

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.8 + phase: 0
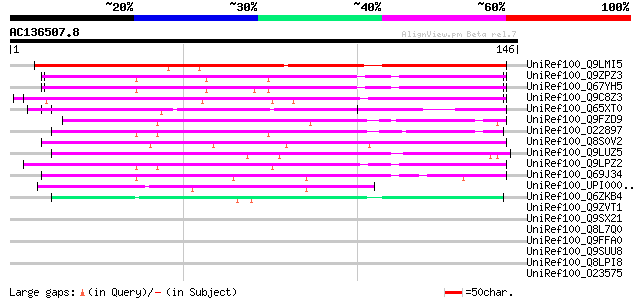
(146 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LMI5 T2D23.4 protein [Arabidopsis thaliana] 113 8e-25
UniRef100_Q9ZPZ3 T31J12.4 [Arabidopsis thaliana] 82 3e-15
UniRef100_Q67YH5 Hypothetical protein At1g09320 [Arabidopsis tha... 82 3e-15
UniRef100_Q9C8Z3 Hypothetical protein F5E6.15 [Arabidopsis thali... 80 9e-15
UniRef100_Q65XT0 Hypothetical protein P0033D06.18 [Oryza sativa] 72 3e-12
UniRef100_Q9FZD9 T1K7.9 protein [Arabidopsis thaliana] 67 8e-11
UniRef100_O22897 Hypothetical protein At2g47230 [Arabidopsis tha... 67 1e-10
UniRef100_Q8S0V2 OJ1014_G12.23 protein [Oryza sativa] 64 7e-10
UniRef100_Q9LUZ5 Gb|AAC80581.1 [Arabidopsis thaliana] 62 3e-09
UniRef100_Q9LPZ2 T23J18.9 [Arabidopsis thaliana] 60 1e-08
UniRef100_Q69J34 Putative agenet domain-containing protein / bro... 54 5e-07
UniRef100_UPI00002902E2 UPI00002902E2 UniRef100 entry 51 5e-06
UniRef100_Q6ZKB4 Hypothetical protein OJ1124_B05.16 [Oryza sativa] 47 7e-05
UniRef100_Q9ZVT1 F15K9.10 protein [Arabidopsis thaliana] 42 0.004
UniRef100_Q9SX21 F24J5.18 protein [Arabidopsis thaliana] 41 0.005
UniRef100_Q8L7Q0 Hypothetical protein At1g68580 [Arabidopsis tha... 41 0.005
UniRef100_Q9FFA0 Gb|AAF16647.1 [Arabidopsis thaliana] 37 0.091
UniRef100_Q9SUU8 Hypothetical protein F8B4.140 [Arabidopsis thal... 36 0.20
UniRef100_Q8LPI8 G2484-1 protein [Arabidopsis thaliana] 35 0.35
UniRef100_O23575 G2484-1 protein [Arabidopsis thaliana] 35 0.35
>UniRef100_Q9LMI5 T2D23.4 protein [Arabidopsis thaliana]
Length = 134
Score = 113 bits (283), Expect = 8e-25
Identities = 59/138 (42%), Positives = 86/138 (61%), Gaps = 8/138 (5%)
Query: 8 VDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYV-IRYKNLLKD-DESELLMETLFP 65
+++ GD+VEVCS+++GF+GSYF AT+VS G Y I+YKNL+ D D+S+ L+E +
Sbjct: 1 MEFVKGDQVEVCSKEDGFLGSYFGATVVSKTPEGSYYKIKYKNLVSDTDQSKRLVEVISA 60
Query: 66 KDLRPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIY 125
+LRP+PP + L + KVD FD DGWWVG++ + + YSVYF + +
Sbjct: 61 DELRPMPPKSLHVL-IRCGDKVDAFDKDGWWVGEVTA-----VRRNIYSVYFSTTDEELE 114
Query: 126 YPCDQIRVHQELVWGDWI 143
YP +R H E V G W+
Sbjct: 115 YPLYSLRKHHEWVNGSWV 132
>UniRef100_Q9ZPZ3 T31J12.4 [Arabidopsis thaliana]
Length = 514
Score = 81.6 bits (200), Expect = 3e-15
Identities = 46/134 (34%), Positives = 70/134 (51%), Gaps = 4/134 (2%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ +G +EV E+EGF S+F A ++ K ++ Y NL +D E L E + +R
Sbjct: 378 FSIGTPIEVSPEEEGFEDSWFLAKLIEYRGKDKCLVEYDNLKAEDGKEPLREEVNVSRIR 437
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P+P F+ + KV+ NDGWWVG I K+L + S Y V F + + +
Sbjct: 438 PLPLESVMVSPFERHDKVNALYNDGWWVGVI--RKVLAKSS--YLVLFKNTQELLKFHHS 493
Query: 130 QIRVHQELVWGDWI 143
Q+R+HQE + G WI
Sbjct: 494 QLRLHQEWIDGKWI 507
Score = 74.3 bits (181), Expect = 5e-13
Identities = 41/133 (30%), Positives = 68/133 (50%), Gaps = 5/133 (3%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ G VEV S++EGF G +F A +V + K+++ Y++L + D E L E +R
Sbjct: 219 FSSGTVVEVSSDEEGFQGCWFAAKVVEPVGEDKFLVEYRDLREKDGIEPLKEETDFLHIR 278
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P PP + + F + K++ F NDGWWVG + K +YF + + +
Sbjct: 279 PPPPRDED-IDFAVGDKINAFYNDGWWVGVVIDGM----KHGTVGIYFRQSQEKMRFGRQ 333
Query: 130 QIRVHQELVWGDW 142
+R+H++ V G W
Sbjct: 334 GLRLHKDWVDGTW 346
Score = 64.3 bits (155), Expect = 5e-10
Identities = 44/141 (31%), Positives = 71/141 (50%), Gaps = 13/141 (9%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIV-----SCLESGKYVIRYKNLLKDDE-SELLMETLF 64
K G VE+ S++ GF GS++ ++ S +S K + Y L D E ++ L E +
Sbjct: 15 KPGSAVEISSDEIGFRGSWYMGKVITIPSSSDKDSVKCQVEYTTLFFDKEGTKPLKEVVD 74
Query: 65 PKDLRPIPP---HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCH 121
LRP P + K + ++VD F NDGWW G + ++L + +SV+F
Sbjct: 75 MSQLRPPAPPMSEIEKKKKIVVGEEVDAFYNDGWWEGDVT--EVLDDGK--FSVFFRSSK 130
Query: 122 QTIYYPCDQIRVHQELVWGDW 142
+ I + D++R H+E V G W
Sbjct: 131 EQIRFRKDELRFHREWVDGAW 151
>UniRef100_Q67YH5 Hypothetical protein At1g09320 [Arabidopsis thaliana]
Length = 517
Score = 81.6 bits (200), Expect = 3e-15
Identities = 46/134 (34%), Positives = 70/134 (51%), Gaps = 4/134 (2%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ +G +EV E+EGF S+F A ++ K ++ Y NL +D E L E + +R
Sbjct: 380 FSIGTPIEVSPEEEGFEDSWFLAKLIEYRGKDKCLVEYDNLKAEDGKEPLREEVNVSRIR 439
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P+P F+ + KV+ NDGWWVG I K+L + S Y V F + + +
Sbjct: 440 PLPLESVMVSPFERHDKVNALYNDGWWVGVI--RKVLAKSS--YLVLFKNTQELLKFHHS 495
Query: 130 QIRVHQELVWGDWI 143
Q+R+HQE + G WI
Sbjct: 496 QLRLHQEWIDGKWI 509
Score = 74.3 bits (181), Expect = 5e-13
Identities = 41/133 (30%), Positives = 68/133 (50%), Gaps = 5/133 (3%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ G VEV S++EGF G +F A +V + K+++ Y++L + D E L E +R
Sbjct: 221 FSSGTVVEVSSDEEGFQGCWFAAKVVEPVGEDKFLVEYRDLREKDGIEPLKEETDFLHIR 280
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P PP + + F + K++ F NDGWWVG + K +YF + + +
Sbjct: 281 PPPPRDED-IDFAVGDKINAFYNDGWWVGVVIDGM----KHGTVGIYFRQSQEKMRFGRQ 335
Query: 130 QIRVHQELVWGDW 142
+R+H++ V G W
Sbjct: 336 GLRLHKDWVDGTW 348
Score = 63.9 bits (154), Expect = 7e-10
Identities = 45/141 (31%), Positives = 72/141 (50%), Gaps = 13/141 (9%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIV-----SCLESGKYVIRYKNLLKDDE-SELLMETLF 64
K G VE+ S++ GF GS++ ++ S +S K + Y L D E ++ L E +
Sbjct: 40 KPGSAVEISSDEIGFRGSWYMGKVITIPSSSDKDSVKCQVEYTTLFFDKEGTKPLKEVVD 99
Query: 65 PKDLR-PIPP--HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCH 121
LR P PP + K + ++VD F NDGWW G + ++L + +SV+F
Sbjct: 100 MSQLRSPAPPMSEIEKKKKIVVGEEVDAFYNDGWWEGDVT--EVLDDGK--FSVFFRSSK 155
Query: 122 QTIYYPCDQIRVHQELVWGDW 142
+ I + D++R H+E V G W
Sbjct: 156 EQIRFRKDELRFHREWVDGAW 176
>UniRef100_Q9C8Z3 Hypothetical protein F5E6.15 [Arabidopsis thaliana]
Length = 466
Score = 80.1 bits (196), Expect = 9e-15
Identities = 47/150 (31%), Positives = 76/150 (50%), Gaps = 12/150 (8%)
Query: 2 RPPEKRVD--------YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKD 53
+PP K++ + G +VEV S++ G+ S+F A IVS L +Y + Y+ L D
Sbjct: 317 KPPLKKLKSCERAEKVFNNGMEVEVRSDEPGYEASWFSAKIVSYLGENRYTVEYQTLKTD 376
Query: 54 DESELLMETLFPKDLRPIPPH-VRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCY 112
DE ELL E D+RP PP + +++L + VD + N+GWW G++ +
Sbjct: 377 DERELLKEEARGSDIRPPPPPLIPKGYRYELYELVDAWYNEGWWSGRVYK---INNNKTR 433
Query: 113 YSVYFDYCHQTIYYPCDQIRVHQELVWGDW 142
Y VYF +++ + + +R Q G W
Sbjct: 434 YGVYFQTTDESLEFAYNDLRPCQVWRNGKW 463
Score = 68.6 bits (166), Expect = 3e-11
Identities = 42/141 (29%), Positives = 75/141 (52%), Gaps = 4/141 (2%)
Query: 5 EKRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDD-ESELLMETL 63
++R Y+ G VEV SE++ + GS++ A I+ L KY++ + +DD ES L + +
Sbjct: 164 KRRDQYEKGALVEVRSEEKAYKGSWYCARILCLLGDDKYIVEHLKFSRDDGESIPLRDVV 223
Query: 64 FPKDLRPIPPHVRNPLK-FKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQ 122
KD+RP+PP +P+ ++ VD + N WW +++ K+L S YSV+ +
Sbjct: 224 EAKDIRPVPPSELSPVVCYEPGVIVDAWFNKRWWTSRVS--KVLGGGSNKYSVFIISTGE 281
Query: 123 TIYYPCDQIRVHQELVWGDWI 143
+R H++ + G W+
Sbjct: 282 ETTILNFNLRPHKDWINGQWV 302
>UniRef100_Q65XT0 Hypothetical protein P0033D06.18 [Oryza sativa]
Length = 448
Score = 71.6 bits (174), Expect = 3e-12
Identities = 44/131 (33%), Positives = 68/131 (51%), Gaps = 11/131 (8%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLRPIP 72
G KVEV S D+GF G++F+ T + + + K ++ Y L DDE L E + + +RP P
Sbjct: 321 GSKVEVTSNDDGFHGAWFQGTALKYVNN-KILVEYDALKADDEITPLTEAIEVQHVRPCP 379
Query: 73 PHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCDQIR 132
P + F L +VD NDGWWVG + S+ ++S F + +Q+R
Sbjct: 380 PDIPVTSGFNLLDEVDACWNDGWWVG-VISKVNSGDRSSTEETEFGH---------EQLR 429
Query: 133 VHQELVWGDWI 143
+H + V G W+
Sbjct: 430 LHCDWVGGRWM 440
Score = 53.5 bits (127), Expect = 9e-07
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query: 6 KRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFP 65
K + YK G +VEV + V ++F A + + ++ Y N K+D S L E +
Sbjct: 162 KTMPYKKGTQVEVAKLEGNSVVAWFSAAVEKAIWKNSLLVDY-NCSKNDGSVLPKEIVDL 220
Query: 66 KDLRPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKI 100
K +RP P H + + F +N +V+ F +GWW+G I
Sbjct: 221 KHIRPHPQHA-SAIIFCINDEVEGFQGNGWWLGVI 254
Score = 50.4 bits (119), Expect = 8e-06
Identities = 35/135 (25%), Positives = 59/135 (42%), Gaps = 7/135 (5%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGK-YVIRYKNLLKDDESELLMETLFPKDL 68
++ GD VEV ++ G+ G++F A++ + + + Y + Y D L E + L
Sbjct: 25 FRPGDLVEVLPDEPGYRGAHFPASVTASHANPRGYTVAY-----DGSGCPLGEVVAASQL 79
Query: 69 RPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPC 128
RP PP + + VD F WWVG +A +V F + + +
Sbjct: 80 RPRPPDAPRGVPPAEHAVVDAFKGGAWWVG-VALGGGRAAAGGRVAVCFPETREVVEFDA 138
Query: 129 DQIRVHQELVWGDWI 143
+R H E V G+W+
Sbjct: 139 ADVRPHLEWVAGEWL 153
>UniRef100_Q9FZD9 T1K7.9 protein [Arabidopsis thaliana]
Length = 695
Score = 67.0 bits (162), Expect = 8e-11
Identities = 48/135 (35%), Positives = 65/135 (47%), Gaps = 13/135 (9%)
Query: 16 VEVCSEDEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFPKDLRPIP 72
VEV SE+EGF G++F A + + K +RY LL D S L+E + + +RP+P
Sbjct: 14 VEVSSEEEGFEGAWFRAVLEENPGNSSRRKLRVRYSTLLDMDGSSPLIEHIEQRFIRPVP 73
Query: 73 PHVRNPLKFKLNQ--KVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCDQ 130
P L + VD DGWW G + + ME Y VYFD I + Q
Sbjct: 74 PEENQQKDVVLEEGLLVDADHKDGWWTGVVVKK---MEDD-NYLVYFDLPPDIIQFERKQ 129
Query: 131 IRVHQELVW--GDWI 143
+R H L+W G WI
Sbjct: 130 LRTH--LIWTGGTWI 142
>UniRef100_O22897 Hypothetical protein At2g47230 [Arabidopsis thaliana]
Length = 701
Score = 66.6 bits (161), Expect = 1e-10
Identities = 45/134 (33%), Positives = 69/134 (50%), Gaps = 10/134 (7%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLMETLFPKDLR 69
G +VEV S +EGF ++F + + +SG K +RY LL DD L+E + P+ +R
Sbjct: 8 GSEVEVSSTEEGFADAWFRGILQENPTKSGRKKLRVRYLTLLNDDALSPLIENIEPRFIR 67
Query: 70 PIPP-HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPC 128
P+PP + N + + VD DGWW G I + +E ++ VY+D I +
Sbjct: 68 PVPPENEYNGIVLEEGTVVDADHKDGWWTGVIIKK---LENGKFW-VYYDSPPDIIEFER 123
Query: 129 DQIRVHQELVWGDW 142
+Q+R H L W W
Sbjct: 124 NQLRPH--LRWSGW 135
>UniRef100_Q8S0V2 OJ1014_G12.23 protein [Oryza sativa]
Length = 380
Score = 63.9 bits (154), Expect = 7e-10
Identities = 43/154 (27%), Positives = 71/154 (45%), Gaps = 20/154 (12%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLE-SGKYVIRYKNLLKDDESE------LLMET 62
Y VGD+VEV +D GF G+++EAT+ + L SG+Y + Y L++ L ET
Sbjct: 90 YAVGDEVEVRMDDPGFHGAFYEATVSARLPCSGRYEVMYSTLVEGGGGRGRRGGGPLRET 149
Query: 63 LFPKDLRPIPPHVRNP----------LKFKLNQKVDVFDNDGWWVGKIAS---EKILMEK 109
+ D+RP PP P + + V+ + +GWW G +++ +
Sbjct: 150 VAACDVRPRPPPPPPPPLAEDGAAPGRELNVFDMVEAYHREGWWPGVVSAAWPARGRKAA 209
Query: 110 SCYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
+ Y+V F C + P +R + V G W+
Sbjct: 210 AAMYTVSFPSCREEAKLPASLVRRRRAFVRGRWM 243
>UniRef100_Q9LUZ5 Gb|AAC80581.1 [Arabidopsis thaliana]
Length = 1095
Score = 62.0 bits (149), Expect = 3e-09
Identities = 48/159 (30%), Positives = 70/159 (43%), Gaps = 30/159 (18%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKD----- 67
GDK+EV S +EG +GS++ T+ S + + IRY N+L DD S L+ET+ D
Sbjct: 29 GDKIEVRSLEEGSLGSWYLGTVTSAKKRRRRCIRYDNILSDDGSGNLVETVDVSDIVEGL 88
Query: 68 -------------LRPIPPHVR-NPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYY 113
LRP+PP + L VDVF +D WW G + + EK
Sbjct: 89 DDCTDVSDTFRGRLRPVPPKLDVAKLNLAYGLCVDVFFSDAWWEGVLFDHENGSEKR--- 145
Query: 114 SVYFDYCHQTIYYPCDQIRVHQEL-----VW---GDWIF 144
V+F + +R+ Q+ W G W+F
Sbjct: 146 RVFFPDLGDELDADLQSLRITQDWNEATETWECRGSWLF 184
>UniRef100_Q9LPZ2 T23J18.9 [Arabidopsis thaliana]
Length = 636
Score = 59.7 bits (143), Expect = 1e-08
Identities = 44/143 (30%), Positives = 70/143 (48%), Gaps = 8/143 (5%)
Query: 5 EKRVDYKVGDKVEVCSEDEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLME 61
E+ + + DKVEV SE+E GSY+ A + + +SG K +RY L + L E
Sbjct: 2 EQMIFIRKDDKVEVFSEEEELKGSYYRAILEDNPTKSGHNKLKVRYLTQLNEHRLAPLTE 61
Query: 62 TLFPKDLRPIPPH-VRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYC 120
+ + +RP+P V + + F VD + DGWW G + K + ++ + VYFD
Sbjct: 62 FVDQRFIRPVPSEDVNDGVVFVEGLMVDAYLKDGWWTGVVV--KTMEDEK--FLVYFDCP 117
Query: 121 HQTIYYPCDQIRVHQELVWGDWI 143
I + ++RVH + WI
Sbjct: 118 PDIIQFEKKKLRVHLDWTGFKWI 140
>UniRef100_Q69J34 Putative agenet domain-containing protein / bromo-adjacent homology
(BAH) domain-containing protein [Oryza sativa]
Length = 636
Score = 54.3 bits (129), Expect = 5e-07
Identities = 43/155 (27%), Positives = 67/155 (42%), Gaps = 25/155 (16%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIV-SCLESGKYVIRYKNLLKDDESELLMETL----- 63
Y VGD++EV S+D G VG +F T++ SC K ++Y +L D+S L E +
Sbjct: 380 YNVGDRIEVLSQDSGIVGCWFRCTVLKSCTNHNKLKVQYDDLQNADDSGRLEEWVPVSTL 439
Query: 64 ---------FPKDLRPIPPHVRNPLKFKLN----QKVDVFDNDGWWVGKIASEKILMEKS 110
P+ R P +N L N VDV+ GWW G + S + S
Sbjct: 440 ARPDKLGLRCPERRRVRPRPQQNSLADGTNLLPGAAVDVWQFSGWWEGVLVSADNISADS 499
Query: 111 CYYSVYFDYCHQTIYYPCD--QIRVHQELVWGDWI 143
+YF + + C +R+ ++ V W+
Sbjct: 500 --LQIYFP--GENFFSVCQLKNLRISKDWVKSHWV 530
>UniRef100_UPI00002902E2 UPI00002902E2 UniRef100 entry
Length = 207
Score = 51.2 bits (121), Expect = 5e-06
Identities = 34/100 (34%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query: 9 DYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLL-KDDESELLMETLFPKD 67
++K G +VEV D+GF GS++ A ++S E G I+Y L K +E + ETL
Sbjct: 61 EFKFGTEVEVKGRDKGFEGSWYLAEVISSKE-GVSTIQYDALFEKGEEKTEVKETLKSDH 119
Query: 68 LRPIPPHVRNPLKFKLN--QKVDVFDNDGWWVGKIASEKI 105
LRP PP KL+ +++ DGWW S ++
Sbjct: 120 LRPKPPEPPEGWDEKLSPGTPLELKHEDGWWQVTYVSSRV 159
>UniRef100_Q6ZKB4 Hypothetical protein OJ1124_B05.16 [Oryza sativa]
Length = 684
Score = 47.4 bits (111), Expect = 7e-05
Identities = 39/146 (26%), Positives = 57/146 (38%), Gaps = 21/146 (14%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLF------PK 66
G + EV S+D G G +F ++ K +RY++L DE+ L E + P
Sbjct: 397 GSRAEVLSQDSGIRGCWFRCFVLK-RRGDKIKVRYEDLQDADETGNLEEWVLLTRIAKPD 455
Query: 67 DL----------RPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVY 116
L RP H ++P F VD + N GWW G + + VY
Sbjct: 456 QLGIRIPERPMVRPYHVHSKDPCSFDAGSIVDAWWNSGWWEGIVLQQ----GNDRRLQVY 511
Query: 117 FDYCHQTIYYPCDQIRVHQELVWGDW 142
F Q + D +R +E G W
Sbjct: 512 FPGEKQIADFCEDDLRHSREWAGGKW 537
>UniRef100_Q9ZVT1 F15K9.10 protein [Arabidopsis thaliana]
Length = 670
Score = 41.6 bits (96), Expect = 0.004
Identities = 33/135 (24%), Positives = 58/135 (42%), Gaps = 10/135 (7%)
Query: 15 KVEVCSEDEGFVGSYFEATIVS-----CLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+VE+ SE++GF +++ A + ES K Y + E T+ + +R
Sbjct: 8 EVEIFSEEDGFRNAWYRAILEETPTNPTSESKKLRFSYMTKSLNKEGSSSPPTVEQRFIR 67
Query: 70 PIPP-HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPC 128
P+PP ++ N + F+ VD W G + ++ ME Y V FD I +
Sbjct: 68 PVPPENLYNGVVFEEGTMVDADYKHRWRTGVVINK---MENDSYL-VLFDCPPDIIQFET 123
Query: 129 DQIRVHQELVWGDWI 143
+R H + +W+
Sbjct: 124 KHLRAHLDWTGSEWV 138
>UniRef100_Q9SX21 F24J5.18 protein [Arabidopsis thaliana]
Length = 625
Score = 41.2 bits (95), Expect = 0.005
Identities = 39/153 (25%), Positives = 68/153 (43%), Gaps = 25/153 (16%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLL-KDDESELLMETLFP---- 65
K G +EV SED G G +F+A ++ K ++Y+++ DDES+ L E +
Sbjct: 341 KKGSLIEVLSEDSGIRGCWFKALVLK-KHKDKVKVQYQDIQDADDESKKLEEWILTSRVA 399
Query: 66 -------------KDLRPI--PPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKS 110
K +RP+ P + + VDV+ DGWW G I +++ EK
Sbjct: 400 AGDHLGDLRIKGRKVVRPMLKPSKENDVCVIGVGMPVDVWWCDGWWEG-IVVQEVSEEK- 457
Query: 111 CYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
+ VY + + + +R +E + +W+
Sbjct: 458 --FEVYLPGEKKMSAFHRNDLRQSREWLDDEWL 488
>UniRef100_Q8L7Q0 Hypothetical protein At1g68580 [Arabidopsis thaliana]
Length = 648
Score = 41.2 bits (95), Expect = 0.005
Identities = 39/153 (25%), Positives = 68/153 (43%), Gaps = 25/153 (16%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLL-KDDESELLMETLFP---- 65
K G +EV SED G G +F+A ++ K ++Y+++ DDES+ L E +
Sbjct: 364 KKGSLIEVLSEDSGIRGCWFKALVLK-KHKDKVKVQYQDIQDADDESKKLEEWILTSRVA 422
Query: 66 -------------KDLRPI--PPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKS 110
K +RP+ P + + VDV+ DGWW G I +++ EK
Sbjct: 423 AGDHLGDLRIKGRKVVRPMLKPSKENDVCVIGVGMPVDVWWCDGWWEG-IVVQEVSEEK- 480
Query: 111 CYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
+ VY + + + +R +E + +W+
Sbjct: 481 --FEVYLPGEKKMSAFHRNDLRQSREWLDDEWL 511
>UniRef100_Q9FFA0 Gb|AAF16647.1 [Arabidopsis thaliana]
Length = 552
Score = 37.0 bits (84), Expect = 0.091
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 1/96 (1%)
Query: 4 PEKRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETL 63
P + + G +VE+ ++ G +++A + + S LLKDD S L E
Sbjct: 6 PSESLSLSEGCEVEISYKNNGNESVWYKAILEAKPNSIFKEELSVRLLKDDFSTPLNELR 65
Query: 64 FPKDLRPIPP-HVRNPLKFKLNQKVDVFDNDGWWVG 98
+RPIPP +V+ + ++ VD D WW G
Sbjct: 66 HKVLIRPIPPTNVQACIDIEIGTFVDADYKDAWWAG 101
Score = 36.6 bits (83), Expect = 0.12
Identities = 27/100 (27%), Positives = 53/100 (53%), Gaps = 7/100 (7%)
Query: 10 YKVGDKVEVCSE-DEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFP 65
+ G VE+CS+ DEG V + A + + +Y+++ K L+ +T+
Sbjct: 165 FSPGTIVELCSKRDEGEV-VWVPALVYKEFKENDEYRYIVKDKPLIGRSYKSRPSKTVDL 223
Query: 66 KDLRPIPPHVRNPLKFKLNQKVDVF-DNDGWWVGKIASEK 104
+ LRPIPP +R +++L++ ++V+ D GW G++ +
Sbjct: 224 RSLRPIPPPIR-VKEYRLDEYIEVYHDGIGWRQGRVVKSE 262
>UniRef100_Q9SUU8 Hypothetical protein F8B4.140 [Arabidopsis thaliana]
Length = 377
Score = 35.8 bits (81), Expect = 0.20
Identities = 29/91 (31%), Positives = 45/91 (48%), Gaps = 3/91 (3%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLRPIP 72
G +VEV S E G++ A IVS Y +R+ + + E E +ME + K +RP P
Sbjct: 6 GSRVEVFSNKEAPYGAWRCAEIVSG-NGHTYNVRFYSFQIEHE-EAVMEKVPRKIIRPCP 63
Query: 73 PHVRNPLKFKLNQKVDVFDNDGWWVGKIASE 103
P V + ++ + V+V DN W + E
Sbjct: 64 PLV-DVERWDTGELVEVLDNFSWKAATVREE 93
>UniRef100_Q8LPI8 G2484-1 protein [Arabidopsis thaliana]
Length = 1058
Score = 35.0 bits (79), Expect = 0.35
Identities = 39/162 (24%), Positives = 66/162 (40%), Gaps = 30/162 (18%)
Query: 6 KRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFP 65
K D K G VEV E+ G +++ A ++S LE K + + +L + ++ L E +
Sbjct: 590 KNEDIKEGSNVEVFKEEPGLRTAWYSANVLS-LEDDKAYVLFSDLSVEQGTDKLKEWVAL 648
Query: 66 KDLRPIPPHVRNPLK----------------------FKLNQKVDVFDNDGWWVGKIASE 103
K P +R P + +K+ +VD + +D W G I +
Sbjct: 649 KGEGDQAPKIR-PARSVTALPYEGTRKRRRAALGDHIWKIGDRVDSWVHDSWLEGVITEK 707
Query: 104 KILMEKSCYYSVYFDYCHQTIYYPCDQIRVHQELVW--GDWI 143
E + +V+F +T+ +R LVW G WI
Sbjct: 708 NKKDENT--VTVHFPAEEETLTIKAWNLR--PSLVWKDGKWI 745
>UniRef100_O23575 G2484-1 protein [Arabidopsis thaliana]
Length = 954
Score = 35.0 bits (79), Expect = 0.35
Identities = 39/162 (24%), Positives = 66/162 (40%), Gaps = 30/162 (18%)
Query: 6 KRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFP 65
K D K G VEV E+ G +++ A ++S LE K + + +L + ++ L E +
Sbjct: 486 KNEDIKEGSNVEVFKEEPGLRTAWYSANVLS-LEDDKAYVLFSDLSVEQGTDKLKEWVAL 544
Query: 66 KDLRPIPPHVRNPLK----------------------FKLNQKVDVFDNDGWWVGKIASE 103
K P +R P + +K+ +VD + +D W G I +
Sbjct: 545 KGEGDQAPKIR-PARSVTALPYEGTRKRRRAALGDHIWKIGDRVDSWVHDSWLEGVITEK 603
Query: 104 KILMEKSCYYSVYFDYCHQTIYYPCDQIRVHQELVW--GDWI 143
E + +V+F +T+ +R LVW G WI
Sbjct: 604 NKKDENT--VTVHFPAEEETLTIKAWNLR--PSLVWKDGKWI 641
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.142 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 269,317,009
Number of Sequences: 2790947
Number of extensions: 10594134
Number of successful extensions: 24970
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 24896
Number of HSP's gapped (non-prelim): 72
length of query: 146
length of database: 848,049,833
effective HSP length: 122
effective length of query: 24
effective length of database: 507,554,299
effective search space: 12181303176
effective search space used: 12181303176
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136507.8


