

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.2 + phase: 0
(713 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
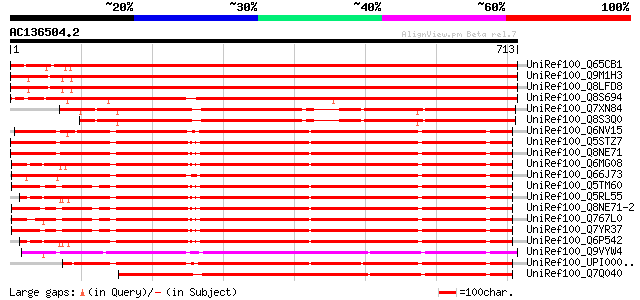
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q65CB1 ABC transporter [Populus tremula x Populus trem... 1159 0.0
UniRef100_Q9M1H3 ABC transporter-like protein [Arabidopsis thali... 1129 0.0
UniRef100_Q8LFD8 Putative ABC transporter [Arabidopsis thaliana] 1125 0.0
UniRef100_Q8S694 Putative ABC transporter [Oryza sativa] 959 0.0
UniRef100_Q7XN84 OSJNBa0011F23.8 protein [Oryza sativa] 575 e-162
UniRef100_Q8S3Q0 Putative ABC transporter protein [Oryza sativa] 566 e-160
UniRef100_Q6NV15 Zgc:85667 [Brachydanio rerio] 553 e-156
UniRef100_Q5STZ7 OTTHUMP00000062376 [Homo sapiens] 547 e-154
UniRef100_Q8NE71 ATP-binding cassette, sub-family F, member 1 [H... 547 e-154
UniRef100_Q6MG08 ATP-binding cassette, sub-family F, member 1 [R... 543 e-153
UniRef100_Q66J73 MGC81714 protein [Xenopus laevis] 540 e-152
UniRef100_Q5TM60 ATP-binding cassette, sub-family F (GCN20), mem... 537 e-151
UniRef100_Q5RL55 ATP-binding cassette, sub-family F (GCN20), mem... 536 e-151
UniRef100_Q8NE71-2 Splice isoform 2 of Q8NE71 [Homo sapiens] 536 e-151
UniRef100_Q767L0 ATP-binding cassette, sub-family F, member 1 [S... 534 e-150
UniRef100_Q7YR37 ATP-binding cassette, sub-family F, member 1 [P... 534 e-150
UniRef100_Q6P542 ATP-binding cassette, sub-family F, member 1 [M... 534 e-150
UniRef100_Q9VYW4 CG1703-PA [Drosophila melanogaster] 533 e-150
UniRef100_UPI0000360FB0 UPI0000360FB0 UniRef100 entry 530 e-149
UniRef100_Q7Q040 ENSANGP00000016545 [Anopheles gambiae str. PEST] 504 e-141
>UniRef100_Q65CB1 ABC transporter [Populus tremula x Populus tremuloides]
Length = 728
Score = 1159 bits (2999), Expect = 0.0
Identities = 593/732 (81%), Positives = 654/732 (89%), Gaps = 23/732 (3%)
Query: 1 MGKKKSEDA-GPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSN-----KAKP 54
MGKK+ EDA G +KAK G+KD +KKEK SV+AMLA MD+K DKPKK SS+ K KP
Sbjct: 1 MGKKQKEDASGAPSKAKAGNKD-AKKEKLSVTAMLASMDQKPDKPKKGSSSTVTSSKPKP 59
Query: 55 KPAPKASAYTDDIDLPPSDDDES----EEEQEE--------KHRPDLKPLEVSIAEKELK 102
K AP +YTD IDLPPSDD+E EEEQ++ + R +LKPL+V+I++KELK
Sbjct: 60 KSAP---SYTDGIDLPPSDDEEEPNGLEEEQQQNDPNKRPSQRRSELKPLDVAISDKELK 116
Query: 103 KREKKDILAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVA 162
KREKK++LAAH E A++EAL+DD DAFTVVIGSRASVLDG+D DANVKDITIENFSV+
Sbjct: 117 KREKKELLAAHAIEHARQEALKDDHDAFTVVIGSRASVLDGEDEGDANVKDITIENFSVS 176
Query: 163 ARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGD 222
ARGKELLKN SVKI+HG+RYGL+GPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEV+GD
Sbjct: 177 ARGKELLKNASVKIAHGRRYGLVGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVIGD 236
Query: 223 DKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLM 282
DKTAL+AVVSAN EL+K+R++VA LQ EG + D +E+DAGE+LAELYE+LQLM
Sbjct: 237 DKTALQAVVSANEELVKLREEVASLQKSDGPAEGENNGDDYDEDDAGERLAELYEKLQLM 296
Query: 283 GSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHL 342
GSDAAESQASKILAGLGFTKDMQGRPT+SFSGGWRMRISLARALFVQPTLLLLDEPTNHL
Sbjct: 297 GSDAAESQASKILAGLGFTKDMQGRPTRSFSGGWRMRISLARALFVQPTLLLLDEPTNHL 356
Query: 343 DLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQ 402
DLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVC++IIHLHD KL YRGNFD FE GYEQ
Sbjct: 357 DLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCNDIIHLHDQKLDSYRGNFDDFEVGYEQ 416
Query: 403 RRREANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKE-SKSKSKGKVDEDETQVE 461
RR+E NKK+EI+DKQ+KAA+R+GN+ QQ+KVKDRAKFAA KE +K+K + KVDED+ E
Sbjct: 417 RRKETNKKFEIYDKQMKAAKRSGNRVQQEKVKDRAKFAATKEAAKNKGRAKVDEDQAAPE 476
Query: 462 VPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGP 521
P KWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDF+LS+VDVGIDMGTRVAIVGP
Sbjct: 477 APRKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFKLSNVDVGIDMGTRVAIVGP 536
Query: 522 NGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQE 581
NGAGKSTLLNLLAGDLVP+EGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQE
Sbjct: 537 NGAGKSTLLNLLAGDLVPTEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQE 596
Query: 582 GLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLD 641
GLSKQEAVR KLGK+GLPSHNHLTPI KLSGGQKARVVFTSISMS+PHILLLDEPTNHLD
Sbjct: 597 GLSKQEAVRGKLGKFGLPSHNHLTPIAKLSGGQKARVVFTSISMSKPHILLLDEPTNHLD 656
Query: 642 MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKE 701
MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEE+S+IWVVEDGTV FPGTFE YKE
Sbjct: 657 MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEEKSEIWVVEDGTVTAFPGTFELYKE 716
Query: 702 DLLKEIKAEVDD 713
+L KEIKAEVDD
Sbjct: 717 ELQKEIKAEVDD 728
>UniRef100_Q9M1H3 ABC transporter-like protein [Arabidopsis thaliana]
Length = 723
Score = 1129 bits (2919), Expect = 0.0
Identities = 576/726 (79%), Positives = 644/726 (88%), Gaps = 16/726 (2%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSK---KEKFSVSAMLAGMDEKADKPKKASSNKAKPKPA 57
MGKKKS+++ +TK KP KD SK KEK SVSAMLAGMD+K DKPKK SS++ K A
Sbjct: 1 MGKKKSDESAATTKVKPSGKDASKDSKKEKLSVSAMLAGMDQKDDKPKKGSSSRTKA--A 58
Query: 58 PKASAYTDDIDLPPSD---DDESEEEQEEKH-----RPDLKPLEVSIAEKELKKREKKDI 109
PK+++YTD IDLPPSD D ES+EE+ +K + + + LE+S+ +KE KKRE K+
Sbjct: 59 PKSTSYTDGIDLPPSDEEDDGESDEEERQKEARRKLKSEQRHLEISVTDKEQKKREAKER 118
Query: 110 LAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELL 169
LA AE AK+EA++DD DAFTVVIGS+ SVL+GDD ADANVKDITIE+FSV+ARGKELL
Sbjct: 119 LALQAAESAKREAMKDDHDAFTVVIGSKTSVLEGDDMADANVKDITIESFSVSARGKELL 178
Query: 170 KNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEA 229
KN SV+ISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGD+K+AL A
Sbjct: 179 KNASVRISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDEKSALNA 238
Query: 230 VVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAES 289
VVSAN EL+K+R++ LQ +SG +G + D +++D GEKLAELY++LQ++GSDAAE+
Sbjct: 239 VVSANEELVKLREEAEALQKSSSGADG-ENVDGEDDDDTGEKLAELYDRLQILGSDAAEA 297
Query: 290 QASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 349
QASKILAGLGFTKDMQ R T+SFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW
Sbjct: 298 QASKILAGLGFTKDMQVRATQSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 357
Query: 350 LEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANK 409
LEEYLCRWKKTLVVVSHDRDFLNTVC+EIIHLHD LHFYRGNFD FESGYEQRR+E NK
Sbjct: 358 LEEYLCRWKKTLVVVSHDRDFLNTVCTEIIHLHDQNLHFYRGNFDGFESGYEQRRKEMNK 417
Query: 410 KYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKE-SKSKSKGK-VDEDETQVEVPHKWR 467
K++++DKQ+KAA+RTGN+ QQ+KVKDRAKF AAKE SKSKSKGK VDE+ E P KWR
Sbjct: 418 KFDVYDKQMKAAKRTGNRGQQEKVKDRAKFTAAKEASKSKSKGKTVDEEGPAPEAPRKWR 477
Query: 468 DYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKS 527
DYSV FHFPEPTELTPPLLQLIEVSFSYPNR DFRLS+VDVGIDMGTRVAIVGPNGAGKS
Sbjct: 478 DYSVVFHFPEPTELTPPLLQLIEVSFSYPNRPDFRLSNVDVGIDMGTRVAIVGPNGAGKS 537
Query: 528 TLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQE 587
TLLNLLAGDLVP+EGE+RRSQKLRIGRYSQHFVDLLTM ETPVQYLLRLHPDQEG SKQE
Sbjct: 538 TLLNLLAGDLVPTEGEMRRSQKLRIGRYSQHFVDLLTMGETPVQYLLRLHPDQEGFSKQE 597
Query: 588 AVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDA 647
AVRAKLGK+GLPSHNHL+PI KLSGGQKARVVFTSISMS+PHILLLDEPTNHLDMQSIDA
Sbjct: 598 AVRAKLGKFGLPSHNHLSPIAKLSGGQKARVVFTSISMSKPHILLLDEPTNHLDMQSIDA 657
Query: 648 LADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
LADALDEFTGGVVLVSHDSRLISRVC +EE+SQIWVVEDGTV FPGTFE+YKEDL +EI
Sbjct: 658 LADALDEFTGGVVLVSHDSRLISRVCAEEEKSQIWVVEDGTVNFFPGTFEEYKEDLQREI 717
Query: 708 KAEVDD 713
KAEVD+
Sbjct: 718 KAEVDE 723
>UniRef100_Q8LFD8 Putative ABC transporter [Arabidopsis thaliana]
Length = 723
Score = 1125 bits (2911), Expect = 0.0
Identities = 575/726 (79%), Positives = 643/726 (88%), Gaps = 16/726 (2%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSK---KEKFSVSAMLAGMDEKADKPKKASSNKAKPKPA 57
MGKKKS+++ +TK KP KD SK KEK SVSAMLAGMD+K DKPKK SS++ K A
Sbjct: 1 MGKKKSDESAATTKVKPSGKDASKDSKKEKLSVSAMLAGMDQKDDKPKKGSSSRTKA--A 58
Query: 58 PKASAYTDDIDLPPSD---DDESEEEQEEKH-----RPDLKPLEVSIAEKELKKREKKDI 109
PK+++YTD IDLPPSD D ES+EE+ +K + + + LE+S+ +KE KKRE K+
Sbjct: 59 PKSTSYTDGIDLPPSDEEDDGESDEEERQKEARRKLKSEQRHLEISVTDKEQKKREAKER 118
Query: 110 LAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELL 169
LA AE AK+EA++DD DAFTVVIGS+ SVL+GDD ADANVKDITIE+FSV+ARGKELL
Sbjct: 119 LALQAAESAKREAMKDDHDAFTVVIGSKTSVLEGDDMADANVKDITIESFSVSARGKELL 178
Query: 170 KNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEA 229
KN SV+ISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGD+K+AL A
Sbjct: 179 KNASVRISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDEKSALNA 238
Query: 230 VVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAES 289
VVSAN EL+K+R++ LQ +SG +G + D +++D GEKLAELY++LQ++GSDAAE+
Sbjct: 239 VVSANEELVKLREEAEALQKSSSGADG-ENVDGEDDDDTGEKLAELYDRLQILGSDAAEA 297
Query: 290 QASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 349
QASKILAGLGFTKDMQ R T+SFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW
Sbjct: 298 QASKILAGLGFTKDMQVRATQSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 357
Query: 350 LEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANK 409
LEEYLCRWKKTLVVVSHDRDFLNTVC+EIIHLHD LHFYRGNFD FESGYEQRR+E NK
Sbjct: 358 LEEYLCRWKKTLVVVSHDRDFLNTVCTEIIHLHDQNLHFYRGNFDGFESGYEQRRKEMNK 417
Query: 410 KYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKE-SKSKSKGK-VDEDETQVEVPHKWR 467
K++++DKQ+KAA+RTGN+ QQ+KVKDRAKF AAKE SKSKSKGK VDE+ E P KWR
Sbjct: 418 KFDVYDKQMKAAKRTGNRGQQEKVKDRAKFTAAKEASKSKSKGKTVDEEGPAPEAPRKWR 477
Query: 468 DYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKS 527
DYSV FHFPEPTELTPPLLQLIEVSFSYPNR DFRLS+VDVGIDMGTRVAIVGPNGAGKS
Sbjct: 478 DYSVVFHFPEPTELTPPLLQLIEVSFSYPNRPDFRLSNVDVGIDMGTRVAIVGPNGAGKS 537
Query: 528 TLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQE 587
TLLNLLAGDLVP+EGE+RRSQKLRIGRYSQHFVDLLTM ETPVQYLLRLHPDQEG SKQE
Sbjct: 538 TLLNLLAGDLVPTEGEMRRSQKLRIGRYSQHFVDLLTMGETPVQYLLRLHPDQEGFSKQE 597
Query: 588 AVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDA 647
AVRAKLGK+GLPSHNHL+PI KLS GQKARVVFTSISMS+PHILLLDEPTNHLDMQSIDA
Sbjct: 598 AVRAKLGKFGLPSHNHLSPIAKLSRGQKARVVFTSISMSKPHILLLDEPTNHLDMQSIDA 657
Query: 648 LADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
LADALDEFTGGVVLVSHDSRLISRVC +EE+SQIWVVEDGTV FPGTFE+YKEDL +EI
Sbjct: 658 LADALDEFTGGVVLVSHDSRLISRVCAEEEKSQIWVVEDGTVNFFPGTFEEYKEDLQREI 717
Query: 708 KAEVDD 713
KAEVD+
Sbjct: 718 KAEVDE 723
>UniRef100_Q8S694 Putative ABC transporter [Oryza sativa]
Length = 710
Score = 959 bits (2479), Expect = 0.0
Identities = 504/732 (68%), Positives = 594/732 (80%), Gaps = 41/732 (5%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADK--PKKASSNKAKPKPAP 58
MG+K D S+ A G KD + SVSA+LA MD A K P KA+S K KP AP
Sbjct: 1 MGRK---DTSSSSSAAGGKKD----KPMSVSAILASMDAPASKAKPSKAAS-KPKPSKAP 52
Query: 59 KASAYTDDIDLPPSDDDESEEE------QEEKHRPDLKPLEVSIAEKELKKREKKDILAA 112
AS+Y DIDLPPSD++E + + + + R + ++ ++K+ KK++K++ +AA
Sbjct: 53 -ASSYMGDIDLPPSDEEEDDADLVAMATKPKAARATVDLNAIAPSQKDAKKKDKREAMAA 111
Query: 113 HVAEQAKKEALRDDRDAFTVVIGSR----ASVLDGDDGA-DANVKDITIENFSVAARGKE 167
AE AK+EALRDDRDAF+VVIG+R A +GD A D N+KDI +ENFSV+ARGKE
Sbjct: 112 AQAEAAKQEALRDDRDAFSVVIGARVAGSAGASEGDSAAADDNIKDIVLENFSVSARGKE 171
Query: 168 LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTAL 227
LLKN S++ISHG+RYGL+GPNGMGKSTLLKLL+WR++PVP++IDVLLVEQE++GD+++AL
Sbjct: 172 LLKNASLRISHGRRYGLVGPNGMGKSTLLKLLSWRQVPVPRSIDVLLVEQEIIGDNRSAL 231
Query: 228 EAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAA 287
EAVV+A+ EL +R + A L+ + + D E+LAE+YE+L L SDAA
Sbjct: 232 EAVVAADEELAALRAEQAKLE-------------ASNDADDNERLAEVYEKLNLRDSDAA 278
Query: 288 ESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAV 347
++ASKILAGLGF + MQ R TKSFSGGWRMRISLARALF+QPTLLLLDEPTNHLDLRAV
Sbjct: 279 RARASKILAGLGFDQAMQARSTKSFSGGWRMRISLARALFMQPTLLLLDEPTNHLDLRAV 338
Query: 348 LWLEEYLC-RWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRRE 406
LWLE+YLC +WKKTL+VVSHDRDFLNTVC+EIIHLHD LH YRGNFD FESGYEQ+R+E
Sbjct: 339 LWLEQYLCSQWKKTLIVVSHDRDFLNTVCNEIIHLHDKNLHVYRGNFDDFESGYEQKRKE 398
Query: 407 ANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKES-KSKSKGKV----DEDETQVE 461
N+K+E+F+KQ+KAA++TG+KA QDKVK +A A KE+ KSK KGK D+D +
Sbjct: 399 MNRKFEVFEKQMKAAKKTGSKAAQDKVKGQALSKANKEAAKSKGKGKNVANDDDDMKPAD 458
Query: 462 VPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGP 521
+P KW DY VEFHFPEPT LTPPLLQLIEV FSYPNR DF+LS VDVGIDMGTRVAIVGP
Sbjct: 459 LPQKWLDYKVEFHFPEPTLLTPPLLQLIEVGFSYPNRPDFKLSGVDVGIDMGTRVAIVGP 518
Query: 522 NGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQE 581
NGAGKSTLLNLLAGDL P++GEVRRSQKLRIGRYSQHFVDLLTM+E VQYLLRLHPDQE
Sbjct: 519 NGAGKSTLLNLLAGDLTPTKGEVRRSQKLRIGRYSQHFVDLLTMEENAVQYLLRLHPDQE 578
Query: 582 GLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLD 641
G+SK EAVRAKLGK+GLP HNHLTPIVKLSGGQKARVVFTSISMS PHILLLDEPTNHLD
Sbjct: 579 GMSKAEAVRAKLGKFGLPGHNHLTPIVKLSGGQKARVVFTSISMSHPHILLLDEPTNHLD 638
Query: 642 MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKE 701
MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDE+RS+IWVVEDGTV F GTFEDYK+
Sbjct: 639 MQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEQRSEIWVVEDGTVNKFDGTFEDYKD 698
Query: 702 DLLKEIKAEVDD 713
+LL+EIK EV++
Sbjct: 699 ELLEEIKKEVEE 710
>UniRef100_Q7XN84 OSJNBa0011F23.8 protein [Oryza sativa]
Length = 621
Score = 575 bits (1483), Expect = e-162
Identities = 327/664 (49%), Positives = 422/664 (63%), Gaps = 77/664 (11%)
Query: 70 PPSDDDESEE--EQEEKHRPDLKPLEVSIA-------EKELKKREKKDILAAHVAEQAKK 120
PPSDDD+ +E P++V+ A KE+K ++K++ A A
Sbjct: 10 PPSDDDDDDELAGAATPRAASFAPVDVNAAAPPPPPQRKEVKTKKKEERRKHEAAAAAAN 69
Query: 121 EALRDDRDAFTVVIGSRASVLDGDDGADA---NVKDITIENFSVAARGKELLKNTSVKIS 177
L DD D++ V IG R ADA NV+D+ +E+ V +G L + S++++
Sbjct: 70 --LWDDPDSYVVTIGGRVLGRGAAATADAATDNVRDVVVEDSDVWVQGVALFEGASLRVA 127
Query: 178 HGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVG-DDKTALEAVVSANVE 236
HG+RYGL+GPNG GK+TLLKLL WRK+PVP+ I V LV QE D + +E V++A+ E
Sbjct: 128 HGRRYGLVGPNGKGKTTLLKLLHWRKLPVPRGIRVTLVVQEDDNRDPRPVIEVVLAADEE 187
Query: 237 LIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILA 296
L +R + L+ ++ G +LAE+YE+L G D A ++A+KILA
Sbjct: 188 LATLRAERDQLEASSAAAAAN-----------GARLAEVYEELTQRGWDTAPARAAKILA 236
Query: 297 GLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYL-C 355
GLGF + Q RP SFSGGW RI+LA ALF+QPTLLLLDEPTNHLDLRAVLWLEEYL
Sbjct: 237 GLGFDQASQARPASSFSGGWIKRIALAGALFMQPTLLLLDEPTNHLDLRAVLWLEEYLTA 296
Query: 356 RWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFD 415
+ K TLVVVSH+ FLN +C E++HL D KLH YRG FD+F YEQ++ +A K+ E
Sbjct: 297 QCKSTLVVVSHEEGFLNAICDEVVHLQDKKLHAYRGGFDSFVGSYEQKKAKAMKESE--- 353
Query: 416 KQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHF 475
+ KAAR++G +A P KW DY+VEFHF
Sbjct: 354 RLAKAARKSGRRA----------------------------------PKKWHDYTVEFHF 379
Query: 476 PEPTELTP--PLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLL 533
PTEL PLL+L E F+ R F+LS +D + MG RVA+VGPNGAGKSTLL LL
Sbjct: 380 AAPTELAGGGPLLRLAEAGFT---RGGFQLSAIDADVSMGQRVAVVGPNGAGKSTLLKLL 436
Query: 534 AGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQY------LLRLHPDQEGLSKQE 587
AG+L P+ GE RR+ KLRIG YSQHF D L +++PVQ+ L HP + SK
Sbjct: 437 AGELTPTSGEARRNPKLRIGLYSQHFCDALPEEKSPVQHGQCHRECLDTHPHLK--SKPW 494
Query: 588 AVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDA 647
RAKL ++GL +HLT I KLSGGQKARV S+++ PH+LLLDEPTN+LDMQ+IDA
Sbjct: 495 EARAKLARFGLAKESHLTTIGKLSGGQKARVALASVALGEPHVLLLDEPTNNLDMQNIDA 554
Query: 648 LADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
LADALDEF GGVV+VSHDSRL+SRVCDDEERS +WVV+DGTVR + GTF +Y++DLL +I
Sbjct: 555 LADALDEFAGGVVIVSHDSRLVSRVCDDEERSALWVVQDGTVRPYDGTFAEYRDDLLDDI 614
Query: 708 KAEV 711
+ E+
Sbjct: 615 RKEM 618
>UniRef100_Q8S3Q0 Putative ABC transporter protein [Oryza sativa]
Length = 606
Score = 567 bits (1460), Expect = e-160
Identities = 317/626 (50%), Positives = 407/626 (64%), Gaps = 68/626 (10%)
Query: 99 KELKKREKKDILAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADA---NVKDIT 155
KE+K ++K++ A A L DD D++ V IG R ADA NV+D+
Sbjct: 33 KEVKTKKKEERRKHEAAAAAAN--LWDDPDSYVVTIGGRVLGRGAAATADAATDNVRDVV 90
Query: 156 IENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLV 215
+E+ V +G L + S++++HG+RYGL+GPNG GK+TLLKLL WRK+PVP+ I V LV
Sbjct: 91 VEDSDVWVQGVALFEGASLRVAHGRRYGLVGPNGKGKTTLLKLLHWRKLPVPRGIRVTLV 150
Query: 216 EQEVVG-DDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAE 274
QE D + +E V++A+ EL +R + L+ ++ G +LAE
Sbjct: 151 VQEDDNRDPRPVIEVVLAADEELATLRAERDQLEASSAAAAAN-----------GARLAE 199
Query: 275 LYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLL 334
+YE+L G D A ++A+KILAGLGF + Q RP SFSGGW RI+LA ALF+QPTLLL
Sbjct: 200 VYEELTQRGWDTAPARAAKILAGLGFDQASQARPASSFSGGWIKRIALAGALFMQPTLLL 259
Query: 335 LDEPTNHLDLRAVLWLEEYL-CRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNF 393
LDEPTNHLDLRAVLWLEEYL + K TLVVVSH+ FLN +C E++HL D KLH YRG F
Sbjct: 260 LDEPTNHLDLRAVLWLEEYLTAQCKSTLVVVSHEEGFLNAICDEVVHLQDKKLHAYRGGF 319
Query: 394 DAFESGYEQRRREANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKV 453
D+F YEQ++ +A K+ E + KAAR++G +A
Sbjct: 320 DSFVGSYEQKKAKAMKESE---RLAKAARKSGRRA------------------------- 351
Query: 454 DEDETQVEVPHKWRDYSVEFHFPEPTELTP--PLLQLIEVSFSYPNREDFRLSDVDVGID 511
P KW DY+VEFHF PTEL PLL+L E F+ R F+LS +D +
Sbjct: 352 ---------PKKWHDYTVEFHFAAPTELAGGGPLLRLAEAGFT---RGGFQLSAIDADVS 399
Query: 512 MGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQ 571
MG RVA+VGPNGAGKSTLL LLAG+L P+ GE RR+ KLRIG YSQHF D L +++PVQ
Sbjct: 400 MGQRVAVVGPNGAGKSTLLKLLAGELTPTSGEARRNPKLRIGLYSQHFCDALPEEKSPVQ 459
Query: 572 Y------LLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISM 625
+ L HP + SK RAKL ++GL +HLT I KLSGGQKARV S+++
Sbjct: 460 HGQCHRECLDTHPHLK--SKPWEARAKLARFGLAKESHLTTIGKLSGGQKARVALASVAL 517
Query: 626 SRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVE 685
PH+LLLDEPTN+LDMQ+IDALADALDEF GGVV+VSHDSRL+SRVCDDEERS +WVV+
Sbjct: 518 GEPHVLLLDEPTNNLDMQNIDALADALDEFAGGVVIVSHDSRLVSRVCDDEERSALWVVQ 577
Query: 686 DGTVRNFPGTFEDYKEDLLKEIKAEV 711
DGTVR + GTF +Y++DLL +I+ E+
Sbjct: 578 DGTVRPYDGTFAEYRDDLLDDIRKEM 603
>UniRef100_Q6NV15 Zgc:85667 [Brachydanio rerio]
Length = 877
Score = 553 bits (1425), Expect = e-156
Identities = 306/718 (42%), Positives = 449/718 (61%), Gaps = 50/718 (6%)
Query: 7 EDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASAYTDD 66
+D G K + K K +K + +A + ++ ++ ++ S K K PK
Sbjct: 183 QDEGAEKKDENEKKGGKKGKKAAAAAKPSKEEDDIEEKEQEKSQKKGKKEPPKKGKPARP 242
Query: 67 IDLPPSDDDESEEE-----------------QEEKHRPDLKPLEVSIAEKELKKREKKDI 109
PPSDDDE EEE QE K + + P ++++KE KK++K
Sbjct: 243 ---PPSDDDEEEEEKSDDNDIMMSAEDAIAAQEAKEKQEDDPF-ANMSKKEKKKKKKIME 298
Query: 110 LAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELL 169
VA + A+ D + SR ++L+ N DI +E FS++A GKEL
Sbjct: 299 YERQVASVRAQNAIEGDFSVSQAELSSRQAMLE-------NASDIKLERFSISAHGKELF 351
Query: 170 KNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEA 229
N + I G+RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV DD A++A
Sbjct: 352 VNADLLIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADDTPAVQA 411
Query: 230 VVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAES 289
V+ A+ +K+ ++ LQ+ ++K D + E+L ++YE+L+ +G+ AAE+
Sbjct: 412 VLKADTRRLKLLEEERQLQS------RLEKGD----DSVSERLDKVYEELRAIGAAAAEA 461
Query: 290 QASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 349
+A +ILAGL FT +MQ RPTK FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+W
Sbjct: 462 KARRILAGLSFTPEMQNRPTKKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIW 521
Query: 350 LEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANK 409
L YL WKKTL++VSHD+ FL+ VC++IIHL + KL++YRGN+ F+ Y Q+++E K
Sbjct: 522 LNNYLQSWKKTLLIVSHDQSFLDDVCTDIIHLDNQKLYYYRGNYLTFKKMYIQKQKELQK 581
Query: 410 KYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDY 469
+Y+ +K+LK + G K+ + K + K+ K K K + +E E+ + ++Y
Sbjct: 582 QYDKQEKKLKDL-KAGGKSTKQAEKQTKEALTRKQQKGKKKSQEEESHEATELLKRPKEY 640
Query: 470 SVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTL 529
+V+F FP P L+PP+L L V F Y ++ +VD GIDM +R+ IVGPNG GKSTL
Sbjct: 641 TVKFTFPNPPPLSPPILGLHSVDFGYEGQKPL-FKNVDFGIDMESRICIVGPNGVGKSTL 699
Query: 530 LNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAV 589
L LL G L P++GE+R++ +L++G ++Q + D L MDE +YL R L Q++
Sbjct: 700 LLLLTGKLNPTKGEMRKNHRLKVGFFNQQYADQLNMDEAATEYLQR----NFNLQYQDS- 754
Query: 590 RAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALA 649
R LG++GL SH H I KLSGGQKARVVF+ +S +P +L+LDEPTN+LD++SIDAL+
Sbjct: 755 RKCLGRFGLESHAHTIQISKLSGGQKARVVFSELSCRQPDVLILDEPTNNLDIESIDALS 814
Query: 650 DALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
+A++E+ G V++VSHD+RLI+ E + Q+WVVED ++ G FEDYK ++L+ +
Sbjct: 815 EAINEYKGAVIIVSHDARLIT-----ETQCQLWVVEDQSINQIDGDFEDYKREVLEAL 867
Score = 35.4 bits (80), Expect = 5.9
Identities = 32/136 (23%), Positives = 56/136 (40%), Gaps = 9/136 (6%)
Query: 2 GKKKSEDAGPSTKAKPG--------SKDVSKKEKFSVSAMLAGMDEKADKPKKASS-NKA 52
GK+ + K KPG S+ + + +K SV A +E+ P K NK
Sbjct: 59 GKQPQKKKKDRRKGKPGDDEDDDEDSEVMQRLKKLSVQASDEDEEEEVVVPAKGGKRNKG 118
Query: 53 KPKPAPKASAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAA 112
A + +DD D DD+ +++ + + + K +E K+ K + K I A
Sbjct: 119 GNIFAALSQGQSDDEDDEAGGDDDDDDDDKPRSKKSSKEVEKVAKGKKKDKPKPKPIKEA 178
Query: 113 HVAEQAKKEALRDDRD 128
EQ + +D+ +
Sbjct: 179 SEDEQDEGAEKKDENE 194
>UniRef100_Q5STZ7 OTTHUMP00000062376 [Homo sapiens]
Length = 846
Score = 547 bits (1409), Expect = e-154
Identities = 303/707 (42%), Positives = 446/707 (62%), Gaps = 32/707 (4%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKA 60
+ K SE+ P+ K K G ++ SK + + A +E+ DK ++ K PK +
Sbjct: 162 INKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNEEEDKEEEIIKEKEPPKQGKEK 221
Query: 61 SAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKK 120
+ + ++ E EEE+EE +++KE KK +K+ VA
Sbjct: 222 AKKAEQ---GSEEEGEGEEEEEEGGESKADDPYAHLSKKEKKKLKKQMEYERQVASLKAA 278
Query: 121 EALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGK 180
A +D + SR ++L+ N DI +E FS++A GKEL N + I G+
Sbjct: 279 NAAENDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVAGR 331
Query: 181 RYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKV 240
RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +K+
Sbjct: 332 RYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKL 391
Query: 241 RQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGF 300
++ LQ E+G DDT A E+L ++YE+L+ G+ AAE++A +ILAGLGF
Sbjct: 392 LEEERRLQGQL--EQG---DDT-----AAERLEKVYEELRATGAAAAEAKARRILAGLGF 441
Query: 301 TKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKT 360
+MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+KT
Sbjct: 442 DPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKT 501
Query: 361 LVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKA 420
L++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 502 LLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKE 561
Query: 421 ARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTE 480
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P
Sbjct: 562 L-KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPP 620
Query: 481 LTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPS 540
L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P+
Sbjct: 621 LSPPVLGLHGVTFGYQGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPT 679
Query: 541 EGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPS 600
GE+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL S
Sbjct: 680 HGEMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGLES 734
Query: 601 HNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVV 660
H H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G V+
Sbjct: 735 HAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVI 794
Query: 661 LVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
+VSHD+RLI+ E Q+WVVE+ +V G FEDYK ++L+ +
Sbjct: 795 VVSHDARLIT-----ETNCQLWVVEEQSVSQIDGDFEDYKREVLEAL 836
>UniRef100_Q8NE71 ATP-binding cassette, sub-family F, member 1 [Homo sapiens]
Length = 845
Score = 547 bits (1409), Expect = e-154
Identities = 303/707 (42%), Positives = 446/707 (62%), Gaps = 32/707 (4%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKA 60
+ K SE+ P+ K K G ++ SK + + A +E+ DK ++ K PK +
Sbjct: 161 INKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNEEEDKEEEIIKEKEPPKQGKEK 220
Query: 61 SAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKK 120
+ + ++ E EEE+EE +++KE KK +K+ VA
Sbjct: 221 AKKAEQ---GSEEEGEGEEEEEEGGESKADDPYAHLSKKEKKKLKKQMEYERQVASLKAA 277
Query: 121 EALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGK 180
A +D + SR ++L+ N DI +E FS++A GKEL N + I G+
Sbjct: 278 NAAENDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVAGR 330
Query: 181 RYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKV 240
RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +K+
Sbjct: 331 RYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKL 390
Query: 241 RQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGF 300
++ LQ E+G DDT A E+L ++YE+L+ G+ AAE++A +ILAGLGF
Sbjct: 391 LEEERRLQGQL--EQG---DDT-----AAERLEKVYEELRATGAAAAEAKARRILAGLGF 440
Query: 301 TKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKT 360
+MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+KT
Sbjct: 441 DPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKT 500
Query: 361 LVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKA 420
L++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 501 LLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKE 560
Query: 421 ARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTE 480
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P
Sbjct: 561 L-KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPP 619
Query: 481 LTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPS 540
L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P+
Sbjct: 620 LSPPVLGLHGVTFGYQGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPT 678
Query: 541 EGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPS 600
GE+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL S
Sbjct: 679 HGEMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGLES 733
Query: 601 HNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVV 660
H H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G V+
Sbjct: 734 HAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVI 793
Query: 661 LVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
+VSHD+RLI+ E Q+WVVE+ +V G FEDYK ++L+ +
Sbjct: 794 VVSHDARLIT-----ETNCQLWVVEEQSVSQIDGDFEDYKREVLEAL 835
>UniRef100_Q6MG08 ATP-binding cassette, sub-family F, member 1 [Rattus norvegicus]
Length = 839
Score = 543 bits (1400), Expect = e-153
Identities = 314/722 (43%), Positives = 456/722 (62%), Gaps = 52/722 (7%)
Query: 3 KKKSEDAGPSTK-AKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKAS 61
+K+ E+ P K AKP ++K +V+ G+ K K +K S KAK KP+ S
Sbjct: 143 EKEEEEEKPVLKPAKPEKNRINK----AVAEEPPGLRNKKGKEEK-SKGKAKNKPSATDS 197
Query: 62 AYTDDIDL--------PPSDDDE------SEEEQEEKHRPDLKPLE--VSIAEKELKKRE 105
DD D+ P D D+ SEEE+EEK ++K + +++KE KK +
Sbjct: 198 EGEDDEDMTKEKEPPRPGKDKDKKGAEQGSEEEKEEKEG-EVKANDPYAHLSKKEKKKLK 256
Query: 106 KKDILAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARG 165
K+ V A +D + SR ++L+ N DI +E FS++A G
Sbjct: 257 KQMDYERQVESLKAANAAENDFSVSQAEVSSRQAMLE-------NASDIKLEKFSISAHG 309
Query: 166 KELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKT 225
KEL N + I G+RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+
Sbjct: 310 KELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETP 369
Query: 226 ALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSD 285
A++AV+ A+ + +++ ++ LQ E+G DDT A EKL ++YE+L+ G+
Sbjct: 370 AVQAVLRADTKRLRLLEEEKRLQGQL--EQG---DDT-----AAEKLEKVYEELRATGAA 419
Query: 286 AAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLR 345
AAE++A +ILAGLGF +MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL
Sbjct: 420 AAEAKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLN 479
Query: 346 AVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRR 405
AV+WL YL W+KTL++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++
Sbjct: 480 AVIWLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDTQRLHYYRGNYMTFKKMYQQKQK 539
Query: 406 EANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHK 465
E K+YE +K+LK + G K+ + K + K+ K + K + +E + E+ +
Sbjct: 540 ELLKQYEKQEKKLKEL-KAGGKSTKQAEKQTKEVLTRKQQKCRRKNQDEESQDPPELLKR 598
Query: 466 WRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAG 525
R+Y+V F FP+P L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG G
Sbjct: 599 PREYTVRFTFPDPPPLSPPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVG 657
Query: 526 KSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSK 585
KSTLL LL G L P+ GE+R++ +L+IG ++Q + + L M+ETP +YL R L
Sbjct: 658 KSTLLLLLTGKLTPTNGEMRKNHRLKIGFFNQQYAEQLHMEETPTEYLQR----GFNLPY 713
Query: 586 QEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSI 645
Q+A R LG++GL SH H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SI
Sbjct: 714 QDA-RKCLGRFGLESHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESI 772
Query: 646 DALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLK 705
DAL +A++E+ G V++VSHD+RLI+ E Q+WVVE+ +V G F+DYK ++L+
Sbjct: 773 DALGEAINEYKGAVIVVSHDARLIT-----ETNCQLWVVEEQSVSQIDGDFDDYKREVLE 827
Query: 706 EI 707
+
Sbjct: 828 AL 829
>UniRef100_Q66J73 MGC81714 protein [Xenopus laevis]
Length = 888
Score = 540 bits (1390), Expect = e-152
Identities = 304/719 (42%), Positives = 444/719 (61%), Gaps = 43/719 (5%)
Query: 3 KKKSEDAGPSTKAKPGSKDV---------SKKEKFSVSAM--LAGMDEKADKPKKASSNK 51
K+KS+ P KA P D SKK K S + + +D A++ KK
Sbjct: 187 KEKSKGKKPVPKAAPVDSDAEDEKKPEKNSKKSKPSQKPVPKASSVDSDAEEEKKPEKGS 246
Query: 52 AKPKPAPKASAYTD---DIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKD 108
K KP+ D D S++ +EE E + + + ++++KE KK++K
Sbjct: 247 KKSKPSQNRFGALDIEEDRVSGESNEGSGDEEGETEEKTAAEDPFANLSKKEKKKKKKML 306
Query: 109 ILAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKEL 168
VA K A +D + SR ++L+ N DI +E FS++A GKEL
Sbjct: 307 EYEKQVATMKKATAAENDFSVSQAELSSRQAMLE-------NASDIKLEKFSISAHGKEL 359
Query: 169 LKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALE 228
N + + G+RYGL+GPNG GK+TLLK +A + + +P NIDVLL EQEV+ D+ A++
Sbjct: 360 FVNADLYVVAGRRYGLVGPNGKGKTTLLKHIANKALNIPPNIDVLLCEQEVIADETPAVQ 419
Query: 229 AVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAE 288
AV+ A+ + +K+ ++ LQ ++K D N A E+L ++YE+L+ G+ +AE
Sbjct: 420 AVLKADKKRLKLLEEEKRLQ------ARLEKGDDN----AAERLEKVYEELRASGAASAE 469
Query: 289 SQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVL 348
++A +ILAGL FT +MQ R T+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+
Sbjct: 470 AKARRILAGLSFTPEMQDRETRRFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVI 529
Query: 349 WLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREAN 408
WL YL WKKTL++VSHD+ FL+ VC++I+HL KL++YRGN+ F+ Y+Q+++E
Sbjct: 530 WLNNYLQGWKKTLLIVSHDQGFLDDVCTDIMHLDSQKLYYYRGNYMTFKKMYQQKQKEMQ 589
Query: 409 KKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRD 468
K+Y+ +K+LK + G A+Q + K + K+ K + K ED E+ + ++
Sbjct: 590 KQYDKQEKKLKDLKAGGKSAKQAE-KQTKEVLTRKQQKCQKKNPDKEDHEPAELLKRPKE 648
Query: 469 YSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKST 528
Y+V+F FP P L+PP+L L V F Y ++ +++ GIDM +RV IVGPNG GKST
Sbjct: 649 YTVKFTFPNPPPLSPPILGLHGVDFGYSGQKPL-FRNLEFGIDMDSRVCIVGPNGVGKST 707
Query: 529 LLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEA 588
LL LL G L P++GE+R++ +L+IG ++Q + D LTM+ET +YL R L Q+A
Sbjct: 708 LLLLLTGKLTPTKGEMRKNHRLKIGFFNQQYADQLTMEETATEYLQR----NFNLPYQDA 763
Query: 589 VRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDAL 648
R LG++GL SH H I KLSGGQKARVVF + P +L+LDEPTN+LD++SIDAL
Sbjct: 764 -RKCLGRFGLESHAHTIQICKLSGGQKARVVFAELCCREPDVLILDEPTNNLDIESIDAL 822
Query: 649 ADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
A+A++E+ G V+ VSHD+RLI+ E +WVVED TV G F+DYK ++L+ +
Sbjct: 823 AEAVNEYKGAVITVSHDARLIT-----ETNCHLWVVEDRTVNQIDGDFDDYKREVLESL 876
>UniRef100_Q5TM60 ATP-binding cassette, sub-family F (GCN20), member 1 [Macaca
mulatta]
Length = 807
Score = 537 bits (1383), Expect = e-151
Identities = 303/705 (42%), Positives = 444/705 (62%), Gaps = 49/705 (6%)
Query: 3 KKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASA 62
++ E+ P AKP ++K +L G K +K +K K KP K +A
Sbjct: 142 EEDEEEKHPPKPAKPEKNRINKAVSEEQQPVLKGKKGKEEK------SKGKAKPQNKFAA 195
Query: 63 YTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKKEA 122
++ ++DE EE +EK P + AE+ +R+ + AA+ AE
Sbjct: 196 LDNE------EEDEEEEIIKEKEPPKQGKEKARKAEQMEYERQVASLKAANAAE------ 243
Query: 123 LRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGKRY 182
+D + SR ++L+ N DI +E FS++A GKEL N + I G+RY
Sbjct: 244 --NDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVAGRRY 294
Query: 183 GLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQ 242
GL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +K+ +
Sbjct: 295 GLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLE 354
Query: 243 KVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTK 302
+ LQ E+G DDT A E+L ++YE+L+ G+ AAE++A +ILAGLGF
Sbjct: 355 EERLLQGQL--EQG---DDT-----AAERLEKVYEELRATGAAAAEAKARRILAGLGFDP 404
Query: 303 DMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLV 362
+MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+KTL+
Sbjct: 405 EMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLL 464
Query: 363 VVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAAR 422
+VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 465 IVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKEL- 523
Query: 423 RTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELT 482
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P L+
Sbjct: 524 KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLS 583
Query: 483 PPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEG 542
PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P+ G
Sbjct: 584 PPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHG 642
Query: 543 EVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHN 602
E+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL SH
Sbjct: 643 EMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGLESHA 697
Query: 603 HLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLV 662
H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G V++V
Sbjct: 698 HTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVV 757
Query: 663 SHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
SHD+RLI+ E Q+WVVE+ +V G FEDYK ++L+ +
Sbjct: 758 SHDARLIT-----ETNCQLWVVEEQSVSQIDGDFEDYKREVLEAL 797
>UniRef100_Q5RL55 ATP-binding cassette, sub-family F (GCN20), member 1 [Mus musculus]
Length = 836
Score = 536 bits (1382), Expect = e-151
Identities = 307/708 (43%), Positives = 447/708 (62%), Gaps = 49/708 (6%)
Query: 15 AKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASAYTDDIDL----- 69
AKP ++K +V+ G+ K K +K S KAK KPA S ++ D
Sbjct: 153 AKPEKNRINK----AVAEEPPGLRSKKGKEEK-SKGKAKSKPAAADSEGEEEEDTAKEKE 207
Query: 70 PPS---DDDESEEEQ-----EEKHRPDLKPLE--VSIAEKELKKREKKDILAAHVAEQAK 119
PP D D+ E EQ +E+ DLK + ++++KE KK +K+ V
Sbjct: 208 PPQQGKDRDKKEAEQGSGEEKEEKEGDLKANDPYANLSKKEKKKLKKQMDYERQVESLKA 267
Query: 120 KEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHG 179
A +D + SR ++L+ N DI +E FS++A GKEL N + I G
Sbjct: 268 ANAAENDFSVSQAEVSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVAG 320
Query: 180 KRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIK 239
+RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + ++
Sbjct: 321 RRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLR 380
Query: 240 VRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLG 299
+ ++ LQ E+G DDT A EKL ++YE+L+ G+ AAE++A +ILAGLG
Sbjct: 381 LLEEERRLQGQL--EQG---DDT-----AAEKLEKVYEELRATGAAAAEAKARRILAGLG 430
Query: 300 FTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKK 359
F +MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+K
Sbjct: 431 FDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRK 490
Query: 360 TLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLK 419
TL++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 491 TLLIVSHDQGFLDDVCTDIIHLDTQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLK 550
Query: 420 AARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPT 479
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P
Sbjct: 551 EL-KAGGKSTKQAEKQTKEVLTRKQQKCRRKNQDEESQEPPELLKRPKEYTVRFTFPDPP 609
Query: 480 ELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVP 539
L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P
Sbjct: 610 PLSPPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTP 668
Query: 540 SEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLP 599
+ GE+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL
Sbjct: 669 TNGEMRKNHRLKIGFFNQQYAEQLHMEETPTEYLQR----SFNLPYQDA-RKCLGRFGLE 723
Query: 600 SHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGV 659
SH H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++++ G V
Sbjct: 724 SHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINDYKGAV 783
Query: 660 VLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
++VSHD+RLI+ E Q+WVVE+ V G F+DYK ++L+ +
Sbjct: 784 IVVSHDARLIT-----ETNCQLWVVEEQGVSQIDGDFDDYKREVLEAL 826
>UniRef100_Q8NE71-2 Splice isoform 2 of Q8NE71 [Homo sapiens]
Length = 807
Score = 536 bits (1382), Expect = e-151
Identities = 302/705 (42%), Positives = 444/705 (62%), Gaps = 49/705 (6%)
Query: 3 KKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASA 62
+++ E+ P AKP ++K L G K +K +K K KP K +A
Sbjct: 142 EEEEEEKHPPKPAKPEKNRINKAVSEEQQPALKGKKGKEEK------SKGKAKPQNKFAA 195
Query: 63 YTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKKEA 122
++ ++D+ EE +EK P + AE+ +R+ + AA+ AE
Sbjct: 196 LDNE------EEDKEEEIIKEKEPPKQGKEKAKKAEQMEYERQVASLKAANAAE------ 243
Query: 123 LRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGKRY 182
+D + SR ++L+ N DI +E FS++A GKEL N + I G+RY
Sbjct: 244 --NDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVAGRRY 294
Query: 183 GLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQ 242
GL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +K+ +
Sbjct: 295 GLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLE 354
Query: 243 KVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTK 302
+ LQ E+G DDT A E+L ++YE+L+ G+ AAE++A +ILAGLGF
Sbjct: 355 EERRLQGQL--EQG---DDT-----AAERLEKVYEELRATGAAAAEAKARRILAGLGFDP 404
Query: 303 DMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLV 362
+MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+KTL+
Sbjct: 405 EMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLL 464
Query: 363 VVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAAR 422
+VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 465 IVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKEL- 523
Query: 423 RTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELT 482
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P L+
Sbjct: 524 KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLS 583
Query: 483 PPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEG 542
PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P+ G
Sbjct: 584 PPVLGLHGVTFGYQGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHG 642
Query: 543 EVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHN 602
E+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL SH
Sbjct: 643 EMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGLESHA 697
Query: 603 HLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLV 662
H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G V++V
Sbjct: 698 HTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVV 757
Query: 663 SHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
SHD+RLI+ E Q+WVVE+ +V G FEDYK ++L+ +
Sbjct: 758 SHDARLIT-----ETNCQLWVVEEQSVSQIDGDFEDYKREVLEAL 797
>UniRef100_Q767L0 ATP-binding cassette, sub-family F, member 1 [Sus scrofa]
Length = 807
Score = 534 bits (1376), Expect = e-150
Identities = 302/709 (42%), Positives = 445/709 (62%), Gaps = 58/709 (8%)
Query: 3 KKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPK----KASSNKAKPKPAP 58
+++ E+ P AKP ++K A E+ PK K +K K KP
Sbjct: 143 EEEEEEKHPPKPAKPEKNRINK----------AVSQEQQPGPKGRKGKEEKSKGKAKPQN 192
Query: 59 KASAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQA 118
K +A D+ +++ EEE +EK P + AE+ +R+ + AA+ AE
Sbjct: 193 KFAALDDE-------EEQDEEEIKEKEPPKQGKEKAKKAEQMEYERQVASLKAANAAE-- 243
Query: 119 KKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISH 178
+D + SR ++L+ N DI +E FS++A GKEL N + I
Sbjct: 244 ------NDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVA 290
Query: 179 GKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELI 238
G+RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +
Sbjct: 291 GRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRL 350
Query: 239 KVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGL 298
K+ ++ LQ E+G DDT A ++L ++YE+L+ G+ AAE++A +ILAGL
Sbjct: 351 KLLEEERRLQGQL--EQG---DDT-----AADRLEKVYEELRATGAAAAEAKARRILAGL 400
Query: 299 GFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWK 358
GF +MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+
Sbjct: 401 GFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWR 460
Query: 359 KTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQL 418
KTL++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+L
Sbjct: 461 KTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKL 520
Query: 419 KAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEP 478
K + G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P
Sbjct: 521 KEL-KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDP 579
Query: 479 TELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLV 538
L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L
Sbjct: 580 PPLSPPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLT 638
Query: 539 PSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGL 598
P+ GE+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL
Sbjct: 639 PTRGEMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGL 693
Query: 599 PSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGG 658
SH H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G
Sbjct: 694 ESHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGA 753
Query: 659 VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
V++VSHD+RLI+ E Q+WVVE+ +V G F+DYK ++L+ +
Sbjct: 754 VIVVSHDARLIT-----ETNCQLWVVEEQSVSQIDGDFDDYKREVLEAL 797
>UniRef100_Q7YR37 ATP-binding cassette, sub-family F, member 1 [Pan troglodytes]
Length = 807
Score = 534 bits (1376), Expect = e-150
Identities = 301/705 (42%), Positives = 443/705 (62%), Gaps = 49/705 (6%)
Query: 3 KKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASA 62
+++ E+ P AKP ++K L G K +K +K K KP K +A
Sbjct: 142 EEEEEEKHPPKPAKPEKNRINKAVSEEQQPALKGKKGKEEK------SKGKAKPQNKFAA 195
Query: 63 YTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKKEA 122
++ ++D+ EE +EK P + AE+ +R+ + AA+ AE
Sbjct: 196 LDNE------EEDKEEEIIKEKEPPKQGKEKAKKAEQMEYERQVASLKAANAAE------ 243
Query: 123 LRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGKRY 182
+D + SR ++L+ N DI +E FS++A GKEL N + I +RY
Sbjct: 244 --NDFSVSQAEMSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVASRRY 294
Query: 183 GLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQ 242
GL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +K+ +
Sbjct: 295 GLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLE 354
Query: 243 KVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTK 302
+ LQ E+G DDT A E+L ++YE+L+ G+ AAE++A +ILAGLGF
Sbjct: 355 EERRLQGQL--EQG---DDT-----AAERLEKVYEELRATGAAAAEAKARRILAGLGFDP 404
Query: 303 DMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLV 362
+MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+KTL+
Sbjct: 405 EMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLL 464
Query: 363 VVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAAR 422
+VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+LK
Sbjct: 465 IVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKEL- 523
Query: 423 RTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELT 482
+ G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P L+
Sbjct: 524 KAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLS 583
Query: 483 PPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEG 542
PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L P+ G
Sbjct: 584 PPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHG 642
Query: 543 EVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHN 602
E+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL SH
Sbjct: 643 EMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQR----GFNLPYQDA-RKCLGRFGLESHA 697
Query: 603 HLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLV 662
H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++E+ G V++V
Sbjct: 698 HTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVV 757
Query: 663 SHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
SHD+RLI+ E Q+WVVE+ +V G FEDYK ++L+ +
Sbjct: 758 SHDARLIT-----ETNCQLWVVEEQSVSQIDGDFEDYKREVLEAL 797
>UniRef100_Q6P542 ATP-binding cassette, sub-family F, member 1 [Mus musculus]
Length = 837
Score = 534 bits (1375), Expect = e-150
Identities = 306/709 (43%), Positives = 447/709 (62%), Gaps = 50/709 (7%)
Query: 15 AKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASAYTDDID------ 68
AKP ++K +V+ G+ K K +K S KAK KPA S ++ +
Sbjct: 153 AKPEKNRINK----AVAEEPPGLRSKKGKEEK-SKGKAKSKPAAADSEGEEEEEDTAKEK 207
Query: 69 LPPS---DDDESEEEQ-----EEKHRPDLKPLE--VSIAEKELKKREKKDILAAHVAEQA 118
PP D D+ E EQ +E+ DLK + ++++KE KK +K+ V
Sbjct: 208 EPPQQGKDRDKKEAEQGSGEEKEEKEGDLKANDPYANLSKKEKKKLKKQMDYERQVESLK 267
Query: 119 KKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISH 178
A +D + SR ++L+ N DI +E FS++A GKEL N + I
Sbjct: 268 AANAAENDFSVSQAEVSSRQAMLE-------NASDIKLEKFSISAHGKELFVNADLYIVA 320
Query: 179 GKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELI 238
G+RYGL+GPNG GK+TLLK +A R + +P NIDVLL EQEVV D+ A++AV+ A+ + +
Sbjct: 321 GRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRL 380
Query: 239 KVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGL 298
++ ++ LQ E+G DDT A EKL ++YE+L+ G+ AAE++A +ILAGL
Sbjct: 381 RLLEEERRLQGQL--EQG---DDT-----AAEKLEKVYEELRATGAAAAEAKARRILAGL 430
Query: 299 GFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWK 358
GF +MQ RPT+ FSGGWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL W+
Sbjct: 431 GFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWR 490
Query: 359 KTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQL 418
KTL++VSHD+ FL+ VC++IIHL +LH+YRGN+ F+ Y+Q+++E K+YE +K+L
Sbjct: 491 KTLLIVSHDQGFLDDVCTDIIHLDTQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKL 550
Query: 419 KAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEP 478
K + G K+ + K + K+ K + K + +E + E+ + ++Y+V F FP+P
Sbjct: 551 KEL-KAGGKSTKQAEKQTKEVLTRKQQKCRRKNQDEESQEPPELLKRPKEYTVRFTFPDP 609
Query: 479 TELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLV 538
L+PP+L L V+F Y ++ ++D GIDM +R+ IVGPNG GKSTLL LL G L
Sbjct: 610 PPLSPPVLGLHGVTFGYEGQKPL-FKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLT 668
Query: 539 PSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGL 598
P+ GE+R++ +L+IG ++Q + + L M+ETP +YL R L Q+A R LG++GL
Sbjct: 669 PTNGEMRKNHRLKIGFFNQQYAEQLHMEETPTEYLQR----SFNLPYQDA-RKCLGRFGL 723
Query: 599 PSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGG 658
SH H I KLSGGQKARVVF ++ P +L+LDEPTN+LD++SIDAL +A++++ G
Sbjct: 724 ESHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINDYKGA 783
Query: 659 VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
V++VSHD+RLI+ E Q+WVVE+ V G F+DYK ++L+ +
Sbjct: 784 VIVVSHDARLIT-----ETNCQLWVVEEQGVSQIDGDFDDYKREVLEAL 827
>UniRef100_Q9VYW4 CG1703-PA [Drosophila melanogaster]
Length = 901
Score = 533 bits (1373), Expect = e-150
Identities = 293/716 (40%), Positives = 436/716 (59%), Gaps = 46/716 (6%)
Query: 17 PGSKDVSKKEKFSVSAMLAGMDEKADKPKK-----------------ASSNKAKPKPAPK 59
P + +KE+ S A E+ +P+K S + P+P PK
Sbjct: 202 PNDSEEEEKEQLSNKAAKESKKEQQKQPEKEVEEQKVEQVLSEKVAAVSLKEKTPEPVPK 261
Query: 60 ASAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAK 119
A + P D+E++ E + K + E + KE KK++K+ + K
Sbjct: 262 AEPEPES---EPQPDEEADAEVDSKFEESKEAKEKKLTHKEKKKQKKQQEYERQMELMTK 318
Query: 120 KEAL--RDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKIS 177
K D + FT+ S+ G A DI IENF+++A+G +L N ++ I+
Sbjct: 319 KGGAGHSDLDNNFTM---SQVQKSAGQKAALEQAVDIKIENFTISAKGNDLFVNANLLIA 375
Query: 178 HGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVEL 237
HG+RYGL+GPNG GK+TLL+ +A R +P NIDVLL EQEVV DKTA+ ++ A+V
Sbjct: 376 HGRRYGLVGPNGHGKTTLLRHIATRAFAIPPNIDVLLCEQEVVATDKTAINTILEADVRR 435
Query: 238 IKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAG 297
++ +K +L+ ++ + D T +EE L + + +L+ +G+ +AE++A +ILAG
Sbjct: 436 TEMLKKADELE-----KQFVAGDLTVQEE-----LNDTFAELKAIGAYSAEARARRILAG 485
Query: 298 LGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRW 357
LGF+K+MQ RPT FSGGWRMR+SLARAL+++PTLL+LDEPTNHLDL AV+WL+ YL W
Sbjct: 486 LGFSKEMQDRPTNKFSGGWRMRVSLARALYLEPTLLMLDEPTNHLDLNAVIWLDNYLQGW 545
Query: 358 KKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQ 417
KKTL++VSHD+ FL+ VC+EIIHL KL +Y+GN+ F+ Y Q+RRE K+YE +K+
Sbjct: 546 KKTLLIVSHDQSFLDNVCNEIIHLDQKKLQYYKGNYSMFKKMYVQKRREMIKEYEKQEKR 605
Query: 418 LKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPE 477
L+ + G + + K + +E + K DEDE E+ + ++Y V+F FPE
Sbjct: 606 LRELKAHGQSKKAAEKKQKESLTRKQEKNKSKQQKQDEDEGPQELLARPKEYIVKFRFPE 665
Query: 478 PTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDL 537
P++L PP+L + V+F++P+++ + VD GID+ +RVAIVGPNG GKST L LL G+L
Sbjct: 666 PSQLQPPILGVHNVTFAFPSQKPLFIK-VDFGIDLTSRVAIVGPNGVGKSTFLKLLLGEL 724
Query: 538 VPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYG 597
P EGE R++ +L +GR+ QH + LT +E+ +YL RL E R LG +G
Sbjct: 725 EPQEGEQRKNHRLHVGRFDQHSGEHLTAEESAAEYLQRLFN-----LPHEKARKALGSFG 779
Query: 598 LPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTG 657
L SH H + LSGGQKARV + +S P +L+LDEPTN+LD++SIDALA+A++E+ G
Sbjct: 780 LVSHAHTIKMKDLSGGQKARVALAELCLSAPDVLILDEPTNNLDIESIDALAEAINEYEG 839
Query: 658 GVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEIKAEVDD 713
GV++VSHD RLI E ++V+ED T+ G F+DY++++L + V++
Sbjct: 840 GVIIVSHDERLIR-----ETGCTLYVIEDQTINEIVGEFDDYRKEVLDSLGEVVNN 890
>UniRef100_UPI0000360FB0 UPI0000360FB0 UniRef100 entry
Length = 613
Score = 530 bits (1364), Expect = e-149
Identities = 286/633 (45%), Positives = 416/633 (65%), Gaps = 32/633 (5%)
Query: 75 DESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKKEALRDDRDAFTVVI 134
+++ EQ +K D P ++++KE KK++K+ +A + AL D +
Sbjct: 6 EDAIAEQAKKEEED--PFH-NLSKKEKKKKKKQMEYERQLASVRAQNALEGDFSISQAEL 62
Query: 135 GSRASVLDGDDGADANVKDITIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKST 194
SR ++L+ N DI +E FS++A GKEL N + I G+RYGL+GPNG GK+T
Sbjct: 63 SSRQAMLE-------NASDIKLERFSISAHGKELFVNADLLIVAGRRYGLVGPNGKGKTT 115
Query: 195 LLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGE 254
LLK +A R + +P NIDVLL EQEVV DD A++AV+ A+ +K+ + LQ
Sbjct: 116 LLKHIANRALSIPPNIDVLLCEQEVVADDTPAVQAVLKADTRRLKLLEDERRLQ------ 169
Query: 255 EGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSG 314
+DK E++ E+L ++YE+L +G+ AAE++A +ILAGL FT +MQ RPTK FSG
Sbjct: 170 ARLDKG----EDNVSEELEKVYEELAAIGAAAAEAKARRILAGLSFTPEMQNRPTKRFSG 225
Query: 315 GWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTV 374
GWRMR+SLARALF++PTLL+LDEPTNHLDL AV+WL YL WKKTL++VSHD+ FL+ V
Sbjct: 226 GWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWKKTLLIVSHDQSFLDEV 285
Query: 375 CSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAARRTGNKAQQDKVK 434
C++IIH + KL++Y GN+ F+ Y Q+++E K+Y+ +K+LK + G K+ + K
Sbjct: 286 CTDIIHXDNQKLYYYGGNYLTFKKMYVQKQKEMQKQYDKQEKKLKDL-KAGGKSTKQAEK 344
Query: 435 DRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFS 494
K+ K + KG+ +E + E+ + R+Y+V+F FP P L+PP+L L V F
Sbjct: 345 QTKDALTRKQQKGRKKGQEEESQDAQELLKRPREYTVKFTFPNPPPLSPPVLGLHSVDFD 404
Query: 495 YPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGR 554
Y +R+ +VD GIDM +R+ IVGPNG GKSTLL LL G L P++GE+R++ +L++G
Sbjct: 405 Y-DRQKPLFKNVDFGIDMESRICIVGPNGVGKSTLLLLLTGKLNPTKGEMRKNHRLKVGF 463
Query: 555 YSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQ 614
++Q + D L MDET +YL R L Q++ R LG++GL SH H I KLSGGQ
Sbjct: 464 FNQQYADQLNMDETATEYLTR----NFNLPYQDS-RKCLGRFGLESHAHTIQISKLSGGQ 518
Query: 615 KARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCD 674
KARVVF +S +P +L+LDEPTN+LD++SIDAL+ A++++ G V++VSHD+RLI+
Sbjct: 519 KARVVFAELSCRQPDVLILDEPTNNLDIESIDALSQAINDYKGAVIIVSHDARLIT---- 574
Query: 675 DEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
E + Q+WVVED T+ G F+DYK ++L+ +
Sbjct: 575 -ETQCQLWVVEDKTINQIDGNFDDYKREVLESL 606
>UniRef100_Q7Q040 ENSANGP00000016545 [Anopheles gambiae str. PEST]
Length = 587
Score = 504 bits (1298), Expect = e-141
Identities = 256/557 (45%), Positives = 382/557 (67%), Gaps = 24/557 (4%)
Query: 153 DITIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDV 212
DI IENF+++A+G +L N ++ I++G+ YGL+GPNG GK+TLL+ +A R +P NIDV
Sbjct: 36 DIKIENFTISAKGNDLFVNANLLIANGRHYGLVGPNGHGKTTLLRHIANRAFAIPPNIDV 95
Query: 213 LLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQN-IASGEEGMDKDDTNEEEDAGEK 271
LL EQEVV D+ +A++ V+ A+V+ + ++ +L+ + SG+ + +K
Sbjct: 96 LLCEQEVVADETSAVDTVLKADVKRTALLKECKELEEAVESGKIELQ-----------DK 144
Query: 272 LAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPT 331
L E+Y +L+ +G+D+AE +A +ILAGLGF+++MQ RPT +FSGGWRMR+SLARALF++PT
Sbjct: 145 LQEVYNELKAIGADSAEPRARRILAGLGFSREMQNRPTNAFSGGWRMRVSLARALFIEPT 204
Query: 332 LLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRG 391
LLLLDEPTNHLDL AV+WL+ YL WKKTL++VSHD+ FL+ VC+EIIHL + KL++Y+G
Sbjct: 205 LLLLDEPTNHLDLNAVIWLDNYLQGWKKTLLIVSHDQSFLDNVCNEIIHLDNKKLYYYKG 264
Query: 392 NFDAFESGYEQRRREANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKE-SKSKSK 450
N+ F+ Y Q+R+E K+YE +++LK + G + + K + +E ++K++
Sbjct: 265 NYTMFKKMYVQKRKEMIKEYEKQERRLKDMKAHGQSKKAAEKKQKENLTRKQEKGRTKNQ 324
Query: 451 GKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGI 510
++D+ VE+ K ++Y V+F FP+P L PP+L L F++P ++ + D GI
Sbjct: 325 KPGEDDDGPVELLSKPKEYIVKFSFPDPPPLQPPILGLHNCHFNFPKQKPLFVG-ADFGI 383
Query: 511 DMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPV 570
D+ +RVAIVGPNG GKST L LL G+L P +GE +R+ +LRIGR+ QH + LT +ETP
Sbjct: 384 DLSSRVAIVGPNGVGKSTFLKLLVGELDPVQGEAKRNHRLRIGRFDQHSGEHLTAEETPA 443
Query: 571 QYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHI 630
+YL RL E R +LG +GL SH H + LSGGQKARV + ++ P +
Sbjct: 444 EYLQRLFN-----LPYEKARKQLGTFGLASHAHTIKMKDLSGGQKARVALAELCLNAPDV 498
Query: 631 LLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVR 690
L+LDEPTN+LD++SIDALA+A++E+ GGV++VSHD RLI E ++V+ED T+
Sbjct: 499 LILDEPTNNLDIESIDALAEAINEYKGGVIIVSHDERLIR-----ETECTLFVIEDQTIN 553
Query: 691 NFPGTFEDYKEDLLKEI 707
G F+DY++++L +
Sbjct: 554 EVDGDFDDYRKEVLDSL 570
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,208,736,861
Number of Sequences: 2790947
Number of extensions: 53858347
Number of successful extensions: 386524
Number of sequences better than 10.0: 26896
Number of HSP's better than 10.0 without gapping: 22203
Number of HSP's successfully gapped in prelim test: 4788
Number of HSP's that attempted gapping in prelim test: 271468
Number of HSP's gapped (non-prelim): 88642
length of query: 713
length of database: 848,049,833
effective HSP length: 135
effective length of query: 578
effective length of database: 471,271,988
effective search space: 272395209064
effective search space used: 272395209064
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC136504.2


