

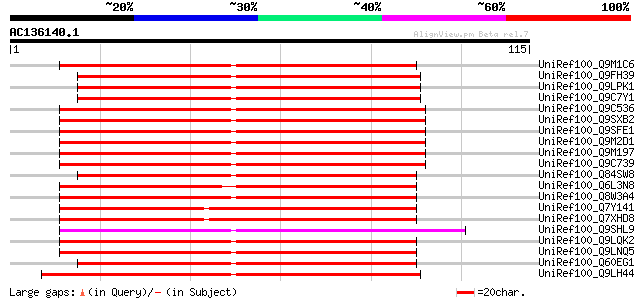
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.1 - phase: 0
(115 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M1C6 Hypothetical protein T2O9.150 [Arabidopsis thal... 98 4e-20
UniRef100_Q9FH39 Copia-type polyprotein [Arabidopsis thaliana] 81 6e-15
UniRef100_Q9LPK1 F6N18.1 [Arabidopsis thaliana] 81 6e-15
UniRef100_Q9C7Y1 Copia-type polyprotein, putative; 28768-32772 [... 81 6e-15
UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis t... 78 4e-14
UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana] 78 4e-14
UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana] 78 4e-14
UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana] 78 4e-14
UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [... 78 4e-14
UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis t... 75 4e-13
UniRef100_Q84SW8 Gag-pol polyprotein [Oryza sativa] 75 5e-13
UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum] 74 8e-13
UniRef100_Q8W3A4 Putative gag-pol polyprotein [Oryza sativa] 74 1e-12
UniRef100_Q7Y141 Putative polyprotein [Oryza sativa] 72 3e-12
UniRef100_Q7XHD8 Putative copia-type polyprotein [Oryza sativa] 72 4e-12
UniRef100_Q9SHL9 Putative retroelement pol polyprotein [Arabidop... 71 5e-12
UniRef100_Q9LQK2 F28G4.2 protein [Arabidopsis thaliana] 71 5e-12
UniRef100_Q9LNQ5 F1L3.20 [Arabidopsis thaliana] 71 5e-12
UniRef100_Q60EG1 Putative polyprotein [Oryza sativa] 70 9e-12
UniRef100_Q9LH44 Copia-like retrotransposable element [Arabidops... 69 2e-11
>UniRef100_Q9M1C6 Hypothetical protein T2O9.150 [Arabidopsis thaliana]
Length = 1339
Score = 98.2 bits (243), Expect = 4e-20
Identities = 48/79 (60%), Positives = 59/79 (73%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V+ TP EAWS +KP V FR+FG I Y HIP +R K LDD S KCV LGVSE+SKA+R
Sbjct: 643 VEGMTPEEAWSGRKPVVEYFRVFGCIGYVHIPDQKRSK-LDDKSKKCVFLGVSEESKAWR 701
Query: 72 FFDRIIKKIVVSEDVAFDE 90
+D ++KKIV+S+DV FDE
Sbjct: 702 LYDPVMKKIVISKDVVFDE 720
>UniRef100_Q9FH39 Copia-type polyprotein [Arabidopsis thaliana]
Length = 1334
Score = 80.9 bits (198), Expect = 6e-15
Identities = 41/76 (53%), Positives = 53/76 (68%), Gaps = 1/76 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP E WS KP V RIFG +AYA +P+ +R K LD+ S KCV+ GVS++SKAYR +D
Sbjct: 648 TPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIK-LDEKSIKCVMFGVSKESKAYRLYDP 706
Query: 76 IIKKIVVSEDVAFDEQ 91
KI++S DV FDE+
Sbjct: 707 ATGKILISRDVQFDEE 722
>UniRef100_Q9LPK1 F6N18.1 [Arabidopsis thaliana]
Length = 1207
Score = 80.9 bits (198), Expect = 6e-15
Identities = 41/76 (53%), Positives = 53/76 (68%), Gaps = 1/76 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP E WS KP V RIFG +AYA +P+ +R K LD+ S KCV+ GVS++SKAYR +D
Sbjct: 553 TPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIK-LDEKSIKCVMFGVSKESKAYRLYDP 611
Query: 76 IIKKIVVSEDVAFDEQ 91
KI++S DV FDE+
Sbjct: 612 ATGKILISRDVQFDEE 627
>UniRef100_Q9C7Y1 Copia-type polyprotein, putative; 28768-32772 [Arabidopsis
thaliana]
Length = 1334
Score = 80.9 bits (198), Expect = 6e-15
Identities = 41/76 (53%), Positives = 53/76 (68%), Gaps = 1/76 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP E WS KP V RIFG +AYA +P+ +R K LD+ S KCV+ GVS++SKAYR +D
Sbjct: 648 TPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIK-LDEKSIKCVMFGVSKESKAYRLYDP 706
Query: 76 IIKKIVVSEDVAFDEQ 91
KI++S DV FDE+
Sbjct: 707 ATGKILISRDVQFDEE 722
>UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1320
Score = 78.2 bits (191), Expect = 4e-14
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana]
Length = 1352
Score = 78.2 bits (191), Expect = 4e-14
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana]
Length = 1291
Score = 78.2 bits (191), Expect = 4e-14
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 614 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 672
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 673 LYNPDTKKTIISRNIVFDEEG 693
>UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana]
Length = 1352
Score = 78.2 bits (191), Expect = 4e-14
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [Arabidopsis
thaliana]
Length = 1272
Score = 78.2 bits (191), Expect = 4e-14
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1352
Score = 75.1 bits (183), Expect = 4e-13
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +K V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>UniRef100_Q84SW8 Gag-pol polyprotein [Oryza sativa]
Length = 1339
Score = 74.7 bits (182), Expect = 5e-13
Identities = 39/75 (52%), Positives = 50/75 (66%), Gaps = 1/75 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP+EAW+ KKP++ R+FG A+A + KK LDD SN V LGV E SKA+R FD
Sbjct: 659 TPFEAWTGKKPHLGHLRVFGCTAHAKVTAPHLKK-LDDRSNPVVYLGVEEGSKAHRLFDP 717
Query: 76 IIKKIVVSEDVAFDE 90
++I+VS DV FDE
Sbjct: 718 RRRQIIVSRDVVFDE 732
>UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum]
Length = 1333
Score = 73.9 bits (180), Expect = 8e-13
Identities = 37/79 (46%), Positives = 53/79 (66%), Gaps = 3/79 (3%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V ++TP EAW+ KKP V RIFG IAYA + F + LD+ S KC+ +G S +SKAYR
Sbjct: 651 VWNTTPLEAWNGKKPRVSHLRIFGCIAYALVNFHSK---LDEKSTKCIFVGYSLQSKAYR 707
Query: 72 FFDRIIKKIVVSEDVAFDE 90
++ I K+++S +V F+E
Sbjct: 708 LYNPISGKVIISRNVVFNE 726
>UniRef100_Q8W3A4 Putative gag-pol polyprotein [Oryza sativa]
Length = 1167
Score = 73.6 bits (179), Expect = 1e-12
Identities = 34/79 (43%), Positives = 51/79 (64%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V + TP+EAW KKP + R+FG I YA +P +R K D+ S++C+ +G ++ K YR
Sbjct: 539 VPNRTPFEAWYGKKPVIGHMRVFGCICYAQVPAQKRVK-FDNKSDRCIFVGYADGIKGYR 597
Query: 72 FFDRIIKKIVVSEDVAFDE 90
++ KKI++S DV FDE
Sbjct: 598 LYNLEKKKIIISRDVIFDE 616
>UniRef100_Q7Y141 Putative polyprotein [Oryza sativa]
Length = 1335
Score = 72.0 bits (175), Expect = 3e-12
Identities = 32/79 (40%), Positives = 51/79 (64%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V + TP+EAW KKP + R+FG I YA +P +Q++ D+ S++C+ +G ++ K YR
Sbjct: 665 VTNRTPFEAWYGKKPVIGHMRVFGCICYAQVP-AQKRVKFDNKSDRCIFVGYADGIKGYR 723
Query: 72 FFDRIIKKIVVSEDVAFDE 90
++ KKI++S D FDE
Sbjct: 724 LYNLEKKKIIISRDAIFDE 742
>UniRef100_Q7XHD8 Putative copia-type polyprotein [Oryza sativa]
Length = 1350
Score = 71.6 bits (174), Expect = 4e-12
Identities = 33/79 (41%), Positives = 51/79 (63%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V + TP+EAW KKP + R+FG I YA +P +Q++ D+ S+ C+ +G ++ K YR
Sbjct: 588 VPNRTPFEAWYGKKPVIGHMRVFGCICYAQVP-AQKRVKFDNKSDWCIFVGYADGIKGYR 646
Query: 72 FFDRIIKKIVVSEDVAFDE 90
++ KKI++S DV FDE
Sbjct: 647 LYNLEKKKIIISRDVIFDE 665
>UniRef100_Q9SHL9 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 822
Score = 71.2 bits (173), Expect = 5e-12
Identities = 37/90 (41%), Positives = 51/90 (56%), Gaps = 1/90 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
+K+ TP EAWS KP V ++FG I Y HIP +R+K DD S + + +G S ++K YR
Sbjct: 367 LKNKTPLEAWSDSKPSVSHMKVFGSICYVHIPDEKRRK-WDDKSKRAIFVGYSSQTKGYR 425
Query: 72 FFDRIIKKIVVSEDVAFDEQGMLSLILKEI 101
+ KI +S DV FDE KE+
Sbjct: 426 VYLLKENKIDISRDVIFDEDSKWDWEKKEV 455
>UniRef100_Q9LQK2 F28G4.2 protein [Arabidopsis thaliana]
Length = 833
Score = 71.2 bits (173), Expect = 5e-12
Identities = 40/79 (50%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
+K TPYE + ++P V RIFG IAYA I Q +K LDD S + LG SKAYR
Sbjct: 204 LKDKTPYECFRDRRPTVEHIRIFGCIAYAKIDKQQLRK-LDDRSRILIHLGTEPGSKAYR 262
Query: 72 FFDRIIKKIVVSEDVAFDE 90
FD +K+VVS DV FDE
Sbjct: 263 LFDPQTQKVVVSRDVVFDE 281
>UniRef100_Q9LNQ5 F1L3.20 [Arabidopsis thaliana]
Length = 1188
Score = 71.2 bits (173), Expect = 5e-12
Identities = 40/79 (50%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
+K TPYE + ++P V RIFG IAYA I Q +K LDD S + LG SKAYR
Sbjct: 586 LKDKTPYECFRDRRPTVEHIRIFGCIAYAKIDKQQLRK-LDDRSRILIHLGTEPGSKAYR 644
Query: 72 FFDRIIKKIVVSEDVAFDE 90
FD +K+VVS DV FDE
Sbjct: 645 LFDPQTQKVVVSRDVVFDE 663
>UniRef100_Q60EG1 Putative polyprotein [Oryza sativa]
Length = 1028
Score = 70.5 bits (171), Expect = 9e-12
Identities = 37/75 (49%), Positives = 49/75 (65%), Gaps = 1/75 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP+EAW+ KKP++ R+FG A+A + KK LDD S + V LG+ E SKA+R FD
Sbjct: 513 TPFEAWTGKKPHLGHLRVFGCTAHAKVTTPNLKK-LDDRSEQFVYLGIEEGSKAHRLFDP 571
Query: 76 IIKKIVVSEDVAFDE 90
++I VS DV FDE
Sbjct: 572 RRRRIHVSRDVVFDE 586
>UniRef100_Q9LH44 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1499
Score = 69.3 bits (168), Expect = 2e-11
Identities = 37/84 (44%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query: 8 SSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKS 67
S + K TP E WS KKP V ++FG + Y HIP +R+K LD + + + +G S +S
Sbjct: 661 SKSLEKGVTPMEIWSGKKPSVDHLKVFGCVCYIHIPDEKRRK-LDTKAKQGIFVGYSNES 719
Query: 68 KAYRFFDRIIKKIVVSEDVAFDEQ 91
K YR F +KI VS+DV FDE+
Sbjct: 720 KGYRVFLLNEEKIEVSKDVTFDEK 743
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 187,047,395
Number of Sequences: 2790947
Number of extensions: 6547285
Number of successful extensions: 18042
Number of sequences better than 10.0: 431
Number of HSP's better than 10.0 without gapping: 265
Number of HSP's successfully gapped in prelim test: 167
Number of HSP's that attempted gapping in prelim test: 17453
Number of HSP's gapped (non-prelim): 447
length of query: 115
length of database: 848,049,833
effective HSP length: 91
effective length of query: 24
effective length of database: 594,073,656
effective search space: 14257767744
effective search space used: 14257767744
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136140.1


