

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.10 + phase: 0
(449 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
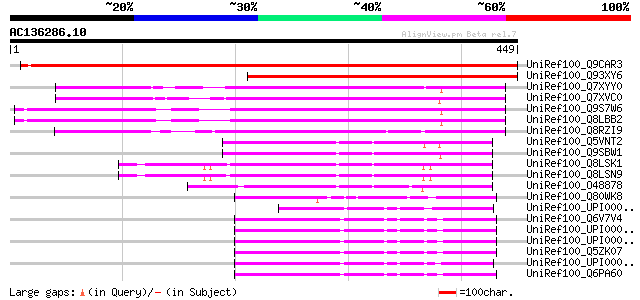
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9CAR3 Hypothetical protein T17F3.17 [Arabidopsis thal... 590 e-167
UniRef100_Q93XY6 Hypothetical protein At1g69800; T17F3.17 [Arabi... 322 1e-86
UniRef100_Q7XYY0 AKIN gamma [Medicago truncatula] 267 4e-70
UniRef100_Q7XVC0 OSJNBa0072D21.10 protein [Oryza sativa] 259 8e-68
UniRef100_Q9S7W6 AKIN gamma [Arabidopsis thaliana] 254 3e-66
UniRef100_Q8LBB2 Hypothetical protein [Arabidopsis thaliana] 252 1e-65
UniRef100_Q8RZI9 OJ1485_B09.7 protein [Oryza sativa] 238 2e-61
UniRef100_Q5VNT2 Putative YZ1 [Oryza sativa] 104 4e-21
UniRef100_Q9SBW1 Putative transcription factor X2 [Oryza sativa] 101 4e-20
UniRef100_Q8LSK1 YZ1 [Zea mays] 91 9e-17
UniRef100_Q8LSN9 YZ1 [Zea mays] 89 2e-16
UniRef100_O48878 Similar to rice gene X [Sorghum bicolor] 83 1e-14
UniRef100_Q80WK8 AMP-activated protein kinase gamma subunit [Mus... 69 3e-10
UniRef100_UPI000029BC63 UPI000029BC63 UniRef100 entry 67 1e-09
UniRef100_Q6V7V4 AMP-activated protein kinase gamma 2 non-cataly... 66 2e-09
UniRef100_UPI00003AB87D UPI00003AB87D UniRef100 entry 65 3e-09
UniRef100_UPI00003AB87C UPI00003AB87C UniRef100 entry 65 3e-09
UniRef100_Q5ZK07 Hypothetical protein [Gallus gallus] 65 3e-09
UniRef100_UPI000024A194 UPI000024A194 UniRef100 entry 65 4e-09
UniRef100_Q6PA60 MGC68503 protein [Xenopus laevis] 65 4e-09
>UniRef100_Q9CAR3 Hypothetical protein T17F3.17 [Arabidopsis thaliana]
Length = 447
Score = 590 bits (1520), Expect = e-167
Identities = 298/441 (67%), Positives = 369/441 (83%), Gaps = 3/441 (0%)
Query: 10 EQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKVVEILAD 69
++E++++ +S C+ YFE +QSRK LP++LQETL +FA IPVSSFP VPGG+V+EI A+
Sbjct: 9 DKEIKSA--VSSCEAYFEKVQSRKNLPKSLQETLNSAFAGIPVSSFPQVPGGRVIEIQAE 66
Query: 70 TPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELAAVALS 129
TPV EAVKILS+S IL+APV + D DWR+RYLGIIDYS+IILWV+ESAELAA+ALS
Sbjct: 67 TPVSEAVKILSDSKILSAPVINTDHESSLDWRERYLGIIDYSSIILWVLESAELAAIALS 126
Query: 130 AGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAA 189
A +ATAAGVGAG VGALG ALG TGP A AGL AAAVGAAV GGVAA++ + KDAP AA
Sbjct: 127 ATSATAAGVGAGAVGALGVAALGMTGPVAAAGLAAAAVGAAVAGGVAAERGIGKDAPTAA 186
Query: 190 NNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPV 249
+ LG+DFY+VILQEEPFKSTTVR+ILKS+RWAPF+PV+ S+ML+V+LLLSKYRLRNVPV
Sbjct: 187 DKLGKDFYEVILQEEPFKSTTVRTILKSFRWAPFLPVSTESSMLSVMLLLSKYRLRNVPV 246
Query: 250 IEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELI 309
I+ G+ DI N++TQSAV+ GLEGC+GRDWFD I+A P++DLGLPFMS ++VISI+S ELI
Sbjct: 247 IKTGEPDIKNYVTQSAVVHGLEGCKGRDWFDHISALPISDLGLPFMSPNEVISIESEELI 306
Query: 310 LEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIV 369
LEAFK MRDN IGGLPVVEG K IVGN+S+RDIRYLLL+PE+FSNFR LTV F KI
Sbjct: 307 LEAFKRMRDNNIGGLPVVEGLNKKIVGNISMRDIRYLLLQPEVFSNFRQLTVKSFATKIA 366
Query: 370 SASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNG-QDQVVGVITLRDVISCFI 428
+A E G ITC+PD+TL SVI++LAS+S+HR+Y G ++++ GVITLRDVISCF+
Sbjct: 367 TAGEEYGLAIPAITCRPDSTLGSVINSLASRSVHRVYVAAGDENELYGVITLRDVISCFV 426
Query: 429 TEPDYHFDDYYGFAVKEMLNQ 449
+EP +F++ GF+VKEMLN+
Sbjct: 427 SEPPNYFENCLGFSVKEMLNR 447
>UniRef100_Q93XY6 Hypothetical protein At1g69800; T17F3.17 [Arabidopsis thaliana]
Length = 248
Score = 322 bits (825), Expect = 1e-86
Identities = 159/240 (66%), Positives = 201/240 (83%), Gaps = 1/240 (0%)
Query: 211 VRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGL 270
VR+ILKS+RWAPF+PV+ S+ML+V+LLLSKYRLRNVPVI+ G+ DI N++TQSAV+ GL
Sbjct: 9 VRTILKSFRWAPFLPVSTESSMLSVMLLLSKYRLRNVPVIKTGEPDIKNYVTQSAVVHGL 68
Query: 271 EGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGP 330
EGC+GRDWFD I+A P++DLGLPFMS ++VISI+S ELILEAFK MRDN IGGLPVVEG
Sbjct: 69 EGCKGRDWFDHISALPISDLGLPFMSPNEVISIESEELILEAFKRMRDNNIGGLPVVEGL 128
Query: 331 AKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRPITCKPDATL 390
K IVGN+S+RDIRYLLL+PE+FSNFR LTV F KI +A E G ITC+PD+TL
Sbjct: 129 NKKIVGNISMRDIRYLLLQPEVFSNFRQLTVKSFATKIATAGEEYGLAIPAITCRPDSTL 188
Query: 391 QSVIHTLASQSIHRIYTVNG-QDQVVGVITLRDVISCFITEPDYHFDDYYGFAVKEMLNQ 449
SVI++LAS+S+HR+Y G ++++ GVITLRDVISCF++EP +F++ GF+VKEMLN+
Sbjct: 189 GSVINSLASRSVHRVYVAAGDENELYGVITLRDVISCFVSEPPNYFENCLGFSVKEMLNR 248
>UniRef100_Q7XYY0 AKIN gamma [Medicago truncatula]
Length = 420
Score = 267 bits (683), Expect = 4e-70
Identities = 147/404 (36%), Positives = 234/404 (57%), Gaps = 36/404 (8%)
Query: 41 ETLTDSFAKIPVSSFPGVPGGKVVEILADTPVGEAVKILSESNILAAPVKDPDAGIGSDW 100
+ L F IPVS+FP + +EI +D + EAVKIL+ NIL+APV D DA + W
Sbjct: 36 DKLNACFESIPVSAFPLPAKNQEIEIKSDATLAEAVKILARHNILSAPVVDVDAPEDATW 95
Query: 101 RDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIA 160
DRY+GI++++ I++W++ +E + S T T+A AIA A G
Sbjct: 96 IDRYIGIVEFAGIVVWILHQSEPPS-PRSPSTPTSA----------SAIAAAANGNTFAR 144
Query: 161 GLTAAAVGAAVVGGVAADKTMAKDAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRW 220
L A +G+A +F++ + E +K+T V+ I ++RW
Sbjct: 145 ELEALGLGSAA-------------------TTSGNFFEDLTSSELYKNTKVQDISGTFRW 185
Query: 221 APFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFD 280
APF+ + ++++ LT+LLLLSKY++++VPV++ G I N ITQ AVI L C G WF+
Sbjct: 186 APFLALERSNSFLTMLLLLSKYKMKSVPVVDLGSGTIDNIITQPAVIHMLAECAGLQWFE 245
Query: 281 CIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSI 340
+ ++D+GLP ++ ++I + +E +L+AFK MR ++GG+PV++ T VGN+S+
Sbjct: 246 SWGTKELSDVGLPLVTPKQIIKVYEDEPVLQAFKEMRKKRVGGVPVIKRGGTTAVGNISL 305
Query: 341 RDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRP-----ITCKPDATLQSVIH 395
RD+++LL PEI+ ++R +TV DF+ V + E K P ITCK D T++ +I
Sbjct: 306 RDVQFLLTAPEIYHDYRTITVKDFLTS-VRSYLEKNKNAFPMSSEFITCKRDCTVKELIQ 364
Query: 396 TLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDYHFDDYY 439
L + IHR+Y V+ + G+ITLRD+IS + EP +F D++
Sbjct: 365 LLDKEQIHRVYVVDDDGNLEGLITLRDIISRLVHEPHGYFGDFF 408
>UniRef100_Q7XVC0 OSJNBa0072D21.10 protein [Oryza sativa]
Length = 425
Score = 259 bits (663), Expect = 8e-68
Identities = 138/403 (34%), Positives = 236/403 (58%), Gaps = 25/403 (6%)
Query: 41 ETLTDSFAKIPVSSFPGVPGGKVVEILADTPVGEAVKILSESNILAAPVKDPDAGIGSDW 100
E L F I V+SFP G +V+EI ++ + + V+ILS++ IL+AP+++ DA + W
Sbjct: 32 EKLNSCFEDIAVASFPRPLGSQVIEIPSNASLADTVEILSKNKILSAPIRNVDAPEDASW 91
Query: 101 RDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIA 160
D+Y+GI++++ I +W++ +E AA +AG+A + V A V LG+ T +
Sbjct: 92 IDKYIGIVEFAGIAMWLLYQSEAAANG-TAGSAVGSPV-ANLVSRLGSFTFRRTSSGRV- 148
Query: 161 GLTAAAVGAAVVGGVAADKTMAKDAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRW 220
+ T ++ + A+ +G F++ + E +K+T V I S+RW
Sbjct: 149 -----------------ETTTDPESDETAS-VGGSFFETLTSSEFYKNTKVGDISGSFRW 190
Query: 221 APFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFD 280
APF+ + + LT+LLLLSKYR++++PV++ G I N ITQS+V+ L C G WF+
Sbjct: 191 APFLALQTSDTFLTMLLLLSKYRMKSLPVVDIGGDKIENIITQSSVVHMLAECVGLPWFE 250
Query: 281 CIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSI 340
+ +++LGLP M K++ + ++ +L+AF++MR+ +GGLPV++ +GN+SI
Sbjct: 251 SWGTKKLSELGLPLMKPCKLVKVNEDQPVLKAFQLMREKGVGGLPVMDTSGTKAIGNISI 310
Query: 341 RDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT----RPITCKPDATLQSVIHT 396
RD++YLL P I+ ++R +T DF+ + E + + ITC+ D ++ +I
Sbjct: 311 RDVQYLLTAPNIYKDYRTITAKDFLTAVRQHLQEQHEASPLLGSVITCRRDDEVKDIILK 370
Query: 397 LASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDYHFDDYY 439
L S+ IHRIY ++ + GVITLRD+IS + EP ++F D++
Sbjct: 371 LDSEKIHRIYVIDDKGNTEGVITLRDIISKLVHEPRHYFGDFF 413
>UniRef100_Q9S7W6 AKIN gamma [Arabidopsis thaliana]
Length = 424
Score = 254 bits (649), Expect = 3e-66
Identities = 147/440 (33%), Positives = 242/440 (54%), Gaps = 45/440 (10%)
Query: 5 QNMAHEQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKVV 64
+++ H +V S+ +K E + +K + E L F IPVS+FP + +
Sbjct: 13 ESLGHRSDV--SSPEAKLGMRVEDLWDEQKPQLSPNEKLNACFESIPVSAFPLSSDSQDI 70
Query: 65 EILADTPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELA 124
EI +DT + EAV+ LS+ +L+APV D DA + W DRY+GI+++ I++W++ E
Sbjct: 71 EIRSDTSLAEAVQTLSKFKVLSAPVVDVDAPEDASWIDRYIGIVEFPGIVVWLLHQLEPP 130
Query: 125 AVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKD 184
+ A V A + T G +A G
Sbjct: 131 SPRSPA------------VAASNGFSHDFTTDVLDNGDSAVTSG---------------- 162
Query: 185 APQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRL 244
+F++V+ E +K+T VR I ++RWAPF+ + K ++ LT+LLLLSKY++
Sbjct: 163 ----------NFFEVLTSSELYKNTKVRDISGTFRWAPFLALQKENSFLTMLLLLSKYKM 212
Query: 245 RNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQ 304
+++PV++ G A I N ITQS VI L C G WF+ + ++++GLP MS D +I I
Sbjct: 213 KSIPVVDLGVAKIENIITQSGVIHMLAECAGLLWFEDWGIKTLSEVGLPIMSKDHIIKIY 272
Query: 305 SNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDF 364
+E +L+AFK+MR +IGG+PV+E ++ VGN+S+RD+++LL PEI+ ++R++T +F
Sbjct: 273 EDEPVLQAFKLMRRKRIGGIPVIERNSEKPVGNISLRDVQFLLTAPEIYHDYRSITTKNF 332
Query: 365 MKKIVSASYESGKVTRP-----ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVIT 419
+ + + G + P I C + TL+ +I L ++ IHRIY V+ + G+IT
Sbjct: 333 LVSVREHLEKCGDTSAPIMSGVIACTKNHTLKELILMLDAEKIHRIYVVDDFGNLEGLIT 392
Query: 420 LRDVISCFITEPDYHFDDYY 439
LRD+I+ + EP +F D++
Sbjct: 393 LRDIIARLVHEPSGYFGDFF 412
>UniRef100_Q8LBB2 Hypothetical protein [Arabidopsis thaliana]
Length = 424
Score = 252 bits (644), Expect = 1e-65
Identities = 146/440 (33%), Positives = 242/440 (54%), Gaps = 45/440 (10%)
Query: 5 QNMAHEQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKVV 64
+++ H +V S+ +K E + +K + E L F IPVS+FP + +
Sbjct: 13 ESLGHRSDV--SSPEAKLGMRVEDLWDEQKPQLSPNEKLNACFESIPVSAFPLSSDSQDI 70
Query: 65 EILADTPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELA 124
EI +DT + EAV+ LS+ +L+APV D DA + W DRY+GI+++ I++W++ E
Sbjct: 71 EIRSDTSLAEAVQTLSKFKVLSAPVVDVDAPEDASWIDRYIGIVEFPGIVVWLLHQLEPP 130
Query: 125 AVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKD 184
+ A V A + T G +A G
Sbjct: 131 SPRSPA------------VAASNGFSHDFTTDVLDNGDSAVTSG---------------- 162
Query: 185 APQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRL 244
+F++V+ E +++T VR I ++RWAPF+ + K ++ LT+LLLLSKY++
Sbjct: 163 ----------NFFEVLTSSELYENTKVRDISGTFRWAPFLALQKENSFLTMLLLLSKYKM 212
Query: 245 RNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQ 304
+++PV++ G A I N ITQS VI L C G WF+ + ++++GLP MS D +I I
Sbjct: 213 KSIPVVDLGVAKIENIITQSGVIHMLAECAGLLWFEDWGIKTLSEVGLPIMSKDHIIKIY 272
Query: 305 SNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDF 364
+E +L+AFK+MR +IGG+PV+E ++ VGN+S+RD+++LL PEI+ ++R++T +F
Sbjct: 273 EDEPVLQAFKLMRRKRIGGIPVIERNSEKPVGNISLRDVQFLLTAPEIYHDYRSITTKNF 332
Query: 365 MKKIVSASYESGKVTRP-----ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVIT 419
+ + + G + P I C + TL+ +I L ++ IHRIY V+ + G+IT
Sbjct: 333 LVSVREHLEKCGDTSAPIMSGVIACTKNHTLKELILMLDAEKIHRIYVVDEFGNLEGLIT 392
Query: 420 LRDVISCFITEPDYHFDDYY 439
LRD+I+ + EP +F D++
Sbjct: 393 LRDIIARLVHEPSGYFGDFF 412
>UniRef100_Q8RZI9 OJ1485_B09.7 protein [Oryza sativa]
Length = 435
Score = 238 bits (608), Expect = 2e-61
Identities = 137/402 (34%), Positives = 227/402 (56%), Gaps = 35/402 (8%)
Query: 40 QETLTDSFAKIPVSSFPGV-PGGKVVEILADTPVGEAVKILSESNILAAPVKDPDAGIGS 98
++ L F IPV+SFP G +VVEI +D + EAV ILS I+ APV++ DA +
Sbjct: 55 RDRLNSCFDAIPVASFPHTFDGAQVVEIPSDATLAEAVDILSRHRIITAPVRNVDAPDDA 114
Query: 99 DWRDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAA 158
W DRY+G+++++ I +W++ +E AA A A +GA +
Sbjct: 115 SWIDRYIGVVEFAGIAVWLLHQSEAAA-------ARADDLGADEL--------------- 152
Query: 159 IAGLTAAAVGAAVVGGVAADKTMAKDAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSY 218
AA +G + G AA + A D Q+A + + + + F T V+ I S+
Sbjct: 153 -----AAKLGTVALEGAAAAR--APDQQQSAEGAVAEAFGALPSSDLFNKTKVKDISGSF 205
Query: 219 RWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDW 278
RWAPF+ + + LT+LLLLSKYR++++PV++ G+ I N ITQ+AV+ L C G W
Sbjct: 206 RWAPFLALQSSDTFLTMLLLLSKYRMKSLPVVDIGEGTISNVITQAAVVHMLAECAGLHW 265
Query: 279 FDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNL 338
F+ A+ + +LGLP + +++ ++ +E L+AF++MR +GG+PVV+ K G++
Sbjct: 266 FEDWGAKSLTELGLPMIRPSRLVKVRHDEPALKAFRLMRKRGVGGIPVVDHAGKP-TGSI 324
Query: 339 SIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESG-KVTRPITCKPDATLQSVIHTL 397
I+D+++LL + ++R LT +F I +A SG K +TCK + +++ +I L
Sbjct: 325 MIKDVKHLLASSDANRDYRTLTAQEF---IANARQSSGEKQMNIVTCKKEESIKEIIFKL 381
Query: 398 ASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDYHFDDYY 439
++ RIY V+ Q + G+ITLRD+I+ + EP +F D++
Sbjct: 382 DAEKRQRIYVVDEQGNLDGLITLRDIIAKLVYEPPGYFGDFF 423
>UniRef100_Q5VNT2 Putative YZ1 [Oryza sativa]
Length = 407
Score = 104 bits (260), Expect = 4e-21
Identities = 70/260 (26%), Positives = 125/260 (47%), Gaps = 24/260 (9%)
Query: 189 ANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN-V 247
A + DF + Q T + + KS+ W PF PV + + +LL SK+R N V
Sbjct: 116 AGDKNSDFLTSLKQHPQIAETKIAWLAKSFLWEPFFPVRSHDTLFHAMLLFSKHRRINVV 175
Query: 248 PVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNE 307
PV+E + ++ F+TQ+AV++ L G +W D IA + +++ F + K +S+ S++
Sbjct: 176 PVVELMNSSVIGFVTQNAVMELLLSSSGLEWLDKIADKQLSE--FRFANTTKPVSVYSDQ 233
Query: 308 LILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMK- 366
+ +A I+ +I G+ VV+ ++G++ D+ LL +F N + L+ +F+K
Sbjct: 234 TLADALHILSKEKI-GVAVVDRKTSCLIGSIQCSDLYQLLDDSSLFRNRKTLSAEEFVKL 292
Query: 367 --KIVSASYESGKVT-----------------RPITCKPDATLQSVIHTLASQSIHRIYT 407
K S E+ + P+T + TL+ + L + +
Sbjct: 293 KSKDEDISTENSSASGGQNVLSLRTGQRITAGLPVTNRKSDTLKQAMEKLTASRSSCSFI 352
Query: 408 VNGQDQVVGVITLRDVISCF 427
V+ +V GV+T RD+IS F
Sbjct: 353 VDEHGRVEGVVTARDIISVF 372
>UniRef100_Q9SBW1 Putative transcription factor X2 [Oryza sativa]
Length = 453
Score = 101 bits (252), Expect = 4e-20
Identities = 66/242 (27%), Positives = 119/242 (48%), Gaps = 6/242 (2%)
Query: 189 ANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRN-V 247
A + DF + Q T + + KS+ W PF PV + + +LL SK+R N V
Sbjct: 180 AGDKNSDFLTSLKQHPQIAETKIAWLAKSFLWEPFFPVRSHDTLFHAMLLFSKHRRINVV 239
Query: 248 PVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNE 307
PV+E + ++ F+TQ+AV++ L G +W D IA + +++ F + K +S+ S++
Sbjct: 240 PVVELMNSSVIGFVTQNAVMELLLSSSGLEWLDKIADKQLSE--FRFANTTKPVSVYSDQ 297
Query: 308 LILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKK 367
+ +A I+ +I G+ VV+ ++G++ D+ LL +F N +
Sbjct: 298 TLADALHILSKEKI-GVAVVDRKTSCLIGSIQCSDLYQLLDDSSLFRNRNTENSSASGGQ 356
Query: 368 IVSASYESGKVTR--PITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVIS 425
V + ++T P+T + TL+ + L + + V+ +V GV+T RD+IS
Sbjct: 357 NVLSLRTGQRITAGLPVTNRKSDTLKQAMEKLTASRSSCSFIVDEHGRVEGVVTARDIIS 416
Query: 426 CF 427
F
Sbjct: 417 VF 418
>UniRef100_Q8LSK1 YZ1 [Zea mays]
Length = 407
Score = 90.5 bits (223), Expect = 9e-17
Identities = 83/363 (22%), Positives = 154/363 (41%), Gaps = 42/363 (11%)
Query: 97 GSDWRDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGP 156
G W + +D+ + S+ A+ +A A + +GAL A+
Sbjct: 21 GECWSEALKSFLDHIPV------SSVPGALQPTASPAVEVKLHGSVLGALDAMYSSNAAG 74
Query: 157 AAIAGLTAAAVGAAV---VGGVA-------ADKTMAKDAPQAANNLGEDFYKVILQEEPF 206
A I + +++G V +G V A + + K + ++ DF + Q
Sbjct: 75 AVIIDVVHSSLGKYVDRDIGFVEFSSLVLWALEELGKVESEPTDSTS-DFLSTLKQHHQI 133
Query: 207 KSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNV-PVIEPGKADIVNFITQSA 265
T + + K + W PF PV + + +LL SK+ NV PV+E + ++ F+TQ A
Sbjct: 134 AETKIAWLAKLFLWEPFFPVRTHDTLFHAMLLFSKHHRLNVAPVVESMNSSVIGFVTQDA 193
Query: 266 VIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLP 325
V++ L G +W D IA + +++ F + K + + S++ + + I+ + G+
Sbjct: 194 VMELLLQSSGLEWLDKIADKQLSE--FRFANVRKPVLVYSDQTLADGLHILSKEKT-GVA 250
Query: 326 VVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFM-------KKIVSA------- 371
V++ ++G+L D+ L +FS T+ + + +K +A
Sbjct: 251 VIDRKTSRLIGSLQCSDLYLFLDDSTLFSKRTTTTLEELISLNNKTDRKCSTAENSCAPG 310
Query: 372 -------SYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVI 424
S +SG V P T TL+ + L + + V+ +V GV+T RD+I
Sbjct: 311 RNILALRSRQSGMVGLPATNLESDTLKQAMEKLTTLRSSCSFIVDEHGRVQGVVTTRDII 370
Query: 425 SCF 427
S F
Sbjct: 371 SVF 373
>UniRef100_Q8LSN9 YZ1 [Zea mays]
Length = 406
Score = 89.4 bits (220), Expect = 2e-16
Identities = 83/363 (22%), Positives = 154/363 (41%), Gaps = 43/363 (11%)
Query: 97 GSDWRDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGP 156
G W + +D+ + S+ A+ +A A + +GAL A+
Sbjct: 21 GECWSEALKSFLDHIPV------SSVPGALQPTASPAVEVKLHGSVLGALDAMYSSNAAG 74
Query: 157 AAIAGLTAAAVGAAV---VGGVA-------ADKTMAKDAPQAANNLGEDFYKVILQEEPF 206
A I + +++G V +G V A + + K ++ ++ DF + Q
Sbjct: 75 AVIIDVVHSSLGKYVDRDIGFVEFSSLVLWALEELGKVESESTDST--DFLSTLKQHHQI 132
Query: 207 KSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNV-PVIEPGKADIVNFITQSA 265
T + + K + W PF PV + + +LL SK+ NV PV+E + + F+TQ A
Sbjct: 133 AETKIAWLAKLFLWEPFFPVRTHDTLFHAMLLFSKHHRLNVAPVVESMNSSAIGFVTQDA 192
Query: 266 VIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLP 325
V++ L G +W D IA + +++ F + K + + S++ + + I+ + G+
Sbjct: 193 VMELLLQSSGLEWLDKIADKQLSE--FRFANVRKPVLVYSDQTLADGLHILSKEKT-GVA 249
Query: 326 VVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFM-------KKIVSA------- 371
V++ ++G+L D+ L +FS T+ + + +K +A
Sbjct: 250 VIDRKTSRLIGSLQCSDLYLFLDDSTLFSKRTTTTLEELISLNNKTDRKCSTAENSSAPG 309
Query: 372 -------SYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVI 424
S +SG V P T TL+ + L + + V+ +V GV+T RD+I
Sbjct: 310 RNILALRSRQSGMVGLPATNLESDTLKQAMEKLTALRSSCSFIVDEHGRVQGVVTTRDII 369
Query: 425 SCF 427
S F
Sbjct: 370 SVF 372
>UniRef100_O48878 Similar to rice gene X [Sorghum bicolor]
Length = 895
Score = 83.2 bits (204), Expect = 1e-14
Identities = 64/276 (23%), Positives = 123/276 (44%), Gaps = 16/276 (5%)
Query: 158 AIAGLTAAAVGAAVVGGVAADKTMAKDAPQAANNLG-EDFYKVILQEEPFKSTTVRSILK 216
++ G A + G V D + ++G +F ++L + + + K
Sbjct: 595 SVLGALDAMYSSNAAGAVIVDVVQSSFGKYVDRDIGFVEFSSLVLW-----ALEIAWLAK 649
Query: 217 SYRWAPFVPVAKNSAMLTVLLLLSKYRLRNV-PVIEPGKADIVNFITQSAVIQGLEGCRG 275
+ W PF PV + +LL SK+ NV PV+E + ++ F+TQ+AV++ L G
Sbjct: 650 LFLWEPFFPVRTQDTLFHTMLLFSKHHRLNVAPVVESINSSVIGFVTQNAVMELLLQSSG 709
Query: 276 RDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIV 335
+W D IA + +++ F + K + + S++ + + I+ ++ G+ V++ ++
Sbjct: 710 LEWLDKIADKQLSE--FRFANVRKPVLVYSDQTLADGLHILSKEKM-GVAVIDRKTSCLI 766
Query: 336 GNLSIRDIRYLLLKPEIFSNFRNLTVMDF----MKKIVSASYESGKVTRPITCKPDATLQ 391
G++ D+ L +FS + T D + + + V P T TL+
Sbjct: 767 GSIQCSDLYLFLDDSSLFS--KRTTAEDSSPPGQNILALRNRQPSMVGLPATNLKSDTLK 824
Query: 392 SVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCF 427
+ L + + V+ Q V GV+T RD+IS F
Sbjct: 825 QAMEKLTTSRSSCSFIVDEQGHVEGVVTTRDIISVF 860
>UniRef100_Q80WK8 AMP-activated protein kinase gamma subunit [Mus musculus]
Length = 490
Score = 68.9 bits (167), Expect = 3e-10
Identities = 54/235 (22%), Positives = 108/235 (44%), Gaps = 16/235 (6%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R I + P V ++ N ++ + L K R+ +PV++P ++
Sbjct: 261 IYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNRIHRLPVLDPVSGTVLY 320
Query: 260 FITQSAVIQGLE---GCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIM 316
+T +++ L R F C R + DLG+ D + +++ +L A I
Sbjct: 321 ILTHKRLLKFLHIFGALLPRPSFLC---RTIQDLGIGTFR-DLAVVLETAP-VLTALDIF 375
Query: 317 RDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESG 376
D ++ LPVV +VG S D+ +L + N +++V + +++
Sbjct: 376 VDRRVSALPVVNESGSQVVGLYSRFDVIHLAAQQTY--NHLDMSVGEALRQRTLC----- 428
Query: 377 KVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
+ ++C+P +L VI +A + +HR+ V+ ++GV++L D++ + P
Sbjct: 429 -LEGVLSCQPHESLGEVIDRIAREQVHRLVLVDETQHLLGVVSLSDILQALVLSP 482
>UniRef100_UPI000029BC63 UPI000029BC63 UniRef100 entry
Length = 329
Score = 67.0 bits (162), Expect = 1e-09
Identities = 45/190 (23%), Positives = 93/190 (48%), Gaps = 11/190 (5%)
Query: 239 LSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSAD 298
L K+++ +PVI+P ++++ +T +++ L + +P+ DL + S
Sbjct: 149 LLKHKIHRLPVIDPESGNVLHILTHKRILRFLHIFGKQIPKPAFTGKPIQDLAIGTFS-- 206
Query: 299 KVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRN 358
V ++Q + +A I + ++ LPVV+ K +V S D+ + L + N +
Sbjct: 207 NVATVQETATLYDALSIFVERRVSALPVVDEQGK-VVALYSRFDV--INLAAQRTYNHLD 263
Query: 359 LTVMDFMKKIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVI 418
+T+ + +++ +G V I C P+ TL +VI + +HR+ V+ D V G+I
Sbjct: 264 MTMQEAVRR------RTGFVEGVIKCYPEETLDTVIERIVEAEVHRLVLVDVADVVKGII 317
Query: 419 TLRDVISCFI 428
+L D++ +
Sbjct: 318 SLSDLLQAMV 327
>UniRef100_Q6V7V4 AMP-activated protein kinase gamma 2 non-catalytic subunit [Rattus
norvegicus]
Length = 326
Score = 66.2 bits (160), Expect = 2e-09
Identities = 54/232 (23%), Positives = 106/232 (45%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ ++++ + L K ++ +PVI+P + +
Sbjct: 95 IYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNKIHRLPVIDPISGNALY 154
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + I N I++A I +
Sbjct: 155 ILTHKRILKFLQLFMSDMPKPAFMKQNLDELGIG--TYHNIAFIHPNTPIIKALNIFVER 212
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
+I LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 213 RISALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALQH--RSQYFEGVVK 267
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C TL++++ + +HR+ VN D +VG+I+L D++ I P
Sbjct: 268 ----CSKLETLETIVDRIVRAEVHRLVVVNEADSIVGIISLSDILQALILTP 315
>UniRef100_UPI00003AB87D UPI00003AB87D UniRef100 entry
Length = 449
Score = 65.5 bits (158), Expect = 3e-09
Identities = 52/232 (22%), Positives = 106/232 (45%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ ++++ + L K ++ +PVI+P + +
Sbjct: 218 IYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNKIHRLPVIDPVSGNALY 277
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + I + I++A I +
Sbjct: 278 ILTHKRILKFLQLFMSEMPKPAFMKKNLDELGIG--TYHNIAFIHPDTPIIKALNIFVER 335
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
+I LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 336 RISALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALQH--RSQYFEGVVK 390
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C TL++++ + +HR+ VN D +VG+I+L D++ + P
Sbjct: 391 ----CSMLETLETIVDRIVKAEVHRLVVVNEADSIVGIISLSDILQALVLTP 438
>UniRef100_UPI00003AB87C UPI00003AB87C UniRef100 entry
Length = 568
Score = 65.5 bits (158), Expect = 3e-09
Identities = 52/232 (22%), Positives = 106/232 (45%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ ++++ + L K ++ +PVI+P + +
Sbjct: 337 IYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNKIHRLPVIDPVSGNALY 396
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + I + I++A I +
Sbjct: 397 ILTHKRILKFLQLFMSEMPKPAFMKKNLDELGIG--TYHNIAFIHPDTPIIKALNIFVER 454
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
+I LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 455 RISALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALQH--RSQYFEGVVK 509
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C TL++++ + +HR+ VN D +VG+I+L D++ + P
Sbjct: 510 ----CSMLETLETIVDRIVKAEVHRLVVVNEADSIVGIISLSDILQALVLTP 557
>UniRef100_Q5ZK07 Hypothetical protein [Gallus gallus]
Length = 328
Score = 65.5 bits (158), Expect = 3e-09
Identities = 52/232 (22%), Positives = 106/232 (45%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ ++++ + L K ++ +PVI+P + +
Sbjct: 97 IYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNKIHRLPVIDPVSGNALY 156
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + I + I++A I +
Sbjct: 157 ILTHKRILKFLQLFMSEMPKPAFMKKNLDELGIG--TYHNIAFIHPDTPIIKALNIFVER 214
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
+I LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 215 RISALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALQH--RSQYFEGVVK 269
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C TL++++ + +HR+ VN D +VG+I+L D++ + P
Sbjct: 270 ----CSMLETLETIVDRIVKAEVHRLVVVNEADSIVGIISLSDILQALVLTP 317
>UniRef100_UPI000024A194 UPI000024A194 UniRef100 entry
Length = 324
Score = 65.1 bits (157), Expect = 4e-09
Identities = 50/229 (21%), Positives = 109/229 (46%), Gaps = 11/229 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V ++ N+++ + L K+++ +PVI+P + +
Sbjct: 100 IYELEEHKIETWREVYLQDSFKPLVSISPNASLYDAVSSLLKHKIHRLPVIDPLTGNALY 159
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ ++ + +L + + D + + S+ + A I +
Sbjct: 160 ILTHKRILKFLKLFISEIPKPAFLSQTLEELNIG--TFDNIAVVHSDTPLYAALGIFVEQ 217
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
++ LPVV+ + +V S D+ + L E N ++TV ++ + Y G
Sbjct: 218 RVSALPVVDENGR-VVDIYSKFDV--INLAAEKTYNNMDITVTKALQH--RSQYFEGV-- 270
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFI 428
+TC+ TLQ++I+ L +HR+ V+ Q+ V G+++L D++ +
Sbjct: 271 --LTCRASETLQAIINRLVEAEVHRLVIVDEQEVVKGIVSLXDILQALV 317
>UniRef100_Q6PA60 MGC68503 protein [Xenopus laevis]
Length = 558
Score = 65.1 bits (157), Expect = 4e-09
Identities = 51/232 (21%), Positives = 106/232 (44%), Gaps = 11/232 (4%)
Query: 200 ILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVN 259
I + E K T R + + P V + ++++ + L K ++ +PVI+P + +
Sbjct: 327 IYELEEHKIETWRELYLQETFKPLVNIFPDASLFDAVYSLIKNKIHRLPVIDPVSGNALY 386
Query: 260 FITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDN 319
+T +++ L+ + + +LG+ + + IQ + I++A I +
Sbjct: 387 ILTHKRILKFLQLFVSEMPKPAFMKQNLEELGIG--TYHNIAFIQPHTPIIKALNIFVER 444
Query: 320 QIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVT 379
++ LPVV+ K +V S D+ + L E N ++TV ++ + Y G V
Sbjct: 445 RVSALPVVDESGK-VVDIYSKFDV--INLAAEKTYNNLDITVTQALEH--RSQYFEGVVK 499
Query: 380 RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEP 431
C TL++++ + +HR+ V+ D +VG+I+L D++ + P
Sbjct: 500 ----CSKPETLETIVDRIVKAEVHRLVVVDEADSIVGIISLSDILQALVLSP 547
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 692,660,201
Number of Sequences: 2790947
Number of extensions: 27928303
Number of successful extensions: 119513
Number of sequences better than 10.0: 1052
Number of HSP's better than 10.0 without gapping: 340
Number of HSP's successfully gapped in prelim test: 727
Number of HSP's that attempted gapping in prelim test: 112747
Number of HSP's gapped (non-prelim): 5263
length of query: 449
length of database: 848,049,833
effective HSP length: 131
effective length of query: 318
effective length of database: 482,435,776
effective search space: 153414576768
effective search space used: 153414576768
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC136286.10


