KIADFGL
Sbjct: 86 LYDVIRERQAAFSEEEIRNFMVQILQGLAYMHNNGYFHRDLKPENLLVTDGTVKIADFGL 145
Query: 123 AREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEI 182
AREVSS PPYT YVSTRWYRAPEVLLQS YTPA+DMWA+GAILAELFTL+P+FPG SE
Sbjct: 146 AREVSSSPPYTDYVSTRWYRAPEVLLQSSAYTPAIDMWAVGAILAELFTLSPLFPGGSET 205
Query: 183 DQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
DQ+YKI +LG PD T + G N R F + + P L ++IPNA++EAIDLI C
Sbjct: 206 DQLYKICAVLGTPDHTVWPEGMNLPRSSSFNFFQ-IPPRNLWELIPNATLEAIDLIQQLC 264
Query: 243 GSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLHDF 302
S+ H N V VPR L K R P LEL L DF
Sbjct: 265 SWDPRRRPTAEQSLQHPFFN-------VGNWVPRPL-HASHTKTIETRPNPRLELNLWDF 316
Query: 303 GPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGENLV 339
G +P+D +L LTL++KPS D N + E ++
Sbjct: 317 GTEPEDNYLDLTLSLKPSFPGTDFSNNVPEHTKEEIL 353
>UniRef100_Q5VRL6 Putative GAMYB-binding protein [Oryza sativa]
Length = 484
Score = 301 bits (772), Expect = 2e-80
Identities = 142/238 (59%), Positives = 189/238 (78%), Gaps = 1/238 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+K++K+K+ WEE NLRE+K+LR+MNH NI+KL+EV+REN+ LFF+FEYM+CNLY
Sbjct: 28 EVVAIKKMKKKYYSWEECINLREVKSLRRMNHPNIVKLKEVIRENDMLFFVFEYMECNLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAR 124
QL+K R KPFSE E+R + Q+ Q LSHMH++G+FHRDLKPENLLVT +++KIADFGLAR
Sbjct: 88 QLMKSRGKPFSETEVRNWCFQIFQALSHMHQRGYFHRDLKPENLLVTKELIKIADFGLAR 147
Query: 125 EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQ 184
E+SS PPYT+YVSTRWYRAPEVLLQ+ Y AVDMWA+GAI+AELF+L P+FPG +E D+
Sbjct: 148 EISSEPPYTEYVSTRWYRAPEVLLQASVYNSAVDMWAMGAIIAELFSLRPLFPGSNEADE 207
Query: 185 MYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
+YKI ILG P+ + G + + F + + LS+++P+AS +AI LI+ C
Sbjct: 208 IYKICSILGTPNQRTWAEGLQLAASIRFQFPQ-SGSIHLSEVVPSASEDAISLISWLC 264
>UniRef100_Q84UD4 GAMYB-binding protein [Hordeum vulgare var. distichum]
Length = 483
Score = 296 bits (759), Expect = 5e-79
Identities = 140/238 (58%), Positives = 188/238 (78%), Gaps = 1/238 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+K++K+K+ WEE NLRE+K+LR+MNH NI+KL+EV+REN+ LFF+FEYM+CNLY
Sbjct: 28 EVVAIKKMKKKYFSWEECINLREVKSLRRMNHPNIVKLKEVIRENDMLFFVFEYMECNLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAR 124
QL+K + KPFSE EIR + Q+ Q LSHMH++G+FHRDLKPENLLVT +++K+ADFGLAR
Sbjct: 88 QLMKSKGKPFSETEIRNWCFQVFQALSHMHQRGYFHRDLKPENLLVTKELIKVADFGLAR 147
Query: 125 EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQ 184
E+ S PPYT+YVSTRWYRAPEVLLQS Y+ AVDMWA+GAI+AELF+ P+FPG SE D+
Sbjct: 148 EIISEPPYTEYVSTRWYRAPEVLLQSSVYSSAVDMWAMGAIIAELFSHRPLFPGSSEADE 207
Query: 185 MYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
+YKI ILG P+ + G + + F + + LS+++P AS +A++LI+ C
Sbjct: 208 IYKICNILGTPNQHTWAGGLQLAASIHFQFPQ-SGSINLSEVVPTASEDALNLISWLC 264
>UniRef100_Q9C5K4 Putative kinase [Arabidopsis thaliana]
Length = 461
Score = 295 bits (755), Expect = 1e-78
Identities = 143/263 (54%), Positives = 198/263 (74%), Gaps = 11/263 (4%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+K++K+K+ W+E NLRE+K+LR+MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTGEVVAIKKMKKKYYSWDECINLREVKSLRRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R+K F+E +I+ + Q+ QGLS+MH++G+FHRDLKPENLLV+ D++KIADFG
Sbjct: 85 NLYQLMKDRQKLFAEADIKNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDIIKIADFG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LAREV+S PP+T+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L PIFPG SE
Sbjct: 145 LAREVNSSPPFTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPIFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + +++ ++ V LS ++P+AS +AI+LI
Sbjct: 205 ADEIYKICSVIGTPTEETWLEGLNLANTINYQFPQLPG-VPLSSLMPSASEDAINLIERL 263
Query: 242 C----------GSVTATSFFPSC 254
C V FF SC
Sbjct: 264 CSWDPSSRPTAAEVLQHPFFQSC 286
>UniRef100_O49669 Kinase-like protein [Arabidopsis thaliana]
Length = 290
Score = 294 bits (753), Expect = 2e-78
Identities = 138/241 (57%), Positives = 193/241 (79%), Gaps = 1/241 (0%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+K++K+K+ W+E NLRE+K+LR+MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTGEVVAIKKMKKKYYSWDECINLREVKSLRRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R+K F+E +I+ + Q+ QGLS+MH++G+FHRDLKPENLLV+ D++KIADFG
Sbjct: 85 NLYQLMKDRQKLFAEADIKNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDIIKIADFG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LAREV+S PP+T+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L PIFPG SE
Sbjct: 145 LAREVNSSPPFTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPIFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + +++ ++ V LS ++P+AS +AI+LI
Sbjct: 205 ADEIYKICSVIGTPTEETWLEGLNLANTINYQFPQLPG-VPLSSLMPSASEDAINLIERL 263
Query: 242 C 242
C
Sbjct: 264 C 264
>UniRef100_Q93Y34 Serine/threonine-protein kinase Mak (Male germ cell-associated
kinase)-like protein [Arabidopsis thaliana]
Length = 499
Score = 292 bits (748), Expect = 9e-78
Identities = 138/241 (57%), Positives = 188/241 (77%), Gaps = 1/241 (0%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+KR+K+K+ WEE NLRE+K+L +MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTNEVVAIKRMKKKYFSWEECVNLREVKSLSRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R K F+E +IR + Q+ QGLS+MH++G+FHRDLKPENLLV+ DV+KIAD G
Sbjct: 85 NLYQLMKDRPKHFAESDIRNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDVIKIADLG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LARE+ S PPYT+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L P+FPG SE
Sbjct: 145 LAREIDSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPLFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + ++++ + V LS ++P AS +A++LI
Sbjct: 205 ADEIYKICSVIGSPTEETWLEGLNLASVINYQFPQFPG-VHLSSVMPYASADAVNLIERL 263
Query: 242 C 242
C
Sbjct: 264 C 264
>UniRef100_Q8H0X4 Serine/threonine-protein kinase Mak (Male germ cell-associated
kinase)-like protein [Arabidopsis thaliana]
Length = 499
Score = 292 bits (748), Expect = 9e-78
Identities = 138/241 (57%), Positives = 188/241 (77%), Gaps = 1/241 (0%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+KR+K+K+ WEE NLRE+K+L +MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTNEVVAIKRMKKKYFSWEECVNLREVKSLSRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R K F+E +IR + Q+ QGLS+MH++G+FHRDLKPENLLV+ DV+KIAD G
Sbjct: 85 NLYQLMKDRPKHFAESDIRNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDVIKIADLG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LARE+ S PPYT+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L P+FPG SE
Sbjct: 145 LAREIDSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPLFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + ++++ + V LS ++P AS +A++LI
Sbjct: 205 ADEIYKICSVIGSPTEETWLEGLNLASVINYQFPQFPG-VHLSSVMPYASADAVNLIERL 263
Query: 242 C 242
C
Sbjct: 264 C 264
>UniRef100_Q6Z8C8 Putative serine/threonine-protein kinase Mak [Oryza sativa]
Length = 459
Score = 290 bits (743), Expect = 4e-77
Identities = 142/263 (53%), Positives = 198/263 (74%), Gaps = 11/263 (4%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VAVK++KRK+ +EE +LRE+K+LR+MNH NI+KL+EV+REN+ L+FI EYM+CNLY
Sbjct: 28 EVVAVKKMKRKYYSFEECMSLREVKSLRRMNHPNIVKLKEVIRENDILYFIMEYMECNLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAR 124
QL+K+R KPFSE E+R + Q+ Q L++MH++G+FHRDLKPENLLV+ DV+K+ADFGLAR
Sbjct: 88 QLMKDRVKPFSEAEVRNWCFQIFQALAYMHQRGYFHRDLKPENLLVSKDVIKLADFGLAR 147
Query: 125 EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQ 184
EV+S+PPYT+YVSTRWYRAPEVLLQS Y AVDMWA+GAI+AEL TL P+FPG SE D+
Sbjct: 148 EVTSVPPYTEYVSTRWYRAPEVLLQSSIYDSAVDMWAMGAIMAELLTLHPLFPGTSEADE 207
Query: 185 MYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC-- 242
+ KI ++G PD + G + + + F + V+ +L++++ + S EA+DLI++ C
Sbjct: 208 ILKICNVIGSPDEQSWPQGLSLAETMKFQFPQ-VSGNQLAEVMTSVSSEAVDLISSLCSW 266
Query: 243 --------GSVTATSFFPSCSIL 257
V +FF C+ +
Sbjct: 267 DPCKRPKAAEVLQHTFFQGCTFV 289
>UniRef100_Q9FHJ3 Serine/threonine-protein kinase Mak (Male germ cell-associated
kinase)-like protein [Arabidopsis thaliana]
Length = 530
Score = 288 bits (737), Expect = 2e-76
Identities = 136/236 (57%), Positives = 184/236 (77%), Gaps = 1/236 (0%)
Query: 7 VAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQL 66
VA+KR+K+K+ WEE NLRE+K+L +MNH NI+KL+EV+REN+ L+F+FEYM+CNLYQL
Sbjct: 38 VAIKRMKKKYFSWEECVNLREVKSLSRMNHPNIVKLKEVIRENDILYFVFEYMECNLYQL 97
Query: 67 IKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAREV 126
+K+R K F+E +IR + Q+ QGLS+MH++G+FHRDLKPENLLV+ DV+KIAD GLARE+
Sbjct: 98 MKDRPKHFAESDIRNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDVIKIADLGLAREI 157
Query: 127 SSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMY 186
S PPYT+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L P+FPG SE D++Y
Sbjct: 158 DSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPLFPGASEADEIY 217
Query: 187 KIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
KI ++G P + G N + ++++ + V LS ++P AS +A++LI C
Sbjct: 218 KICSVIGSPTEETWLEGLNLASVINYQFPQFPG-VHLSSVMPYASADAVNLIERLC 272
>UniRef100_Q62726 Serine/threonine kinase ICK [Rattus norvegicus]
Length = 629
Score = 281 bits (718), Expect = 3e-74
Identities = 138/235 (58%), Positives = 179/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH NI+KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANIVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSRPPYTDYVSTRWYRAPEVLLRSTNYSSPIDVWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F+ + + P L +IPNAS EAI L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFIWPQCI-PNNLKTLIPNASSEAIQLL 261
>UniRef100_Q6Z8C7 Putative serine/threonine-protein kinase Mak [Oryza sativa]
Length = 425
Score = 280 bits (716), Expect = 5e-74
Identities = 137/256 (53%), Positives = 191/256 (74%), Gaps = 11/256 (4%)
Query: 12 LKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKERE 71
+KRK+ +EE +LRE+K+LR+MNH NI+KL+EV+REN+ L+FI EYM+CNLYQL+K+R
Sbjct: 1 MKRKYYSFEECMSLREVKSLRRMNHPNIVKLKEVIRENDILYFIMEYMECNLYQLMKDRV 60
Query: 72 KPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAREVSSMPP 131
KPFSE E+R + Q+ Q L++MH++G+FHRDLKPENLLV+ DV+K+ADFGLAREV+S+PP
Sbjct: 61 KPFSEAEVRNWCFQIFQALAYMHQRGYFHRDLKPENLLVSKDVIKLADFGLAREVTSVPP 120
Query: 132 YTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCI 191
YT+YVSTRWYRAPEVLLQS Y AVDMWA+GAI+AEL TL P+FPG SE D++ KI +
Sbjct: 121 YTEYVSTRWYRAPEVLLQSSIYDSAVDMWAMGAIMAELLTLHPLFPGTSEADEILKICNV 180
Query: 192 LGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC--------- 242
+G PD + G + + + F + V+ +L++++ + S EA+DLI++ C
Sbjct: 181 IGSPDEQSWPQGLSLAETMKFQFPQ-VSGNQLAEVMTSVSSEAVDLISSLCSWDPCKRPK 239
Query: 243 -GSVTATSFFPSCSIL 257
V +FF C+ +
Sbjct: 240 AAEVLQHTFFQGCTFV 255
>UniRef100_Q9JKV2 Serine/threonine kinase ICK [Mus musculus]
Length = 629
Score = 280 bits (716), Expect = 5e-74
Identities = 138/235 (58%), Positives = 179/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH NI+KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANIVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSRPPYTDYVSTRWYRAPEVLLRSTNYSSPIDIWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F+ + + P L +IPNAS EAI L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFLWPQCI-PNNLKTLIPNASSEAIQLL 261
>UniRef100_UPI00003ABC95 UPI00003ABC95 UniRef100 entry
Length = 556
Score = 279 bits (714), Expect = 8e-74
Identities = 135/236 (57%), Positives = 184/236 (77%), Gaps = 2/236 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+F+FEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFVFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLL-VTNDVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL + +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNMMYQILQGLAFIHKHGFFHRDMKPENLLCIGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+TL P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSIYSSPIDIWAVGSIMAELYTLRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT 239
+++KI +LG P + + G + S ++F + V P+ L +IPNAS EAI L++
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYHLSSAMNFRFPQCV-PISLKTLIPNASSEAIQLMS 262
>UniRef100_UPI00003ABC94 UPI00003ABC94 UniRef100 entry
Length = 617
Score = 279 bits (714), Expect = 8e-74
Identities = 135/236 (57%), Positives = 184/236 (77%), Gaps = 2/236 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+F+FEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFVFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLL-VTNDVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL + +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNMMYQILQGLAFIHKHGFFHRDMKPENLLCIGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+TL P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSIYSSPIDIWAVGSIMAELYTLRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT 239
+++KI +LG P + + G + S ++F + V P+ L +IPNAS EAI L++
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYHLSSAMNFRFPQCV-PISLKTLIPNASSEAIQLMS 262
>UniRef100_Q8IXN4 MAK protein [Homo sapiens]
Length = 457
Score = 279 bits (713), Expect = 1e-73
Identities = 148/300 (49%), Positives = 201/300 (66%), Gaps = 17/300 (5%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+ L P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMAELYMLRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT---- 239
+++KI +LG P + + G + ++F + V P+ L +IPNAS EAI L+T
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYQLASSMNFRFPQCV-PINLKTLIPNASNEAIQLMTEMLN 266
Query: 240 ------ARCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKP 293
+F +L SN L + +N ++ PLE K S V+P
Sbjct: 267 WDPKKRPTASQALKHPYFQVGQVLGPSSNHLESKQSLNKQL-----QPLESKPSLVEVEP 321
>UniRef100_P20794 Serine/threonine-protein kinase MAK [Homo sapiens]
Length = 623
Score = 279 bits (713), Expect = 1e-73
Identities = 148/300 (49%), Positives = 201/300 (66%), Gaps = 17/300 (5%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+ L P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMAELYMLRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT---- 239
+++KI +LG P + + G + ++F + V P+ L +IPNAS EAI L+T
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYQLASSMNFRFPQCV-PINLKTLIPNASNEAIQLMTEMLN 266
Query: 240 ------ARCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKP 293
+F +L SN L + +N ++ PLE K S V+P
Sbjct: 267 WDPKKRPTASQALKHPYFQVGQVLGPSSNHLESKQSLNKQL-----QPLESKPSLVEVEP 321
>UniRef100_UPI000013ED70 UPI000013ED70 UniRef100 entry
Length = 634
Score = 278 bits (711), Expect = 2e-73
Identities = 137/235 (58%), Positives = 178/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH N++KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANVVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSKPPYTDYVSTRWYRAPEVLLRSTNYSSPIDVWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F + V P L +IPNAS EA+ L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFRWPQCV-PNNLKTLIPNASSEAVQLL 261
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 608,577,577
Number of Sequences: 2790947
Number of extensions: 24852241
Number of successful extensions: 117555
Number of sequences better than 10.0: 18437
Number of HSP's better than 10.0 without gapping: 9342
Number of HSP's successfully gapped in prelim test: 9095
Number of HSP's that attempted gapping in prelim test: 80918
Number of HSP's gapped (non-prelim): 22165
length of query: 381
length of database: 848,049,833
effective HSP length: 129
effective length of query: 252
effective length of database: 488,017,670
effective search space: 122980452840
effective search space used: 122980452840
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC136841.7


