

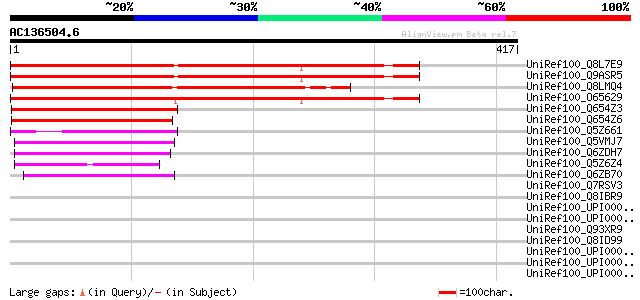
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.6 - phase: 0 /pseudo
(417 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L7E9 Hypothetical protein At4g35920 [Arabidopsis tha... 365 e-99
UniRef100_Q9ASR5 AT4g35920/F4B14_190 [Arabidopsis thaliana] 365 e-99
UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sat... 362 1e-98
UniRef100_O65629 Hypothetical protein T19K4.50 [Arabidopsis thal... 354 2e-96
UniRef100_Q654Z3 Receptor protein kinase-like [Oryza sativa] 139 1e-31
UniRef100_Q654Z6 Receptor protein kinase-like [Oryza sativa] 129 1e-28
UniRef100_Q5Z661 Receptor protein kinase-like [Oryza sativa] 103 9e-21
UniRef100_Q5VMJ7 Cytokinin-regulated kinase-like [Oryza sativa] 103 1e-20
UniRef100_Q6ZDH7 Receptor kinase-like [Oryza sativa] 93 2e-17
UniRef100_Q5Z6Z4 Hypothetical protein B1018E06.42 [Oryza sativa] 86 1e-15
UniRef100_Q6ZB70 Receptor-like kinase CHRK1-like [Oryza sativa] 80 1e-13
UniRef100_Q7RSV3 Hypothetical protein [Plasmodium yoelii yoelii] 42 0.032
UniRef100_Q8IBR9 Exported serine/threonine protein kinase [Plasm... 41 0.072
UniRef100_UPI0000330E48 UPI0000330E48 UniRef100 entry 39 0.27
UniRef100_UPI00004659B1 UPI00004659B1 UniRef100 entry 38 0.47
UniRef100_Q93XR9 Bg55 protein [Bruguiera gymnorrhiza] 38 0.61
UniRef100_Q8ID99 Hypothetical protein PF13_0321 [Plasmodium falc... 38 0.61
UniRef100_UPI00002EA191 UPI00002EA191 UniRef100 entry 37 0.79
UniRef100_UPI00004377F3 UPI00004377F3 UniRef100 entry 37 1.0
UniRef100_UPI0000435ED3 UPI0000435ED3 UniRef100 entry 37 1.0
>UniRef100_Q8L7E9 Hypothetical protein At4g35920 [Arabidopsis thaliana]
Length = 421
Score = 365 bits (938), Expect = e-99
Identities = 204/347 (58%), Positives = 238/347 (67%), Gaps = 20/347 (5%)
Query: 1 MASSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQL 60
M+ SW+ GEIA+VAQLTGLDAVKLIG+IVKAANTA MHKKNCRQFAQHLKLIGNLL+QL
Sbjct: 1 MSHSWDGLGEIASVAQLTGLDAVKLIGLIVKAANTAWMHKKNCRQFAQHLKLIGNLLEQL 60
Query: 61 KISELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDR 120
KISE+KKYPETREPLE LEDALRRSY+LVNSC+DRSYLYLLAMGWNIVYQFRK Q+EIDR
Sbjct: 61 KISEMKKYPETREPLEGLEDALRRSYLLVNSCRDRSYLYLLAMGWNIVYQFRKHQDEIDR 120
Query: 121 YLRLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLK 180
+L+++PLITLVDNAR +RER E I++DQ YTLD+ED+ VQ V +K E +E VLK
Sbjct: 121 FLKIIPLITLVDNAR---IRERFEYIDRDQREYTLDEEDRHVQDVILKQESTREAASVLK 177
Query: 181 KTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNFAAYSLPE-- 238
KTLSCSYPN F EALK ENEKLQ+EL SQ + D+ QCE IQRL+ VT AA P+
Sbjct: 178 KTLSCSYPNLRFCEALKTENEKLQIELQRSQEHYDVAQCEVIQRLIGVTQAAAAVEPDSE 237
Query: 239 --------KLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKKPKFQKRIRCQLEVHI 290
K S + EYS + S S +S + R H
Sbjct: 238 KELTKKASKKSERSSSMKTEYSYDEDSPKKSSTRAASRSTSNVSSGHDLLSRRASQAQHH 297
Query: 291 SRKIGILIYLLVVLNLIFIFSFAGIKTFFYPCGTFSKIATVATNRPI 337
L+ +L F KTFF+PCGT +KIAT A+NR I
Sbjct: 298 EEWHTDLLACCSEPSLCF-------KTFFFPCGTLAKIATAASNRHI 337
>UniRef100_Q9ASR5 AT4g35920/F4B14_190 [Arabidopsis thaliana]
Length = 440
Score = 365 bits (938), Expect = e-99
Identities = 204/347 (58%), Positives = 238/347 (67%), Gaps = 20/347 (5%)
Query: 1 MASSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQL 60
M+ SW+ GEIA+VAQLTGLDAVKLIG+IVKAANTA MHKKNCRQFAQHLKLIGNLL+QL
Sbjct: 1 MSHSWDGLGEIASVAQLTGLDAVKLIGLIVKAANTAWMHKKNCRQFAQHLKLIGNLLEQL 60
Query: 61 KISELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDR 120
KISE+KKYPETREPLE LEDALRRSY+LVNSC+DRSYLYLLAMGWNIVYQFRK Q+EIDR
Sbjct: 61 KISEMKKYPETREPLEGLEDALRRSYLLVNSCRDRSYLYLLAMGWNIVYQFRKHQDEIDR 120
Query: 121 YLRLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLK 180
+L+++PLITLVDNAR +RER E I++DQ YTLD+ED+ VQ V +K E +E VLK
Sbjct: 121 FLKIIPLITLVDNAR---IRERFEYIDRDQREYTLDEEDRHVQDVILKQESTREAASVLK 177
Query: 181 KTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNFAAYSLPE-- 238
KTLSCSYPN F EALK ENEKLQ+EL SQ + D+ QCE IQRL+ VT AA P+
Sbjct: 178 KTLSCSYPNLRFCEALKTENEKLQIELQRSQEHYDVAQCEVIQRLIGVTQAAAAVEPDSE 237
Query: 239 --------KLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKKPKFQKRIRCQLEVHI 290
K S + EYS + S S +S + R H
Sbjct: 238 KELTKKASKKSERSSSMKTEYSYDEDSPKKSSTRAASRSTSNVSSGHDLLSRRASQAQHH 297
Query: 291 SRKIGILIYLLVVLNLIFIFSFAGIKTFFYPCGTFSKIATVATNRPI 337
L+ +L F KTFF+PCGT +KIAT A+NR I
Sbjct: 298 EEWHTDLLACCSEPSLCF-------KTFFFPCGTLAKIATAASNRHI 337
>UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sativa]
Length = 1202
Score = 362 bits (929), Expect = 1e-98
Identities = 183/278 (65%), Positives = 221/278 (78%), Gaps = 10/278 (3%)
Query: 3 SSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKI 62
+SWE G++A V QLTGLDAV+LI MIVKAA+TAR+HK+NCR+FAQHLKLIG LL+QL++
Sbjct: 2 ASWENLGDVATVVQLTGLDAVRLISMIVKAASTARLHKRNCRRFAQHLKLIGGLLEQLRV 61
Query: 63 SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYL 122
SELKKYPETREPLEQLEDALRR+Y+LV+SCQDRSYLYLLAMGWNIVYQFRKAQNEID YL
Sbjct: 62 SELKKYPETREPLEQLEDALRRAYLLVHSCQDRSYLYLLAMGWNIVYQFRKAQNEIDNYL 121
Query: 123 RLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLKKT 182
RLVPLITLVDNA RVRER+E IE+DQC Y+ DDED++VQ + P+ TVVLKKT
Sbjct: 122 RLVPLITLVDNA---RVRERMEYIERDQCEYSFDDEDKEVQDALLNPDPSTNPTVVLKKT 178
Query: 183 LSCSYPNFSFTEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNFAAYSLPEKLSP 242
LSCSYPN F EAL+KE+EKLQ+EL SQ+NMD+ QCE IQ LL VT A S+PEK +
Sbjct: 179 LSCSYPNLPFNEALRKESEKLQVELQRSQSNMDMGQCEVIQHLLGVTKTVASSIPEKCAT 238
Query: 243 EDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKKPKFQK 280
++ E ++++H+ + S + +H PK QK
Sbjct: 239 P---KVSEKADSNHTKVS----EDSAKTYHDDSPKKQK 269
>UniRef100_O65629 Hypothetical protein T19K4.50 [Arabidopsis thaliana]
Length = 447
Score = 354 bits (909), Expect = 2e-96
Identities = 203/370 (54%), Positives = 238/370 (63%), Gaps = 40/370 (10%)
Query: 1 MASSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQL 60
M+ SW+ GEIA+VAQLTGLDAVKLIG+IVKAANTA MHKKNCRQFAQHLKLIGNLL+QL
Sbjct: 1 MSHSWDGLGEIASVAQLTGLDAVKLIGLIVKAANTAWMHKKNCRQFAQHLKLIGNLLEQL 60
Query: 61 KISELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDR 120
KISE+KKYPETREPLE LEDALRRSY+LVNSC+DRSYLYLLAMGWNIVYQFRK Q+EIDR
Sbjct: 61 KISEMKKYPETREPLEGLEDALRRSYLLVNSCRDRSYLYLLAMGWNIVYQFRKHQDEIDR 120
Query: 121 YLRLVPLITLVDNAR-----------------------ATRVRERLEVIEKDQCAYTLDD 157
+L+++PLITLVDNAR ++ER E I++DQ YTLD+
Sbjct: 121 FLKIIPLITLVDNARIRVILPAFWSNKFLCLMLWYIEILIVIQERFEYIDRDQREYTLDE 180
Query: 158 EDQKVQTVFMKPEHDKEDTVVLKKTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDIN 217
ED+ VQ V +K E +E VLKKTLSCSYPN F EALK ENEKLQ+EL SQ + D+
Sbjct: 181 EDRHVQDVILKQESTREAASVLKKTLSCSYPNLRFCEALKTENEKLQIELQRSQEHYDVA 240
Query: 218 QCEFIQRLLDVTNFAAYSLPE----------KLSPEDNYQIVEYSNNSHSDANGSKGHSS 267
QCE IQRL+ VT AA P+ K S + EYS + S S +S
Sbjct: 241 QCEVIQRLIGVTQAAAAVEPDSEKELTKKASKKSERSSSMKTEYSYDEDSPKKSSTRAAS 300
Query: 268 NEKHHKKKPKFQKRIRCQLEVHISRKIGILIYLLVVLNLIFIFSFAGIKTFFYPCGTFSK 327
+ R H L+ +L F KTFF+PCGT +K
Sbjct: 301 RSTSNVSSGHDLLSRRASQAQHHEEWHTDLLACCSEPSLCF-------KTFFFPCGTLAK 353
Query: 328 IATVATNRPI 337
IAT A+NR I
Sbjct: 354 IATAASNRHI 363
>UniRef100_Q654Z3 Receptor protein kinase-like [Oryza sativa]
Length = 541
Score = 139 bits (351), Expect = 1e-31
Identities = 69/138 (50%), Positives = 98/138 (71%), Gaps = 1/138 (0%)
Query: 2 ASSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLK 61
+S W G+ +NVAQL G+DA+ L+ M+V+AA AR H+ CR+ QH++L+G LL +L+
Sbjct: 89 SSLWGVLGQASNVAQLVGVDALGLVSMVVQAALAARRHRDACRRLGQHVELVGGLLWELE 148
Query: 62 ISELKKYPETREPLEQLEDALRRSYILVNSCQ-DRSYLYLLAMGWNIVYQFRKAQNEIDR 120
++EL + TR PLEQL+ ALRR Y LV +CQ DR YL+ L +G + + R AQ+EID
Sbjct: 149 LAELMRREATRRPLEQLQGALRRCYALVTACQEDRGYLHRLLLGARMANELRAAQHEIDM 208
Query: 121 YLRLVPLITLVDNARATR 138
Y+RL+PLI LVDN+ + R
Sbjct: 209 YIRLIPLIALVDNSSSNR 226
>UniRef100_Q654Z6 Receptor protein kinase-like [Oryza sativa]
Length = 567
Score = 129 bits (325), Expect = 1e-28
Identities = 64/134 (47%), Positives = 92/134 (67%), Gaps = 1/134 (0%)
Query: 2 ASSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLK 61
+S W G+ +NVAQL G+DA+ L+ M+ +AA AR H+ CR+ QH +L+G LL +L+
Sbjct: 4 SSLWGVLGQASNVAQLVGVDALGLVSMVAQAALAARRHRDACRRLGQHAELVGGLLRELE 63
Query: 62 ISELKKYPETREPLEQLEDALRRSYILVNSCQ-DRSYLYLLAMGWNIVYQFRKAQNEIDR 120
++EL + TR PLEQL+ ALRR Y L +CQ D YL+ L +G + + R AQ+EID
Sbjct: 64 LAELMRREATRRPLEQLQGALRRCYALATACQEDGGYLHRLLLGARMADELRAAQHEIDM 123
Query: 121 YLRLVPLITLVDNA 134
Y+RL+PLI LVD++
Sbjct: 124 YIRLIPLIALVDSS 137
>UniRef100_Q5Z661 Receptor protein kinase-like [Oryza sativa]
Length = 540
Score = 103 bits (257), Expect = 9e-21
Identities = 59/140 (42%), Positives = 84/140 (59%), Gaps = 22/140 (15%)
Query: 1 MASS-WEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQ 59
MAS W G+ ++VAQL G+DA CR+ QH+ L+G LL +
Sbjct: 1 MASGLWGALGQASSVAQLVGVDA--------------------CRRLGQHVDLVGGLLRE 40
Query: 60 LKISELKKYPETREPLEQLEDALRRSYILVNSCQDR-SYLYLLAMGWNIVYQFRKAQNEI 118
L+++EL + TR PLE+L+ ALRR Y LV +CQD YL+ L +G + + R AQ+EI
Sbjct: 41 LELAELMRREATRRPLERLQGALRRCYALVRACQDDCGYLHRLLLGARMADELRAAQHEI 100
Query: 119 DRYLRLVPLITLVDNARATR 138
D Y+RL+PLI+LVD++ R
Sbjct: 101 DMYIRLIPLISLVDSSNNNR 120
>UniRef100_Q5VMJ7 Cytokinin-regulated kinase-like [Oryza sativa]
Length = 532
Score = 103 bits (256), Expect = 1e-20
Identities = 55/131 (41%), Positives = 76/131 (57%)
Query: 5 WEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKISE 64
W G+ A VAQL G D LI I++AA TAR +K+ C Q A+ + +I +LL L+ E
Sbjct: 4 WGGLGQAATVAQLVGADVGGLISSIIQAAATARQNKRECDQLARRVVMIADLLPHLQDPE 63
Query: 65 LKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRL 124
+ + PE R PL +L D LR ++ LV SCQ RS Y M + +FR Q++ID YL +
Sbjct: 64 VMRRPEVRRPLAELGDTLREAHELVASCQGRSAAYRFVMAGRLADRFRDVQSKIDSYLIV 123
Query: 125 VPLITLVDNAR 135
P I +D R
Sbjct: 124 FPFIAHIDITR 134
>UniRef100_Q6ZDH7 Receptor kinase-like [Oryza sativa]
Length = 171
Score = 92.8 bits (229), Expect = 2e-17
Identities = 51/128 (39%), Positives = 68/128 (52%)
Query: 5 WEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKISE 64
W + G A VAQL G D LI I++AA TAR +K C Q A +I +LL L+ E
Sbjct: 2 WGELGNAATVAQLVGADLGGLISKIIQAAATARRNKAECEQLAVRASMIYDLLPHLQHPE 61
Query: 65 LKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRL 124
+ + PE PL LED LR ++ LV CQ + Y M + + R Q ID YL
Sbjct: 62 VVRRPEVLRPLALLEDTLREAHQLVTCCQHKGPTYRFVMAGRLADKIRSVQARIDSYLLF 121
Query: 125 VPLITLVD 132
+PLI+ +D
Sbjct: 122 LPLISHID 129
>UniRef100_Q5Z6Z4 Hypothetical protein B1018E06.42 [Oryza sativa]
Length = 138
Score = 86.3 bits (212), Expect = 1e-15
Identities = 44/119 (36%), Positives = 67/119 (55%), Gaps = 4/119 (3%)
Query: 5 WEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKISE 64
W +A VAQ+ G+DA LI M+ + A R +K CRQ A+H++ +G LL ++ +
Sbjct: 4 WSSVDPVATVAQIAGVDAYGLISMVTERAEKVRRNKYECRQLAEHVETVGGLLHHVERDD 63
Query: 65 LKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYLR 123
P PLE+LE LR + +LV+SC+ SY G I QF++ + +ID YL+
Sbjct: 64 ----PRIAAPLEKLEGTLREAVVLVSSCEASSYFRRFFRGAKIAEQFQRIKEKIDFYLQ 118
>UniRef100_Q6ZB70 Receptor-like kinase CHRK1-like [Oryza sativa]
Length = 578
Score = 80.1 bits (196), Expect = 1e-13
Identities = 47/125 (37%), Positives = 70/125 (55%), Gaps = 1/125 (0%)
Query: 12 ANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKI-SELKKYPE 70
A VAQL G DA L+ I++A AR +++ CR A+ ++G+LL L SE + PE
Sbjct: 13 ATVAQLAGADAAGLVSAILQAVRAARRNRRECRLLARRAMMVGDLLRLLPPESETMRRPE 72
Query: 71 TREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRLVPLITL 130
R L+ L ALR++ LV SCQ+ + L QFR+ Q EI+ Y+ L P+++
Sbjct: 73 VRRALDGLGGALRQALELVESCQESGAVRGLMTAGRQAEQFREVQGEINDYMLLFPVVSH 132
Query: 131 VDNAR 135
+D R
Sbjct: 133 IDITR 137
>UniRef100_Q7RSV3 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 1456
Score = 42.0 bits (97), Expect = 0.032
Identities = 48/231 (20%), Positives = 100/231 (42%), Gaps = 32/231 (13%)
Query: 66 KKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRLV 125
KKY E ++ + + ++ ++NS + N++Y K + ++ +RL+
Sbjct: 676 KKYSEKQKCCKNTDHEYLKNSDIINSIK------------NLIYSI-KIKEDVKFDIRLL 722
Query: 126 PLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKP-----EHDKEDTVVLK 180
I D+ + + ++E Q D+ + ++ T ++ + D+ED + LK
Sbjct: 723 YYILDEDSCVHPFLNDLEFLVENIQLPKYQDNRNSQINTYSIQELSDFDDSDEEDIIFLK 782
Query: 181 KTLSCSYPNFSFTEALKKENEKL--------QLELHHSQANMDINQCEFIQRLLDVTNFA 232
KT+ +FS + LKKE KL EL + + + C +L +N
Sbjct: 783 KTIFGKPSSFSTDKKLKKEKNKLSKYNELYENYELCSNYESTSNDSCSGYSNIL--SNDK 840
Query: 233 AYSLPEKLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKKPKFQKRIR 283
+K+ +N+Q + NN++ N +K H N++ + + + +IR
Sbjct: 841 KKKKKKKVKQYENFQKI---NNTNLMVNKNK-HKKNDEEIYQMDQLENKIR 887
>UniRef100_Q8IBR9 Exported serine/threonine protein kinase [Plasmodium falciparum]
Length = 2695
Score = 40.8 bits (94), Expect = 0.072
Identities = 23/71 (32%), Positives = 40/71 (55%), Gaps = 2/71 (2%)
Query: 238 EKLSPEDNYQIVEYSNNSHSDANGSKGHSSNE--KHHKKKPKFQKRIRCQLEVHISRKIG 295
EK DN I++ +NNS+ + N + + SN +H+KK F K I Q+ + + K
Sbjct: 2403 EKCHNNDNCYIIDTTNNSNINNNNNNNNCSNNVLRHNKKSDIFNKNIVNQINNNKTIKYI 2462
Query: 296 ILIYLLVVLNL 306
IL+Y +++ N+
Sbjct: 2463 ILMYEIIIKNM 2473
>UniRef100_UPI0000330E48 UPI0000330E48 UniRef100 entry
Length = 519
Score = 38.9 bits (89), Expect = 0.27
Identities = 41/188 (21%), Positives = 82/188 (42%), Gaps = 27/188 (14%)
Query: 55 NLLDQLKISELKKYPETREPLEQLEDALRRSYILVNSCQD------------RSYLYLL- 101
+L + KIS L K E ++ ++QL +++ L + + + Y+ +
Sbjct: 158 SLNQEKKISLLDKIAEYKKDIDQLSPSIKDQSSLFDRKNNEFQKIEGEYRTAQEYVKTIQ 217
Query: 102 AMGWNIVYQFRKAQNEIDRYLRLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQK 161
++ W + ++ +DR + + T V N +R +++++EKDQ Y++D +
Sbjct: 218 SLRWEAQEKIVDERSILDRSISSIEEKTTVIN----HLRSKIKILEKDQKKYSIDQKKID 273
Query: 162 VQTVFMKPEHDKEDTVVLKKTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDI--NQC 219
T E E +K + +S E KE LQ+ELH ++N+ +Q
Sbjct: 274 NNT-----EQQLEKVRKIKSEIDIFQKQYSNNE---KEYSNLQMELHTKKSNLTSLKSQL 325
Query: 220 EFIQRLLD 227
F L++
Sbjct: 326 SFFNELVE 333
>UniRef100_UPI00004659B1 UPI00004659B1 UniRef100 entry
Length = 1037
Score = 38.1 bits (87), Expect = 0.47
Identities = 40/157 (25%), Positives = 68/157 (42%), Gaps = 11/157 (7%)
Query: 189 NFSFTEALKKENEKLQLE--LHHSQANMDINQCEFIQRLLDVTNFAAYSLPEKLSPEDNY 246
N E + K+N+ ++ E L HSQ D Q ++ + + E+++ +
Sbjct: 103 NCEDNENIDKQNQLMENEQILKHSQIKYDNPNDPSNQFSINKKEPHFHRIKEEINESNID 162
Query: 247 QIVEYSNNSHSDANGSKGHSSNEKHHKKKP------KFQKRIRCQLEVHIS--RKIGILI 298
I E SNNS + NE + + KF R + +E+H++ K I+I
Sbjct: 163 NIYENSNNSLLETQRDSEKKENENNENCQSSDENYVKFGNRKKKVVELHLALPTKTHIMI 222
Query: 299 YLLVVLNLIFIFSFAGIKTFFYPCGT-FSKIATVATN 334
L+ N+I I + Y CGT FS+I+ + N
Sbjct: 223 KELMNKNIIKFLITQNIDSLHYRCGTKFSQISEIHGN 259
>UniRef100_Q93XR9 Bg55 protein [Bruguiera gymnorrhiza]
Length = 756
Score = 37.7 bits (86), Expect = 0.61
Identities = 33/131 (25%), Positives = 66/131 (50%), Gaps = 13/131 (9%)
Query: 35 TARMHKKNCRQFAQHLKLIGNLLDQLKISELKKYPETR---EPLEQLEDALRRSYILVNS 91
+ ++H+ CR+ + + I L ++ S P R E L L DAL R+ ++
Sbjct: 17 SCKVHQSICRELRKVVDRIERLFPNIEASR----PRCRLGIEVLCLLNDALDRAKQVLQY 72
Query: 92 CQDRSYLYLLAMGWNIVYQFRKAQNEIDRYL-RLVPLITLVDNARATRVRERLEVIEKDQ 150
C + S LYL G IV + +K++N +++ L ++ ++ ++ A ++V + L V +
Sbjct: 73 CSESSKLYLALNGDVIVSRCQKSRNLLEQSLDQIQTMVPVILAAEISQVIDDLRVAK--- 129
Query: 151 CAYTLDDEDQK 161
+ LD D++
Sbjct: 130 --FVLDHSDEE 138
>UniRef100_Q8ID99 Hypothetical protein PF13_0321 [Plasmodium falciparum]
Length = 547
Score = 37.7 bits (86), Expect = 0.61
Identities = 31/125 (24%), Positives = 60/125 (47%), Gaps = 4/125 (3%)
Query: 172 DKEDTVVLKKTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNF 231
D+ ++ T++ + N + T N+ L+ + NM+ N +L D NF
Sbjct: 314 DQTQQIIPSDTINNTENNNTSTNNNNNNNKNFYLKFEE-RFNMN-NDRIITNKLPDENNF 371
Query: 232 AAYS-LPEKLSPEDNYQIVEY-SNNSHSDANGSKGHSSNEKHHKKKPKFQKRIRCQLEVH 289
A + L ++L N I+ Y +NN++++ N + +++N K +KKK K + I +
Sbjct: 372 QALNELNKELRNAHNLSIINYDNNNNNNNNNNNNNNNNNNKLNKKKKKKKNSITTNINKK 431
Query: 290 ISRKI 294
+ KI
Sbjct: 432 NNNKI 436
>UniRef100_UPI00002EA191 UPI00002EA191 UniRef100 entry
Length = 115
Score = 37.4 bits (85), Expect = 0.79
Identities = 20/69 (28%), Positives = 43/69 (61%), Gaps = 2/69 (2%)
Query: 140 RERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKED-TVVLKKTLSCSYPNFSFTEALKK 198
R++ E+IEKD TL+ ++ +F+ P ++K + ++V++K L+ + N S T + +
Sbjct: 25 RDKFEIIEKDIMTSTLNSNKNRISLIFIDPPYEKYNISLVIEKILNSNLTN-SETLIVIE 83
Query: 199 ENEKLQLEL 207
E+ K++L +
Sbjct: 84 ESNKIKLTI 92
>UniRef100_UPI00004377F3 UPI00004377F3 UniRef100 entry
Length = 741
Score = 37.0 bits (84), Expect = 1.0
Identities = 30/128 (23%), Positives = 57/128 (44%), Gaps = 12/128 (9%)
Query: 184 SCSYPNFSFTEALKKENEKLQLELHHS-----------QANMDINQCEFIQRLLDVTNFA 232
S Y + S + L KE E+++ E+ +A D + + L +N
Sbjct: 18 SSDYSDSSVQQRLDKERERIRREIRKELKIKEGAENLRRATTDKRNAQQVDSQLRCSNRR 77
Query: 233 AYSLPEKLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKK-PKFQKRIRCQLEVHIS 291
SL +L D + +V+ ++++H DA S G S HK++ ++++ +L+V
Sbjct: 78 LDSLHAELQELDAHIVVKGTDDTHDDAPQSPGAESRSSAHKERIAALERQLNIELKVKQG 137
Query: 292 RKIGILIY 299
+ I IY
Sbjct: 138 AENMIPIY 145
>UniRef100_UPI0000435ED3 UPI0000435ED3 UniRef100 entry
Length = 913
Score = 37.0 bits (84), Expect = 1.0
Identities = 30/128 (23%), Positives = 57/128 (44%), Gaps = 12/128 (9%)
Query: 184 SCSYPNFSFTEALKKENEKLQLELHHS-----------QANMDINQCEFIQRLLDVTNFA 232
S Y + S + L KE E+++ E+ +A D + + L +N
Sbjct: 18 SSDYSDSSVQQRLDKERERIRREIRKELKIKEGAENLRRATTDKRNAQQVDSQLRCSNRR 77
Query: 233 AYSLPEKLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHHKKK-PKFQKRIRCQLEVHIS 291
SL +L D + +V+ ++++H DA S G S HK++ ++++ +L+V
Sbjct: 78 LDSLHAELQELDAHIVVKGTDDTHDDAPQSPGAESRSSAHKERIAALERQLNIELKVKQG 137
Query: 292 RKIGILIY 299
+ I IY
Sbjct: 138 AENMIPIY 145
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 656,602,628
Number of Sequences: 2790947
Number of extensions: 27066828
Number of successful extensions: 107955
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 107877
Number of HSP's gapped (non-prelim): 113
length of query: 417
length of database: 848,049,833
effective HSP length: 130
effective length of query: 287
effective length of database: 485,226,723
effective search space: 139260069501
effective search space used: 139260069501
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC136504.6


