

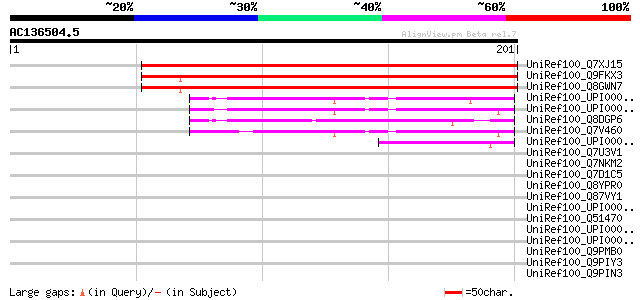
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.5 + phase: 0
(201 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XJ15 Hypothetical protein P0478E02.11 [Oryza sativa] 222 5e-57
UniRef100_Q9FKX3 Arabidopsis thaliana genomic DNA, chromosome 5,... 214 1e-54
UniRef100_Q8GWN7 Hypothetical protein At5g66090/K2A18_17 [Arabid... 211 9e-54
UniRef100_UPI000031D2D2 UPI000031D2D2 UniRef100 entry 52 1e-05
UniRef100_UPI0000269A15 UPI0000269A15 UniRef100 entry 48 1e-04
UniRef100_Q8DGP6 Tlr2269 protein [Synechococcus elongatus] 47 2e-04
UniRef100_Q7V460 Universal stress protein [Prochlorococcus marinus] 47 3e-04
UniRef100_UPI0000340910 UPI0000340910 UniRef100 entry 46 5e-04
UniRef100_Q7U3V1 Hypothetical protein [Synechococcus sp.] 45 0.001
UniRef100_Q7NKM2 Gll1455 protein [Gloeobacter violaceus] 42 0.007
UniRef100_Q7D1C5 AGR_C_878p [Agrobacterium tumefaciens] 37 0.23
UniRef100_Q8YPR0 Alr4132 protein [Anabaena sp.] 36 0.52
UniRef100_Q87VY1 TRNA-i(6)A37 modification enzyme MiaB [Pseudomo... 36 0.68
UniRef100_UPI0000264235 UPI0000264235 UniRef100 entry 35 1.2
UniRef100_Q51470 Hypothetical UPF0004 protein PA3980 [Pseudomona... 35 1.2
UniRef100_UPI000021DCBC UPI000021DCBC UniRef100 entry 35 1.5
UniRef100_UPI000042EED3 UPI000042EED3 UniRef100 entry 34 2.0
UniRef100_Q9PMB0 Putative methyl-accepting chemotaxis signal tra... 34 2.0
UniRef100_Q9PIY3 Methyl-accepting chemotaxis signal transduction... 34 2.0
UniRef100_Q9PIN3 Putative methyl-accepting chemotaxis signal tra... 34 2.0
>UniRef100_Q7XJ15 Hypothetical protein P0478E02.11 [Oryza sativa]
Length = 194
Score = 222 bits (565), Expect = 5e-57
Identities = 105/149 (70%), Positives = 128/149 (85%)
Query: 53 VKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVV 112
V+A+ E A+A D+F Q KH+LLP+TDR PYLSEGT+QA AT+ +LA KYGA+ITVVV
Sbjct: 46 VRAEVNESGSALAADAFAQVKHVLLPVTDRNPYLSEGTRQAAATSASLAKKYGANITVVV 105
Query: 113 IDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVV 172
ID++ KE PEH+TQ+SSIRWHLSEGG ++ L+ERLG+G KPTAIIG+VADEL LDLVV
Sbjct: 106 IDDKPKEEFPEHDTQMSSIRWHLSEGGFTEFGLMERLGEGKKPTAIIGEVADELELDLVV 165
Query: 173 ISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+SMEAIH+KH+D NLLAEFIPCPV+LLPL
Sbjct: 166 LSMEAIHSKHVDGNLLAEFIPCPVLLLPL 194
>UniRef100_Q9FKX3 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K2A18
[Arabidopsis thaliana]
Length = 210
Score = 214 bits (545), Expect = 1e-54
Identities = 106/153 (69%), Positives = 131/153 (85%), Gaps = 4/153 (2%)
Query: 53 VKAKPQEPEVAIAT----DSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADI 108
VKA+ +E E + + D+F+ KHLLLP+ DR PYLSEGT+QA ATT +LA KYGADI
Sbjct: 58 VKAQAKEAEASPSPVGVGDAFSNVKHLLLPVIDRNPYLSEGTRQAAATTTSLAKKYGADI 117
Query: 109 TVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNL 168
TVVVIDE+++ES EHETQ+S+IRWHLSEGG +++KLLERLG+G K TAIIG+VADEL +
Sbjct: 118 TVVVIDEEKRESSSEHETQVSNIRWHLSEGGFEEFKLLERLGEGKKATAIIGEVADELKM 177
Query: 169 DLVVISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+LVV+SMEAIH+K+IDANLLAEFIPCPV+LLPL
Sbjct: 178 ELVVMSMEAIHSKYIDANLLAEFIPCPVLLLPL 210
>UniRef100_Q8GWN7 Hypothetical protein At5g66090/K2A18_17 [Arabidopsis thaliana]
Length = 210
Score = 211 bits (537), Expect = 9e-54
Identities = 105/153 (68%), Positives = 129/153 (83%), Gaps = 4/153 (2%)
Query: 53 VKAKPQEPEVAIAT----DSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADI 108
VKA+ +E E + + D+F+ KHLLLP+ DR PYLS GT+QA ATT LA KYGADI
Sbjct: 58 VKAQAKEAEASPSPVGVGDAFSNVKHLLLPVIDRNPYLSGGTRQAAATTTGLAKKYGADI 117
Query: 109 TVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNL 168
TVVVIDE+++ES EHETQ+S+IRWHLSEGG +++KLLERLG+G K TAIIG+VADEL +
Sbjct: 118 TVVVIDEEKRESSSEHETQVSNIRWHLSEGGFEEFKLLERLGEGKKATAIIGEVADELKM 177
Query: 169 DLVVISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+LVV+SMEAIH+K+IDANLLAEFIPCPV+LLPL
Sbjct: 178 ELVVMSMEAIHSKYIDANLLAEFIPCPVLLLPL 210
>UniRef100_UPI000031D2D2 UPI000031D2D2 UniRef100 entry
Length = 127
Score = 51.6 bits (122), Expect = 1e-05
Identities = 37/135 (27%), Positives = 70/135 (51%), Gaps = 15/135 (11%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQ---L 128
F+ +L PI DR S + A + LA + + + ++ + E ++ + +H L
Sbjct: 2 FETVLFPI-DR----SRQALETAAKALELARSHSSRLVLLSVVEAERPGMQDHGVVAQLL 56
Query: 129 SSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTK---HIDA 185
R + + G+ +++ER G KPT +I DVADE+N+D++V+ ++ A
Sbjct: 57 QQTREQVEQAGVP-CEVVERQG---KPTFVICDVADEMNVDVIVMGTRGVNLSSDGESTA 112
Query: 186 NLLAEFIPCPVMLLP 200
+ +F PCPV+++P
Sbjct: 113 ARVIQFAPCPVLVVP 127
>UniRef100_UPI0000269A15 UPI0000269A15 UniRef100 entry
Length = 127
Score = 48.1 bits (113), Expect = 1e-04
Identities = 36/135 (26%), Positives = 64/135 (46%), Gaps = 15/135 (11%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQ---L 128
F+ +L P S AT + L ++ + + ++ + E ++ + + L
Sbjct: 2 FRTVLFPFDQ-----SRAAMDTAATVLQLVQQHDSKLVLLSVVEAGNGTMGDVDAVAQLL 56
Query: 129 SSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLL 188
+E G+K +L+ER G KP +IGDVADE+ DL+V+ I+ + +
Sbjct: 57 QKAHDTFAEAGVK-CELIEREG---KPAFVIGDVADEIEADLIVMGTRGINLEQAGTSTA 112
Query: 189 AEFI---PCPVMLLP 200
A I PCPV+++P
Sbjct: 113 ARVIQLAPCPVLVVP 127
>UniRef100_Q8DGP6 Tlr2269 protein [Synechococcus elongatus]
Length = 129
Score = 47.4 bits (111), Expect = 2e-04
Identities = 36/140 (25%), Positives = 67/140 (47%), Gaps = 23/140 (16%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
FK +L PI DR S T++A+ T + + + + + ++ ++E + PE E ++
Sbjct: 2 FKTILFPI-DR----SRDTQEAMPTVVEMVKLFQSQLFLLSVEESPEPD-PELEAAIAEF 55
Query: 132 RWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVIS-----------MEAIHT 180
+ E + + E L KP +I DVADE+N L+++ E++ T
Sbjct: 56 LNRVKEAFAQQAIVAETLLRRGKPAFVICDVADEINASLIIMGCRGTALTPEGFQESVST 115
Query: 181 KHIDANLLAEFIPCPVMLLP 200
+ I+ PCPV+++P
Sbjct: 116 RVIN------LAPCPVLVVP 129
>UniRef100_Q7V460 Universal stress protein [Prochlorococcus marinus]
Length = 127
Score = 47.0 bits (110), Expect = 3e-04
Identities = 34/135 (25%), Positives = 67/135 (49%), Gaps = 15/135 (11%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQ---L 128
F+ +L PI + + +K + LA + + + ++ + + ++ + + E L
Sbjct: 2 FETVLFPIDQSREAMETASK-----ALELARSHNSRLIILSVVQAERPEMHDPEAVAVLL 56
Query: 129 SSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLL 188
R + + G+ ++LER G KP +I DVADELN+D++V+ ++ +
Sbjct: 57 KRAREQVKQAGIA-CEVLEREG---KPAFVICDVADELNVDVIVMGTRGVNLDGDSESTA 112
Query: 189 AEFI---PCPVMLLP 200
A I PCPV+++P
Sbjct: 113 ARVIQLAPCPVLVVP 127
>UniRef100_UPI0000340910 UPI0000340910 UniRef100 entry
Length = 62
Score = 46.2 bits (108), Expect = 5e-04
Identities = 23/57 (40%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLLA---EFIPCPVMLLP 200
+RL G +P +I DVADELN+D++V+ + D + A E PCPV+L+P
Sbjct: 6 QRLERGGEPLTVILDVADELNVDVIVMGTGGVSLNSDDGSTAARVIELAPCPVLLVP 62
>UniRef100_Q7U3V1 Hypothetical protein [Synechococcus sp.]
Length = 119
Score = 45.1 bits (105), Expect = 0.001
Identities = 22/57 (38%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLLA---EFIPCPVMLLP 200
+RL G +P +I DVADELN+D++V+ + D + A E PCPV+++P
Sbjct: 63 QRLERGGEPLTVILDVADELNVDVIVMGTGGVSLNSNDGSTAARVIELAPCPVLVVP 119
>UniRef100_Q7NKM2 Gll1455 protein [Gloeobacter violaceus]
Length = 142
Score = 42.4 bits (98), Expect = 0.007
Identities = 33/141 (23%), Positives = 65/141 (45%), Gaps = 18/141 (12%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
F+ +LLP+ S +++A+ ++LA +Y + + ++ + + +E +PE E L+ +
Sbjct: 8 FELILLPLDS-----SPESERALQVALSLARQYSSKLLLLSVVDLPEE-MPEREAHLAEV 61
Query: 132 RWHLSEGGLKDYKLLERLG-------DGSKPTAIIGDVADELNLDLVVISMEAI-----H 179
R L+ G D K +I DVADE+ L+V+ +
Sbjct: 62 RAKAQALAQTVRDRLQGEGYPTDARIDAGKAAFVICDVADEVGAGLIVMGSRGLGLIEEG 121
Query: 180 TKHIDANLLAEFIPCPVMLLP 200
H ++ + PCPV+++P
Sbjct: 122 KHHSTSSRVINLSPCPVLVVP 142
>UniRef100_Q7D1C5 AGR_C_878p [Agrobacterium tumefaciens]
Length = 160
Score = 37.4 bits (85), Expect = 0.23
Identities = 35/120 (29%), Positives = 51/120 (42%), Gaps = 25/120 (20%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVID----------------- 114
FKH+L+P TD P + AI ALA + GA +TVV +
Sbjct: 17 FKHILIP-TDGSPL----AQIAIDQGFALAREAGAKVTVVTVSEPFHVIASDVEDIAAIA 71
Query: 115 EQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVIS 174
E++ E E L + H + GL LL R G +P I ++AD DL+ ++
Sbjct: 72 EEEFHRCEEAEHLLRDTQAHAAAMGLDCEALLARAG---RPDEAIIEIADRTGCDLIAMA 128
>UniRef100_Q8YPR0 Alr4132 protein [Anabaena sp.]
Length = 135
Score = 36.2 bits (82), Expect = 0.52
Identities = 34/142 (23%), Positives = 58/142 (39%), Gaps = 21/142 (14%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
FK +L PI S ++A + KYG+ + ++ + E ES P+ + +
Sbjct: 2 FKTVLFPIDQ-----SREAREAADVVTNVVQKYGSRLILLSVVE---ESSPDESSADPMV 53
Query: 132 RWHLSEGGLKDYKLL--------ERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHI 183
L+D + L E + KP I DVADE+ DL+++ +
Sbjct: 54 SPEAVAKLLRDAQALFLQQGIPSEIVEKQGKPAFTICDVADEIGADLIIMGCRGLGLTEE 113
Query: 184 DA-----NLLAEFIPCPVMLLP 200
A + PCPV+++P
Sbjct: 114 GATDSVTTRVINLSPCPVLIVP 135
>UniRef100_Q87VY1 TRNA-i(6)A37 modification enzyme MiaB [Pseudomonas syringae]
Length = 442
Score = 35.8 bits (81), Expect = 0.68
Identities = 35/138 (25%), Positives = 58/138 (41%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I + P+V ++ +F HL P D A + +
Sbjct: 105 PQTLHRLPEMIDAARITRLPQVDVSFPEIEKFDHLPEPRVD--------GPSAYVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY T V+ + E + + + HL+E G+++ LL + +G + T G
Sbjct: 157 CSKY---CTFCVVPYTRGEEVSRPFDDVLTEVTHLAEHGVREVTLLGQNVNGYRGTTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
VAD +L V ++ I
Sbjct: 214 RVADLADLIRAVAVVDGI 231
>UniRef100_UPI0000264235 UPI0000264235 UniRef100 entry
Length = 297
Score = 35.0 bits (79), Expect = 1.2
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 2/31 (6%)
Query: 100 LANKYGADITVVVIDEQ--QKESLPEHETQL 128
L N YGADIT + +D Q ++LPE+ETQL
Sbjct: 106 LFNYYGADITSIAVDNHGLQTDNLPEYETQL 136
>UniRef100_Q51470 Hypothetical UPF0004 protein PA3980 [Pseudomonas aeruginosa]
Length = 446
Score = 35.0 bits (79), Expect = 1.2
Identities = 35/138 (25%), Positives = 58/138 (41%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I ++P+V ++ +F DR P A + +
Sbjct: 105 PQTLHRLPEMIDAARSTRKPQVDVSFPEIEKF--------DRLPEPRVDGPTAFVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY VV ++ S P + I HL+E G+++ LL + +G + G
Sbjct: 157 CSKY-CSFCVVPYTRGEEVSRPFDDVIAEVI--HLAENGVREVTLLGQNVNGFRGLTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
+AD L VV +++ I
Sbjct: 214 RLADFAELLRVVAAVDGI 231
>UniRef100_UPI000021DCBC UPI000021DCBC UniRef100 entry
Length = 738
Score = 34.7 bits (78), Expect = 1.5
Identities = 22/56 (39%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 12 LVKPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKPQEPEVAIATD 67
L +P L SP P L ++ P S +S +PPST PSG + P PEV D
Sbjct: 589 LSEPLLNSPDPLLSSSSRPDSPSSS--SPPSTQEASPSGDVAVGSPSMPEVGSPRD 642
>UniRef100_UPI000042EED3 UPI000042EED3 UniRef100 entry
Length = 2269
Score = 34.3 bits (77), Expect = 2.0
Identities = 21/54 (38%), Positives = 30/54 (54%), Gaps = 7/54 (12%)
Query: 15 PSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKPQEPEVAIATDS 68
P++++ PS NPP SIT+ NPPS++ T P IV P IAT++
Sbjct: 1211 PNIVTTLPSNITTNPPSSITT---NPPSSIKTNPPSSIVTT----PSSNIATNT 1257
>UniRef100_Q9PMB0 Putative methyl-accepting chemotaxis signal transduction protein
[Campylobacter jejuni]
Length = 662
Score = 34.3 bits (77), Expect = 2.0
Identities = 27/91 (29%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Query: 91 KQAIATTIALANKYGADITVV----VIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLL 146
KQ+ ALAN+ G T V Q +SL E L I + +K ++
Sbjct: 469 KQSSDFANALANESGKLQTAVQSLTTSSNSQAQSLEETAAALEEITSSMQNVSVKTSDVI 528
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEA 177
+ + T IIGD+AD++NL + ++EA
Sbjct: 529 TQSEEIKNVTGIIGDIADQINLLALNAAIEA 559
>UniRef100_Q9PIY3 Methyl-accepting chemotaxis signal transduction protein
[Campylobacter jejuni]
Length = 659
Score = 34.3 bits (77), Expect = 2.0
Identities = 27/91 (29%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Query: 91 KQAIATTIALANKYGADITVV----VIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLL 146
KQ+ ALAN+ G T V Q +SL E L I + +K ++
Sbjct: 466 KQSSDFANALANESGKLQTAVQSLTTSSNSQAQSLEETAAALEEITSSMQNVSVKTSDVI 525
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEA 177
+ + T IIGD+AD++NL + ++EA
Sbjct: 526 TQSEEIKNVTGIIGDIADQINLLALNAAIEA 556
>UniRef100_Q9PIN3 Putative methyl-accepting chemotaxis signal transduction protein
[Campylobacter jejuni]
Length = 665
Score = 34.3 bits (77), Expect = 2.0
Identities = 27/91 (29%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Query: 91 KQAIATTIALANKYGADITVV----VIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLL 146
KQ+ ALAN+ G T V Q +SL E L I + +K ++
Sbjct: 472 KQSSDFANALANESGKLQTAVQSLTTSSNSQAQSLEETAAALEEITSSMQNVSVKTSDVI 531
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEA 177
+ + T IIGD+AD++NL + ++EA
Sbjct: 532 TQSEEIKNVTGIIGDIADQINLLALNAAIEA 562
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 346,529,238
Number of Sequences: 2790947
Number of extensions: 14488648
Number of successful extensions: 39525
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 39495
Number of HSP's gapped (non-prelim): 66
length of query: 201
length of database: 848,049,833
effective HSP length: 121
effective length of query: 80
effective length of database: 510,345,246
effective search space: 40827619680
effective search space used: 40827619680
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC136504.5


