

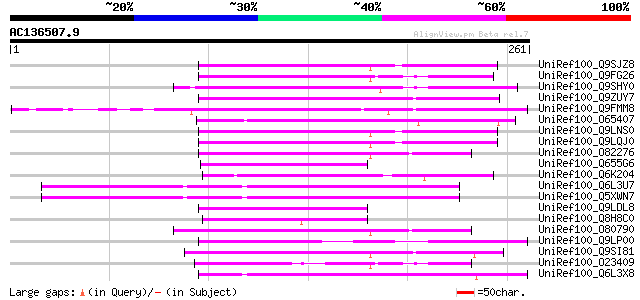
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.9 + phase: 0 /pseudo
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 81 3e-14
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 77 3e-13
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 74 5e-12
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 73 8e-12
UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like... 71 2e-11
UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thal... 67 5e-10
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 65 2e-09
UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana] 65 2e-09
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 65 2e-09
UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa] 61 3e-08
UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa] 60 7e-08
UniRef100_Q6L3U7 Putative RNase H domain containing protein [Sol... 59 2e-07
UniRef100_Q5XWN7 Hypothetical protein [Solanum tuberosum] 59 2e-07
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 58 2e-07
UniRef100_Q8H8C0 Hypothetical protein OJ1134F05.9 [Oryza sativa] 58 3e-07
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 57 5e-07
UniRef100_Q9LP00 F9C16.18 [Arabidopsis thaliana] 56 8e-07
UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana] 55 2e-06
UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 ... 55 2e-06
UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum] 52 2e-05
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 80.9 bits (198), Expect = 3e-14
Identities = 56/155 (36%), Positives = 77/155 (49%), Gaps = 8/155 (5%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W+ P W+K+NTDGA NP A+ GGV RDHN +IG F N+ SA A+L GV
Sbjct: 151 VSWVSPEDGWVKLNTDGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGV 210
Query: 156 ITAINIAGENNWRNIWLETDLQLAV----LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLI 211
+ IA R + LE D ++ V + + + + LR L Y + I
Sbjct: 211 YYGLFIAWGRGARRVELEVDSKMVVGFLTTGIADSHPLSFLLR---LCYDFLSKGWIVRI 267
Query: 212 THIYIEGNSCADCMANVGLTLS-SFVWLPSLPDCI 245
+H+Y E N AD +AN +LS L S PD +
Sbjct: 268 SHVYREANRLADGLANYAFSLSLGLHLLESRPDVV 302
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 77.4 bits (189), Expect = 3e-13
Identities = 55/161 (34%), Positives = 76/161 (47%), Gaps = 24/161 (14%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W P W+ +NTDGA NP +A+ GGV RD + ++ F N+ SA A+L GV
Sbjct: 506 IAWRKPAEGWVTMNTDGASHGNPGQATAGGVIRDEHGSWLVGFALNIGVCSAPLAELWGV 565
Query: 156 ITAINIAGENNWRNIWLETDLQLAV------------LAFKNLNMVPWSLRNKWLNYFEH 203
+ +A E WR + LE D L V LAF + + + W+
Sbjct: 566 YYGLVVAWERGWRRVRLEVDSALVVGFLQSGIGDSHPLAFL-VRLCHGFISKDWI----- 619
Query: 204 VRSMNFLITHIYIEGNSCADCMANVGLTLS-SFVWLPSLPD 243
VR ITH+Y E N AD +AN TL F+ L S P+
Sbjct: 620 VR-----ITHVYREANRLADGLANYAFTLPFGFLLLDSCPE 655
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 73.6 bits (179), Expect = 5e-12
Identities = 59/185 (31%), Positives = 80/185 (42%), Gaps = 24/185 (12%)
Query: 83 INIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL 142
+ I PR + + W+ P W+KVNTDGA NP AS GGV RD + G F N+
Sbjct: 548 VGITQPRVE--RMIGWVSPCVGWVKVNTDGASRGNPGLASAGGVLRDCTGAWCGGFSLNI 605
Query: 143 NTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKN-----------LNMVPW 191
SA A+L GV + A E + LE D ++ V K + +
Sbjct: 606 GRCSAPQAELWGVYYGLYFAWEKKVPRVELEVDSEVIVGFLKTGISDSHPLSFLVRLCHG 665
Query: 192 SLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANVGLTLS-SFVWLPSLPDCIKHDYG 250
L+ WL VR I H+Y E N AD +AN +LS F +PD +
Sbjct: 666 FLQKDWL-----VR-----IVHVYREANRLADGLANYAFSLSLGFHSFDLVPDAMSSLLR 715
Query: 251 RNRLG 255
+ LG
Sbjct: 716 EDTLG 720
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 72.8 bits (177), Expect = 8e-12
Identities = 54/154 (35%), Positives = 74/154 (47%), Gaps = 4/154 (2%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W P W K+NTDGA NP AS GGV RD + G F N+ SA A+L GV
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWGV 203
Query: 156 ITAINIAGENNWRNIWLETDLQLAVLAFK-NLNMV-PWSLRNKWLNYFEHVRSMNFLITH 213
+ IA E + +E D ++ V K +N V P S + + F R I+H
Sbjct: 204 YYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFIS-RDWRVRISH 262
Query: 214 IYIEGNSCADCMANVGLTLS-SFVWLPSLPDCIK 246
+Y E N AD +AN +L F L +PD ++
Sbjct: 263 VYREANRLADGLANYAFSLPLGFHSLSLVPDSLR 296
>UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 308
Score = 71.2 bits (173), Expect = 2e-11
Identities = 73/273 (26%), Positives = 112/273 (40%), Gaps = 44/273 (16%)
Query: 2 IYGKLCDKRWTPQCKVVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTG 61
+Y LCD+ C+ + + I ++ IW+ WK NI G
Sbjct: 62 LYDNLCDRS---SCEDIPWSTIFAVI--IWW------------GWKWRCSNIF------G 98
Query: 62 NRTKLTSFVNMDEFVFLKAFKINIHPPRA---------PLIKEVM-WIPPIPPWIKVNTD 111
TK D F+K + + ++ P ++ ++ W+ P W+KVNTD
Sbjct: 99 ENTKCR-----DRVKFVKEWVVEVYRAHLGNALVGSTQPRVERLIGWVLPCVGWVKVNTD 153
Query: 112 GALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIW 171
GA NP AS GGV RD + G F N+ SA +A+L GV + A E +
Sbjct: 154 GASRGNPGLASAGGVLRDCEGAWCGGFSLNIGRCSAQHAELWGVYYGLYFAWEKKVPRVE 213
Query: 172 LETDLQLAVLAFKNLNMV---PWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANV 228
LE D + A++ F + P S + + F + I ++Y E N AD +AN
Sbjct: 214 LEVDSE-AIVGFLKTGISDSHPLSFLVRLCHNFLQ-KDWLVRIVYVYREANCLADGLANY 271
Query: 229 GLTLS-SFVWLPSLPDCIKHDYGRNRLGMHSFR 260
+ LS F +PD + + LG R
Sbjct: 272 TILLSLGFHSFDFVPDAMSSLLKEDTLGSTQLR 304
>UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thaliana]
Length = 229
Score = 67.0 bits (162), Expect = 5e-10
Identities = 46/166 (27%), Positives = 82/166 (48%), Gaps = 7/166 (4%)
Query: 95 EVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLG 154
+V+W P IK+N DG+ + +AS GGVFR+H F+ + +++ ++ A+
Sbjct: 60 KVVWKKPENGRIKLNFDGSRGREG-QASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAA 118
Query: 155 VITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHV--RSMNFLIT 212
+ + +A EN ++WLE D ++ + + NK +NY + V N +++
Sbjct: 119 LKRGLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLS 178
Query: 213 HIYIEGNSCADCMANVGLTLSS-FVWLPSLPDC---IKHDYGRNRL 254
H+Y EGN AD +A +G VW P+ I HD + ++
Sbjct: 179 HVYREGNRVADKLAKLGHQFQDPKVWRVQPPEIVLPIMHDDAKGKI 224
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 64.7 bits (156), Expect = 2e-09
Identities = 48/155 (30%), Positives = 72/155 (45%), Gaps = 8/155 (5%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W PP W K+NTDGA NP A+ GGV RD + + F N+ SA A+L G
Sbjct: 83 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 142
Query: 156 ITAINIAGENNWRNIWLETDLQLAV----LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLI 211
+NIA E + +E D ++ V + + + + +R L + + + I
Sbjct: 143 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVR---LCHGLLSKDWSVRI 199
Query: 212 THIYIEGNSCADCMANVGLTLS-SFVWLPSLPDCI 245
+H+Y E N AD +AN L F S PD +
Sbjct: 200 SHVYREANRLADGLANYAFFLPLGFHLFNSTPDIV 234
>UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana]
Length = 272
Score = 64.7 bits (156), Expect = 2e-09
Identities = 48/155 (30%), Positives = 72/155 (45%), Gaps = 8/155 (5%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W PP W K+NTDGA NP A+ GGV RD + + F N+ SA A+L G
Sbjct: 102 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 161
Query: 156 ITAINIAGENNWRNIWLETDLQLAV----LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLI 211
+NIA E + +E D ++ V + + + + +R L + + + I
Sbjct: 162 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVR---LCHGLLSKDWSVRI 218
Query: 212 THIYIEGNSCADCMANVGLTLS-SFVWLPSLPDCI 245
+H+Y E N AD +AN L F S PD +
Sbjct: 219 SHVYREANRLADGLANYAFFLPLGFHLFNSTPDIV 253
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 64.7 bits (156), Expect = 2e-09
Identities = 43/139 (30%), Positives = 66/139 (46%), Gaps = 3/139 (2%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W P W+K+ TDGA N A+ GG R+ ++G F N+ + +A A+L G
Sbjct: 1061 IRWQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGA 1120
Query: 156 ITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITH 213
+ IA + +R + L+ D +L V L+ N P S + F R ++H
Sbjct: 1121 YYGLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGF-FTRDWLVRVSH 1179
Query: 214 IYIEGNSCADCMANVGLTL 232
+Y E N AD +AN TL
Sbjct: 1180 VYREANRLADGLANYAFTL 1198
>UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa]
Length = 240
Score = 61.2 bits (147), Expect = 3e-08
Identities = 27/84 (32%), Positives = 46/84 (54%)
Query: 97 MWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVI 156
+W P W+K+N DG+ + AS GGV+RDH+ F+ + + + T ++ A+L +
Sbjct: 78 IWRKPEEGWMKLNFDGSSKHSTGIASIGGVYRDHDGAFLLGYAERIGTATSSVAELAALR 137
Query: 157 TAINIAGENNWRNIWLETDLQLAV 180
+ +A N WR +W E D + V
Sbjct: 138 RGLELAVRNGWRRVWAEGDSKAVV 161
>UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa]
Length = 212
Score = 59.7 bits (143), Expect = 7e-08
Identities = 41/154 (26%), Positives = 72/154 (46%), Gaps = 13/154 (8%)
Query: 98 WIPPIPPWIKVNTDGALTKNPTK-ASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVI 156
W P WIK+N DG+ +K+ TK AS GGV+RDH F+ + + + ++ A+L +
Sbjct: 51 WRKPEAGWIKLNFDGS-SKHATKIASIGGVYRDHEGAFVLGYAERIGRATSSVAELAALR 109
Query: 157 TAINIAGENNWRNIWLETDLQLAV-LAFKNLNMVPWSLRNKWLNYFEHVRSM-----NFL 210
+ + N WR +W E D + V + N+ + L + ++ +
Sbjct: 110 RGLELVVRNGWRRVWAEGDSKTVVDVVCDRANV----RSEEDLRQCREIAALLPLIDDMA 165
Query: 211 ITHIYIEGNSCADCMANVG-LTLSSFVWLPSLPD 243
++H+Y GN A A +G + VW + P+
Sbjct: 166 VSHVYRSGNKVAHGFARLGHKAVRPRVWRAAPPE 199
>UniRef100_Q6L3U7 Putative RNase H domain containing protein [Solanum demissum]
Length = 722
Score = 58.5 bits (140), Expect = 2e-07
Identities = 54/210 (25%), Positives = 86/210 (40%), Gaps = 4/210 (1%)
Query: 17 VVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFV 76
V++ + I I IW R ++ D+K W ++N + R SF + +
Sbjct: 287 VIIHTLPILIFWEIWKRRCACKYGDQKKMWYRTMENHVWWNLKMSLRMTFPSFEIGNSWR 346
Query: 77 FLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIG 136
L ++ P P K V W P +K+NTDG+ + A G + RDH I
Sbjct: 347 DLLNKVESLRP--YPKWKIVHWNTPNINCVKINTDGSFSSG--NAGLGWIVRDHTRRMIM 402
Query: 137 AFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNK 196
AF + +S A+ L I + + N +LE D +L V +N ++
Sbjct: 403 AFSIPSSCSSNNLAEALAARFGILWCLQQGFHNCYLELDSKLVVDMVRNGQATNLKIKGV 462
Query: 197 WLNYFEHVRSMNFLITHIYIEGNSCADCMA 226
+ + V MN + H Y E N AD +A
Sbjct: 463 VEDIIQVVAKMNCEVNHCYREANQVADALA 492
>UniRef100_Q5XWN7 Hypothetical protein [Solanum tuberosum]
Length = 1095
Score = 58.5 bits (140), Expect = 2e-07
Identities = 54/210 (25%), Positives = 86/210 (40%), Gaps = 4/210 (1%)
Query: 17 VVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFV 76
V++ + I I IW R ++ D+K W ++N + R SF + +
Sbjct: 465 VIIHTLPILIFWEIWKRRCACKYGDQKKMWYRTMENHVWWNLKMSLRMTFPSFEIGNSWR 524
Query: 77 FLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIG 136
L ++ P P K V W P +K+NTDG+ + A G + RDH I
Sbjct: 525 DLLNKVESLRP--YPKWKIVHWNTPNINCVKINTDGSFSSG--NAGLGWIVRDHTRRMIM 580
Query: 137 AFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNK 196
AF + +S A+ L I + + N +LE D +L V +N ++
Sbjct: 581 AFSIPSSCSSNNLAEALAARFGILWCLQQGFHNCYLELDSKLVVDMVRNGQATNLKIKGV 640
Query: 197 WLNYFEHVRSMNFLITHIYIEGNSCADCMA 226
+ + V MN + H Y E N AD +A
Sbjct: 641 VEDIIQVVAKMNCEVNHCYREANQVADALA 670
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 58.2 bits (139), Expect = 2e-07
Identities = 33/85 (38%), Positives = 45/85 (52%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W PP W+K+NTDGA N A+ GGV RD + G F ++ SA A+L GV
Sbjct: 839 VAWKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGV 898
Query: 156 ITAINIAGENNWRNIWLETDLQLAV 180
+ +A E + + LE D +L V
Sbjct: 899 YYGLYMAWERRFTRVELEVDSELVV 923
>UniRef100_Q8H8C0 Hypothetical protein OJ1134F05.9 [Oryza sativa]
Length = 210
Score = 57.8 bits (138), Expect = 3e-07
Identities = 27/85 (31%), Positives = 41/85 (47%), Gaps = 2/85 (2%)
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTN--SAFNAKLLGV 155
W P W+K+N DG+ AS GG FRDH F+ + + + S+F A+L +
Sbjct: 47 WRKPEVGWMKLNFDGSRNDATGAASIGGAFRDHEGAFVAGYAERMIVGGASSFTAELAAL 106
Query: 156 ITAINIAGENNWRNIWLETDLQLAV 180
+ +A WR +W E D + V
Sbjct: 107 RRGLELAARYGWRRVWAEGDSRAVV 131
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 57.0 bits (136), Expect = 5e-07
Identities = 42/152 (27%), Positives = 67/152 (43%), Gaps = 3/152 (1%)
Query: 83 INIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL 142
+N H RA + + + W P W+K+ TDGA + A+ G + ++G F N+
Sbjct: 787 LNNHVKRARVERMIRWKAPSDRWVKLTTDGASRGHQGLAAASGAILNLQGEWLGGFALNI 846
Query: 143 NTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNY 200
+ A A+L G + IA + +R + L D +L V L+ P S +
Sbjct: 847 GSCDAPLAELWGAYYGLLIAWDKGFRRVELNLDSELVVGFLSTGISKAHPLSFLVRLCQG 906
Query: 201 FEHVRSMNFLITHIYIEGNSCADCMANVGLTL 232
F R ++H+Y E N AD +AN L
Sbjct: 907 F-FTRDWLVRVSHVYREANRLADGLANYAFFL 937
>UniRef100_Q9LP00 F9C16.18 [Arabidopsis thaliana]
Length = 196
Score = 56.2 bits (134), Expect = 8e-07
Identities = 45/166 (27%), Positives = 67/166 (40%), Gaps = 32/166 (19%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W P W+K+NTDGA NPT ++ GGV RD ++G F N+ +A A+L GV
Sbjct: 56 VGWSVPSEGWLKLNTDGASRGNPTLSTAGGVLRDREGEWLGGFSLNIGRRTAPVAELWGV 115
Query: 156 ITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIY 215
L V + W +R I+H+Y
Sbjct: 116 SE---------------RHPLSFLVRLCHGFLLKDWIVR----------------ISHVY 144
Query: 216 IEGNSCADCMANVGLTLSSFVWL-PSLPDCIKHDYGRNRLGMHSFR 260
E N AD +AN L + + S+PD ++ + + +G R
Sbjct: 145 REANRLADGLANYAFLLPIGIHVFVSVPDVVEPLFSDDVIGSARLR 190
>UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana]
Length = 233
Score = 54.7 bits (130), Expect = 2e-06
Identities = 49/166 (29%), Positives = 72/166 (42%), Gaps = 7/166 (4%)
Query: 89 RAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAF 148
R+ + ++V W P W K+NTDGA NP A+ GG R+ + F N+ A
Sbjct: 56 RSRMERQVRWSKPSLGWCKLNTDGASHGNPGLATAGGALRNEYGEWCFGFALNIGRCLAP 115
Query: 149 NAKLLGVITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNYFEHVRS 206
A+L GV + +A + + LE D ++ V L + P S + + F R
Sbjct: 116 LAELWGVYYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLS-RD 174
Query: 207 MNFLITHIYIEGNSCADCMANVGLTL----SSFVWLPSLPDCIKHD 248
I H+Y E N AD +AN L +F P+ D I D
Sbjct: 175 WIVRIGHVYREANRLADGLANYAFDLPLGYHAFASPPNSLDSILRD 220
>UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 [Arabidopsis
thaliana]
Length = 559
Score = 54.7 bits (130), Expect = 2e-06
Identities = 44/151 (29%), Positives = 66/151 (43%), Gaps = 37/151 (24%)
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLL 153
+ + W P W+KV+TDGA NP A+ GGV RD + +++G F L AF
Sbjct: 24 RHIAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQL----AF----- 74
Query: 154 GVITAINIAGENNWRNIWLETDLQLAV------------LAFKNLNMVPWSLRNKWLNYF 201
+ + ++ + LE D +L V LAF + + L WL
Sbjct: 75 -----VRLRWQSCGGRVELEVDSELVVGFLQSGISEAHPLAFL-VRLCHGLLSKDWL--- 125
Query: 202 EHVRSMNFLITHIYIEGNSCADCMANVGLTL 232
VR ++H+Y E N AD +AN +L
Sbjct: 126 --VR-----VSHVYREANRLADGLANYAFSL 149
>UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum]
Length = 1155
Score = 52.0 bits (123), Expect = 2e-05
Identities = 42/168 (25%), Positives = 71/168 (42%), Gaps = 5/168 (2%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V WI P + K+NTDG+ + GG+ R+ I AF L ++ A+ + +
Sbjct: 990 VNWIKPPTMFAKLNTDGSCVNG--RCGGGGILRNALGQVIMAFTIKLGEGTSSWAEAMSM 1047
Query: 156 ITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIY 215
+ + + + I ETD L A +PW + + V F+I H
Sbjct: 1048 LHGMQLCIQRGVNMIIGETDSILLAKAITENWSIPWRMYIPVKKIQKMVEEHGFIINHCL 1107
Query: 216 IEGNSCADCMANVGLTLS---SFVWLPSLPDCIKHDYGRNRLGMHSFR 260
E N AD +A++ L+ F +LP +K +R+ + +FR
Sbjct: 1108 REANQPADKLASISLSTDVNHVFKSYANLPSLVKGLVNLDRMNLPTFR 1155
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 465,505,870
Number of Sequences: 2790947
Number of extensions: 19094944
Number of successful extensions: 52527
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 52444
Number of HSP's gapped (non-prelim): 98
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC136507.9


