

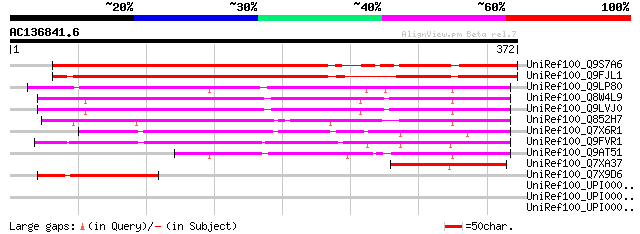
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.6 + phase: 0
(372 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9S7A6 T1B9.2 protein [Arabidopsis thaliana] 328 1e-88
UniRef100_Q9FJL1 Gb|AAF02142.1 [Arabidopsis thaliana] 292 1e-77
UniRef100_Q9LP80 T1N15.6 [Arabidopsis thaliana] 180 5e-44
UniRef100_Q8W4L9 Hypothetical protein At3g17800; MEB5.2 [Arabido... 171 3e-41
UniRef100_Q9LVJ0 Gb|AAF02142.1 [Arabidopsis thaliana] 171 3e-41
UniRef100_Q852H7 Hypothetical protein OSJNBa0078A17.10 [Oryza sa... 169 1e-40
UniRef100_Q7X6R1 OSJNBa0006A01.17 protein [Oryza sativa] 163 6e-39
UniRef100_Q9FVR1 Hypothetical protein F3C3.6 [Arabidopsis thaliana] 157 4e-37
UniRef100_Q9AT51 Hypothetical protein [Brassica napus] 134 3e-30
UniRef100_Q7XA37 Hypothetical protein [Solanum bulbocastanum] 78 5e-13
UniRef100_Q7X9D6 ABRH7 [Marsilea quadrifolia] 48 4e-04
UniRef100_UPI0000287803 UPI0000287803 UniRef100 entry 37 1.2
UniRef100_UPI00002DCCDB UPI00002DCCDB UniRef100 entry 36 1.5
UniRef100_UPI00002CBF8F UPI00002CBF8F UniRef100 entry 35 4.4
>UniRef100_Q9S7A6 T1B9.2 protein [Arabidopsis thaliana]
Length = 368
Score = 328 bits (842), Expect = 1e-88
Identities = 182/343 (53%), Positives = 238/343 (69%), Gaps = 30/343 (8%)
Query: 32 LIKASAGASSHCE-SSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDA 90
++ +A S CE SSLN PL PR+ G+FL VL N R LFH A +ELK LADDR+A
Sbjct: 42 MVVVAAAGQSRCEPGSSLNAPLEPRSAQGRFLRSVLLNKRQLFHYAAADELKQLADDREA 101
Query: 91 ANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLY 150
A +RM L+S SDEA LHRRIAE+KE C+ AV+DIM +LIF+K+SEIR PLVPKLSRC+Y
Sbjct: 102 ALARMSLSSGSDEASLHRRIAELKERYCKTAVQDIMYMLIFYKYSEIRVPLVPKLSRCIY 161
Query: 151 NGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIY 210
NGRLEI PSKDWELESI++ + L++I+EHV+ V GL+ VT++WATT++++ L ++Y
Sbjct: 162 NGRLEIWPSKDWELESIYSCDTLEIIKEHVSAVIGLRVNSCVTDNWATTQIQKLHLRKVY 221
Query: 211 VASILYGYFLKSVSLRYHLERNLNLANHDVH-PGHRTNLSFKDMCPYGFEDDIFGHLSNM 269
ASILYGYFLKS SLR+ LE +L+ D+H G+ + IFG
Sbjct: 222 AASILYGYFLKSASLRHQLECSLS----DIHGSGY-------------LKSPIFGCSFT- 263
Query: 270 KPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSV 329
G I +++ L+ Y+ F P +LQRCAK R++EA NL+ S ALF +E
Sbjct: 264 --TGTAQISNKQQ---LRHYISDFDPETLQRCAKPRTEEARNLIEKQSLALFGTE----- 313
Query: 330 DSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVYKLKDH 372
+SD+ I+TSFSSLKRLVLEAVAFG+FLW+TE Y+D YKLK++
Sbjct: 314 ESDETIVTSFSSLKRLVLEAVAFGTFLWDTELYVDGAYKLKEN 356
>UniRef100_Q9FJL1 Gb|AAF02142.1 [Arabidopsis thaliana]
Length = 344
Score = 292 bits (747), Expect = 1e-77
Identities = 163/341 (47%), Positives = 217/341 (62%), Gaps = 50/341 (14%)
Query: 32 LIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAA 91
L+ SA +S S++ PL+PR+ G+FLS VL R LFH AV + LK LADD++A+
Sbjct: 43 LVVVSAASSGQ----SIDAPLVPRSPQGRFLSSVLVKKRQLFHFAVADLLKQLADDKEAS 98
Query: 92 NSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYN 151
SRM L+ SDEA LHRRIA++KE+ C++A+EDIM +LI +KFSEIR PLVPKL C+YN
Sbjct: 99 LSRMFLSYGSDEASLHRRIAQLKESDCQIAIEDIMYMLILYKFSEIRVPLVPKLPSCIYN 158
Query: 152 GRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYV 211
GRLEI PSKDWELESIH+ +VL++I+EH V L+ S+T+ ATT++ + L ++Y
Sbjct: 159 GRLEISPSKDWELESIHSFDVLELIKEHSNAVISLRVNSSLTDDCATTEIDKNRLSKVYT 218
Query: 212 ASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKP 271
AS+LYGYFLKS SLR+ LE +L+ H G T
Sbjct: 219 ASVLYGYFLKSASLRHQLECSLS-----QHHGSFT------------------------- 248
Query: 272 IGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSVDS 331
+ L+ Y+ F P L+RCAK RS EA +L+ S ALF E S
Sbjct: 249 ------------KQLRHYISEFDPKILRRCAKPRSHEAKSLIEKQSLALFGPE----ESS 292
Query: 332 DDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVYKLKDH 372
+ I+TSFSSLKRL+LEAVAFG+FLW+TE+Y+D +KLK++
Sbjct: 293 KESIVTSFSSLKRLLLEAVAFGTFLWDTEEYVDGAFKLKEN 333
>UniRef100_Q9LP80 T1N15.6 [Arabidopsis thaliana]
Length = 423
Score = 180 bits (457), Expect = 5e-44
Identities = 121/375 (32%), Positives = 203/375 (53%), Gaps = 28/375 (7%)
Query: 14 FTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLF 73
F+RP ++ R ++KASA + ES + PL ++ VG+FLS +L +H +L
Sbjct: 51 FSRPFQSGTTCVKSRRSFVVKASASGDASTESIA---PLQLKSPVGQFLSQILVSHPHLV 107
Query: 74 HVAVQEELKLLADDRDAANSRMLLASE-SDEALLHRRIAEMKENQCEVAVEDIMSLLIFH 132
AV+++L+ L DRDA +S + +L+RRIAE+KE + A+E+I+ L+
Sbjct: 108 PAAVEQQLEQLQIDRDAEEQSKDASSVLGTDIVLYRRIAEVKEKERRRALEEILYALVVQ 167
Query: 133 KFSEIRAPLVPKL--SRCLYNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKP 190
KF + LVP + S +GR++ P+ D ELE +H+ EV +MI+ H++ + K
Sbjct: 168 KFMDANVTLVPSITSSSADPSGRVDTWPTLDGELERLHSPEVYEMIQNHLSIIL----KN 223
Query: 191 SVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRT-NLS 249
+ A ++ + +G++Y AS++YGYFLK + R+ LE+ + + G + +
Sbjct: 224 RTDDLTAVAQISKLGVGQVYAASVMYGYFLKRIDQRFQLEKTMRILPGGSDEGETSIEQA 283
Query: 250 FKDMCPYGFED--DIFGHLSNMKPIGQ--------GLIRQEEEIEDLKCYVMRFHPGSLQ 299
+D+ +E+ + + +S+ + +G G + + LK YVM F +LQ
Sbjct: 284 GRDVERNFYEEAEETYQAVSSNQDVGSFVGGINASGGFSSDMKQSRLKTYVMSFDGETLQ 343
Query: 300 RCAKLRSKEAVNLVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAF 352
R A +RS+E+V ++ ++ ALF +G D+ I SF LKRLVLEAV F
Sbjct: 344 RYATIRSRESVGIIEKHTEALFGRPEIVITPQGTIDSSKDEHIKISFKGLKRLVLEAVTF 403
Query: 353 GSFLWETEDYIDNVY 367
GSFLW+ E ++D+ Y
Sbjct: 404 GSFLWDVESHVDSRY 418
>UniRef100_Q8W4L9 Hypothetical protein At3g17800; MEB5.2 [Arabidopsis thaliana]
Length = 421
Score = 171 bits (433), Expect = 3e-41
Identities = 113/363 (31%), Positives = 189/363 (51%), Gaps = 23/363 (6%)
Query: 21 SSSLSFRSRVPLIKASAGASSHCESSSLNTPLLP---RTQVGKFLSGVLQNHRNLFHVAV 77
S + + ++R + ++ AS+ S S P+ P ++ G+FLS +L +H +L AV
Sbjct: 61 SLTTTAKTRRSFVVRASSASNDASSGSSPKPIAPLQLQSPAGQFLSQILVSHPHLVPAAV 120
Query: 78 QEELKLLADDRDAANSRMLLAS-ESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSE 136
+++L+ L DRD+ AS + +L+RRIAE+KEN+ +E+I+ L+ KF E
Sbjct: 121 EQQLEQLQTDRDSQGQNKDSASVPGTDIVLYRRIAELKENERRRTLEEILYALVVQKFME 180
Query: 137 IRAPLVPKLSRCL-YNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTES 195
LVP +S +GR++ P+K +LE +H+ E+ +MI H+ + G + + +
Sbjct: 181 ANVSLVPSVSPSSDPSGRVDTWPTKVEKLERLHSPEMYEMIHNHLALILGSR----MGDL 236
Query: 196 WATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP 255
+ ++ + +G++Y AS++YGYFLK V R+ LE+ + + + +
Sbjct: 237 NSVAQISKLRVGQVYAASVMYGYFLKRVDQRFQLEKTMKILPGGSDESKTSVEQAEGTAT 296
Query: 256 Y----GFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVN 311
Y ++ + G G E + L+ YVM F +LQR A +RS+EAV
Sbjct: 297 YQAAVSSHPEVGAFAGGVSAKGFG---SEIKPSRLRSYVMSFDAETLQRYATIRSREAVG 353
Query: 312 LVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
++ ++ ALF EG D+ I SF +KRLVLEAV FGSFLW+ E ++D
Sbjct: 354 IIEKHTEALFGKPEIVITPEGTVDSSKDEQIKISFGGMKRLVLEAVTFGSFLWDVESHVD 413
Query: 365 NVY 367
Y
Sbjct: 414 ARY 416
>UniRef100_Q9LVJ0 Gb|AAF02142.1 [Arabidopsis thaliana]
Length = 421
Score = 171 bits (433), Expect = 3e-41
Identities = 113/363 (31%), Positives = 189/363 (51%), Gaps = 23/363 (6%)
Query: 21 SSSLSFRSRVPLIKASAGASSHCESSSLNTPLLP---RTQVGKFLSGVLQNHRNLFHVAV 77
S + + ++R + ++ AS+ S S P+ P ++ G+FLS +L +H +L AV
Sbjct: 61 SLTTTAKTRRSFVVRASSASNDASSGSSPKPIAPLQLQSPAGQFLSQILVSHPHLVPAAV 120
Query: 78 QEELKLLADDRDAANSRMLLAS-ESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSE 136
+++L+ L DRD+ AS + +L+RRIAE+KEN+ +E+I+ L+ KF E
Sbjct: 121 EQQLEQLQTDRDSQGQNKDSASVPGTDIVLYRRIAELKENERRRTLEEILYALVVQKFME 180
Query: 137 IRAPLVPKLSRCL-YNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTES 195
LVP +S +GR++ P+K +LE +H+ E+ +MI H+ + G + + +
Sbjct: 181 ANVSLVPSVSPSSDPSGRVDTWPTKVEKLERLHSPEMYEMIHNHLALILGSR----MGDL 236
Query: 196 WATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP 255
+ ++ + +G++Y AS++YGYFLK V R+ LE+ + + + +
Sbjct: 237 NSVAQISKLRVGQVYAASVMYGYFLKRVDQRFQLEKTMKILPGGSDESKTSVEQAEGTAT 296
Query: 256 Y----GFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVN 311
Y ++ + G G E + L+ YVM F +LQR A +RS+EAV
Sbjct: 297 YQAAVSSHPEVGAFAGGVSAKGFG---SEIKPSRLRSYVMSFDAETLQRYATIRSREAVG 353
Query: 312 LVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
++ ++ ALF EG D+ I SF +KRLVLEAV FGSFLW+ E ++D
Sbjct: 354 IIEKHTEALFGKPEIVITPEGTVDSSKDEQIKISFGGMKRLVLEAVTFGSFLWDVESHVD 413
Query: 365 NVY 367
Y
Sbjct: 414 ARY 416
>UniRef100_Q852H7 Hypothetical protein OSJNBa0078A17.10 [Oryza sativa]
Length = 413
Score = 169 bits (427), Expect = 1e-40
Identities = 123/375 (32%), Positives = 195/375 (51%), Gaps = 48/375 (12%)
Query: 24 LSFRSRVPLIKASAGASSHCES-----SSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQ 78
L+ R R +++A++ + S ES SS PL + VG+FLS +L H +L A +
Sbjct: 51 LTCRRRGVVVRAASWSPSGPESLPPPPSSSIAPLQMESPVGQFLSQILATHPHLLPAAAE 110
Query: 79 EELKLLADDRDAA--NSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSE 136
++L+ L DRDAA N A + +L+RRIAE+KE + + A+E+I+ L+ KF E
Sbjct: 111 QQLEQLQTDRDAAKDNGGDKPAPSDGDIVLYRRIAEVKEKERKRALEEILYALVVQKFVE 170
Query: 137 IRAPLVPKLSRCLYN-GRLEI-LPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTE 194
LVP LS + + GR++ S + +LE +H+ E +MI H+ + G + +
Sbjct: 171 AGVSLVPALSHSISSSGRVDQWAESVEGKLEKMHSQEAYEMIENHLALILGQRQADATVA 230
Query: 195 SWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLN--------------LANHDV 240
A +K+R +G++Y AS++YGYFLK V R+ LE+++ + D
Sbjct: 231 --AISKLR---VGQVYAASVMYGYFLKRVDQRFQLEKSMKTLPWGSEEEDKLNQVMTTDS 285
Query: 241 HPGHRTNLSFKDMCPYGFEDDIFGHLS-NMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQ 299
P +T+ S +M + + G S ++KP L+ YVM F +LQ
Sbjct: 286 RPSPQTSTSHPEMASWTSPNFSAGGPSQSVKPCR------------LRSYVMSFDSETLQ 333
Query: 300 RCAKLRSKEAVNLVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAF 352
A +RSKEA ++ ++ ALF EG D+ + SF+ L+RL+LEAV F
Sbjct: 334 SYATIRSKEAFGIIEKHTEALFGKPEIVITPEGTVDSSKDEHVRISFAGLRRLILEAVTF 393
Query: 353 GSFLWETEDYIDNVY 367
GSFLW+ E ++D Y
Sbjct: 394 GSFLWDVESFVDTRY 408
>UniRef100_Q7X6R1 OSJNBa0006A01.17 protein [Oryza sativa]
Length = 405
Score = 163 bits (413), Expect = 6e-39
Identities = 109/327 (33%), Positives = 174/327 (52%), Gaps = 22/327 (6%)
Query: 51 PLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRI 110
PL+ T G+ L +LQ+H +L V ++L+ L ++DA AS+ + LL++RI
Sbjct: 86 PLMFETPSGQLLVQILQSHPHLLPATVDQQLENLQSEKDAQEKE---ASKVPQDLLYKRI 142
Query: 111 AEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGR-LEILPSKDWELESIHT 169
AE+KE + + +E+I+ I +KF E + P L+ R + LP+++ L+SIH+
Sbjct: 143 AEVKEKERQNTLEEIIYCWIIYKFMENDISMTPALAPLGGPVRDISSLPNQEDRLQSIHS 202
Query: 170 LEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHL 229
+ L+MI+ H+ + G K + ++ LG++Y ASI+YGYFLK V R+ L
Sbjct: 203 PDALEMIQNHLNLIMGEKVAAPLD---TVVEISNLNLGKLYAASIMYGYFLKRVDERFQL 259
Query: 230 ERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQEEEIED--LK 287
E+N+ + P + + +++ P F D L + P G+ + +EE L+
Sbjct: 260 EKNMKT----LPPNPKQQIVLENLKPNPFWD--MESLVQITPDGEEIDLDDEESNPNKLR 313
Query: 288 CYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSVDSDDV-------ILTSFS 340
YV R +LQR A +RSKEAV+L+ + ALF +D V I +F
Sbjct: 314 SYVSRLDADTLQRYATIRSKEAVSLIEKQTQALFGRPDIKVLDDGSVNAKDGQMITITFI 373
Query: 341 SLKRLVLEAVAFGSFLWETEDYIDNVY 367
L LVLEA AFGSFLWE E ++++ Y
Sbjct: 374 ELTHLVLEAAAFGSFLWEAESHVESKY 400
>UniRef100_Q9FVR1 Hypothetical protein F3C3.6 [Arabidopsis thaliana]
Length = 406
Score = 157 bits (397), Expect = 4e-37
Identities = 111/363 (30%), Positives = 192/363 (52%), Gaps = 24/363 (6%)
Query: 19 SFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQ 78
S S++ + R R ++AS S+ E+ + P+ + VG+ L +L+ H +L V V
Sbjct: 49 SSSTNENGRGRSVTVRASGDEDSN-ENFAPLAPVELESPVGQLLEQILRTHPHLLPVTVD 107
Query: 79 EELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIR 138
E+L+ A + ++ + S S + +L +RI+E+++ + + +I+ L+ H+F E
Sbjct: 108 EQLEKFAAESESRKAD----SSSTQDILQKRISEVRDKERRKTLAEIIYCLVVHRFVEKG 163
Query: 139 APLVPKLSRCLYN-GRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWA 197
++P++ GR+++ P+++ +LE IH+ + +MI+ H+++V G P+V +
Sbjct: 164 ISMIPRIKPTSDPAGRIDLWPNQEEKLEVIHSADAFEMIQSHLSSVLG--DGPAVGPLSS 221
Query: 198 TTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP-Y 256
++ + LG++Y AS +YGYFL+ V RY LER +N +T F++ P Y
Sbjct: 222 IVQIGKIKLGKLYAASAMYGYFLRRVDQRYQLERTMNTLPKRPE---KTRERFEEPSPPY 278
Query: 257 GFEDD---IFGHLSNMKPIGQGLIRQEEEIED--LKCYVMRFHPGSLQRCAKLRSKEAVN 311
D I P + R E+E L+ YV +LQR A +RSKEA+
Sbjct: 279 PLWDPDSLIRIQPEEYDPDEYAIQRNEDESSSYGLRSYVTYLDSDTLQRYATIRSKEAMT 338
Query: 312 LVRSYSSALFN-------SEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
L+ + ALF +G +D+V+ S S L LVLEAVAFGSFLW++E Y++
Sbjct: 339 LIEKQTQALFGRPDIRILEDGKLDTSNDEVLSLSVSGLAMLVLEAVAFGSFLWDSESYVE 398
Query: 365 NVY 367
+ Y
Sbjct: 399 SKY 401
>UniRef100_Q9AT51 Hypothetical protein [Brassica napus]
Length = 256
Score = 134 bits (338), Expect = 3e-30
Identities = 86/260 (33%), Positives = 138/260 (53%), Gaps = 24/260 (9%)
Query: 122 VEDIMSLLIFHKFSEIRAPLVPKL--SRCLYNGRLEILPSKDWELESIHTLEVLDMIREH 179
+E+I+ L+ KF E LVP + S +GR++ P+K +LE +H+ E+ +MI H
Sbjct: 2 LEEILYALVVQKFMEANVSLVPSITPSSSDPSGRVDTWPTKVEKLEKLHSSEMYEMIHNH 61
Query: 180 VTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHD 239
+ + G P + + + ++ + +G++Y AS++YGYFLK V R+ LE+++ L
Sbjct: 62 LALILG----PRMGDLASVAQISKLRVGQVYAASVMYGYFLKRVDQRFQLEKSMKLLTGG 117
Query: 240 VHPGHRT-----NLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFH 294
+ + N++FK + + G +S K G E + L+ YVM F
Sbjct: 118 LDESKTSVEQPENVTFKAVSSHPEVGSFAGGVS-AKGFGS-----EIKPSRLRTYVMSFD 171
Query: 295 PGSLQRCAKLRSKEAVNLVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVL 347
+LQR A +RS+EAV ++ ++ ALF EG D+ I SF +KRLVL
Sbjct: 172 SETLQRYATIRSREAVGIIEKHTEALFGKPEIVITPEGTVDSSKDEQIKISFGGMKRLVL 231
Query: 348 EAVAFGSFLWETEDYIDNVY 367
EAV FGSFLW+ E ++D Y
Sbjct: 232 EAVTFGSFLWDVESHVDARY 251
>UniRef100_Q7XA37 Hypothetical protein [Solanum bulbocastanum]
Length = 157
Score = 77.8 bits (190), Expect = 5e-13
Identities = 43/92 (46%), Positives = 58/92 (62%), Gaps = 7/92 (7%)
Query: 280 EEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALF-------NSEGFDSVDSD 332
E + E L+ YVM +LQR A LRSKEAV ++ + ALF + +G V +D
Sbjct: 24 EGKPERLRSYVMYLDADTLQRYATLRSKEAVTVIEKQTQALFGRPDIKVSGDGTLDVSND 83
Query: 333 DVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
+V+ ++S L LVLEAVAFGSFLWETE Y++
Sbjct: 84 EVLSVTYSGLTMLVLEAVAFGSFLWETESYVE 115
>UniRef100_Q7X9D6 ABRH7 [Marsilea quadrifolia]
Length = 176
Score = 48.1 bits (113), Expect = 4e-04
Identities = 30/90 (33%), Positives = 57/90 (63%), Gaps = 4/90 (4%)
Query: 21 SSSLSFRSRVPL-IKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQE 79
+S++S R + L I A AS+ S++ PL+P + G+FL+ +LQ+H +L A ++
Sbjct: 89 ASTVSNRRKTRLTINAEMTASA---STTPLAPLIPESPSGQFLTELLQSHPHLVPAAAEQ 145
Query: 80 ELKLLADDRDAANSRMLLASESDEALLHRR 109
+L+ LA+ R+AA+S+ + +E +L++R
Sbjct: 146 QLETLAEAREAASSQEQPTPDGNELVLYKR 175
>UniRef100_UPI0000287803 UPI0000287803 UniRef100 entry
Length = 333
Score = 36.6 bits (83), Expect = 1.2
Identities = 29/130 (22%), Positives = 60/130 (45%), Gaps = 15/130 (11%)
Query: 41 SHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDR-DAANSRMLLAS 99
+H +++++ + T++GKFLS + + RN FH+A + +K + + N+ L S
Sbjct: 48 NHIDTANMYGMGVSETRIGKFLSSIGKKDRNFFHIATKGGIKTKPGGKPEFDNTEDYLTS 107
Query: 100 ESDEALLHRRIAE-----MKENQCEVAVEDIMSLLIFHK---------FSEIRAPLVPKL 145
E D++L + + + +E+++ L+ K FSEI + K
Sbjct: 108 ELDKSLKRLGVENIGLYYIHRRDGKTPIEEVVETLLKFKKAGKINQFGFSEISPVSLEKA 167
Query: 146 SRCLYNGRLE 155
S + G ++
Sbjct: 168 SNIHHVGAVQ 177
>UniRef100_UPI00002DCCDB UPI00002DCCDB UniRef100 entry
Length = 281
Score = 36.2 bits (82), Expect = 1.5
Identities = 19/66 (28%), Positives = 38/66 (56%), Gaps = 1/66 (1%)
Query: 41 SHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLL-ADDRDAANSRMLLAS 99
+H +++++ + T++GKFLS + + RN FH+A + +K + N++ L S
Sbjct: 48 NHIDTANMYGMGISETRIGKFLSSIGKKDRNFFHIATKGGIKTKPGGPPEFDNTKDYLTS 107
Query: 100 ESDEAL 105
E D++L
Sbjct: 108 ELDKSL 113
>UniRef100_UPI00002CBF8F UPI00002CBF8F UniRef100 entry
Length = 147
Score = 34.7 bits (78), Expect = 4.4
Identities = 18/66 (27%), Positives = 38/66 (57%), Gaps = 1/66 (1%)
Query: 41 SHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLL-ADDRDAANSRMLLAS 99
+H +++++ + T++G+FLS + + RN FH+A + +K + N++ L S
Sbjct: 48 NHIDTANMYGMGISETRIGEFLSSIGKKDRNFFHIATKGGIKTKPGGPPEFDNTKDYLTS 107
Query: 100 ESDEAL 105
E D++L
Sbjct: 108 ELDKSL 113
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,239,810
Number of Sequences: 2790947
Number of extensions: 23590870
Number of successful extensions: 55798
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 55746
Number of HSP's gapped (non-prelim): 19
length of query: 372
length of database: 848,049,833
effective HSP length: 129
effective length of query: 243
effective length of database: 488,017,670
effective search space: 118588293810
effective search space used: 118588293810
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC136841.6


