

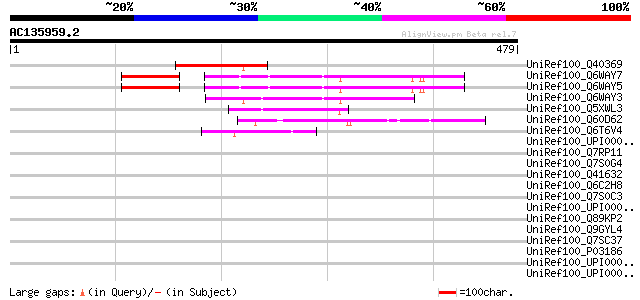
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135959.2 - phase: 0
(479 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40369 RPE15 protein [Medicago sativa] 133 1e-29
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 104 5e-21
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 104 6e-21
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 100 7e-20
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 69 2e-10
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 68 7e-10
UniRef100_Q6T6V4 Hypothetical protein [Vitis cinerea x Vitis rup... 57 1e-06
UniRef100_UPI000024B6BB UPI000024B6BB UniRef100 entry 46 0.002
UniRef100_Q7RP11 Hypothetical protein [Plasmodium yoelii yoelii] 46 0.002
UniRef100_Q7S0G4 Hypothetical protein [Neurospora crassa] 46 0.003
UniRef100_Q41632 Alpha/beta-type gliadin [Triticum urartu] 45 0.004
UniRef100_Q6C2H8 Similar to tr|Q95ST9 Drosophila melanogaster CG... 45 0.006
UniRef100_Q7S0C3 Hypothetical protein [Neurospora crassa] 45 0.006
UniRef100_UPI00002F95FC UPI00002F95FC UniRef100 entry 44 0.008
UniRef100_Q89KP2 Bll4862 protein [Bradyrhizobium japonicum] 44 0.010
UniRef100_Q9GYL4 Hypothetical protein R04E5.8 [Caenorhabditis el... 44 0.010
UniRef100_Q7SC37 Predicted protein [Neurospora crassa] 44 0.010
UniRef100_P03186 Large tegument protein [Epstein-Barr virus] 44 0.010
UniRef100_UPI000042EE1F UPI000042EE1F UniRef100 entry 44 0.013
UniRef100_UPI00003189DF UPI00003189DF UniRef100 entry 43 0.017
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 133 bits (334), Expect = 1e-29
Identities = 64/91 (70%), Positives = 68/91 (74%), Gaps = 4/91 (4%)
Query: 157 INAAQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQA 216
+NA QLP Y Y Q SQHPFFPPFY QYPLP GQP VPVNA+ QQ+Q QPPAQQQQ QQ
Sbjct: 108 VNATQLPLPYQYMQISQHPFFPPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQQQ 167
Query: 217 RPT----FPPIPMLYAELLPTLLLRGHYTTR 243
P FPPIPM YAELLPTLL +GH TR
Sbjct: 168 GPRPKTYFPPIPMRYAELLPTLLAKGHCVTR 198
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 104 bits (260), Expect = 5e-21
Identities = 86/271 (31%), Positives = 128/271 (46%), Gaps = 29/271 (10%)
Query: 185 PLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQ 244
P P Q Q V QQ QQQ P Q +Q R F +PM YAELLP LL G R
Sbjct: 380 PQPRQQQQQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT 438
Query: 245 GKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLINQGKLTFENNIPHVLDN 304
PP LPP + ++++CDFH GA GH E C AL++ V+ LI+ + F +P+V++N
Sbjct: 439 -MAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFA-PVPNVVNN 496
Query: 305 PLPNHAA--VNMIEVYE-EAPRLDVRNVTTPLVPLHIKLCQASLFDHDHASCPECFCNPL 361
P+P H VN IE E E ++V +V T L+ + +L ++ C C +
Sbjct: 497 PMPQHGGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESEN 556
Query: 362 GYCVVQNDIQSLMNDNYL----------TVSDVCV-----------IVS--VFHDPPVKS 398
G ++ IQ +M++ L TVS + + +VS + PV
Sbjct: 557 GCDQLRTGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTI 616
Query: 399 MLLKENVEPLVIRLPGPVPYTSAKAIPYKYN 429
+ P I G +++A+P+KY+
Sbjct: 617 SVPVTISAPTTIAASGRRAVENSRAVPWKYD 647
Score = 49.7 bits (117), Expect = 2e-04
Identities = 25/55 (45%), Positives = 35/55 (63%)
Query: 106 LDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPETFLITFQVMDINAA 160
L ++ G L S L+V+AF G++++V GE+ LP+ I PE F I F VMDI A
Sbjct: 861 LKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPA 915
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 104 bits (259), Expect = 6e-21
Identities = 86/271 (31%), Positives = 128/271 (46%), Gaps = 29/271 (10%)
Query: 185 PLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQ 244
P P Q Q V QQ QQQ P Q +Q R F +PM YAELLP LL G R
Sbjct: 380 PQPRQQQQQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT 438
Query: 245 GKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLINQGKLTFENNIPHVLDN 304
PP LPP + ++++CDFH GA GH E C AL++ V+ LI+ + F +P+V++N
Sbjct: 439 -MAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFA-PVPNVVNN 496
Query: 305 PLPNHAA--VNMIEVYE-EAPRLDVRNVTTPLVPLHIKLCQASLFDHDHASCPECFCNPL 361
P+P H VN IE E E ++V +V T L+ + +L ++ C C +
Sbjct: 497 PMPQHGGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESEN 556
Query: 362 GYCVVQNDIQSLMNDNYL----------TVSDVCV-----------IVS--VFHDPPVKS 398
G ++ IQ +M++ L TVS + + +VS +D PV
Sbjct: 557 GCDQLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTI 616
Query: 399 MLLKENVEPLVIRLPGPVPYTSAKAIPYKYN 429
+ P I G +++ +P+KY+
Sbjct: 617 SVPVTISAPTTIVASGRRAVENSRVVPWKYD 647
Score = 49.7 bits (117), Expect = 2e-04
Identities = 25/55 (45%), Positives = 35/55 (63%)
Query: 106 LDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPETFLITFQVMDINAA 160
L ++ G L S L+V+AF G++++V GE+ LP+ I PE F I F VMDI A
Sbjct: 861 LKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPA 915
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 100 bits (250), Expect = 7e-20
Identities = 70/202 (34%), Positives = 104/202 (50%), Gaps = 7/202 (3%)
Query: 186 LPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPT--FPPIPMLYAELLPTLLLRGHYTTR 243
+P+ QP+ VQQ QQQ + Q +Q P F +PM YAELLP LL G
Sbjct: 376 IPAPQPRQQQQQRVQQPQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLG-LVEL 434
Query: 244 QGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLINQGKLTFENNIPHVLD 303
PP LPP + ++++CDFH GA GH E C AL++ V+ LI+ + F +P+V++
Sbjct: 435 CTMAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINFA-PVPNVVN 493
Query: 304 NPLPNHAA--VNMIEVYEEAPRLD-VRNVTTPLVPLHIKLCQASLFDHDHASCPECFCNP 360
NP+P H VN IE E +D V +V T L+ + +L ++ C C +
Sbjct: 494 NPMPQHGGHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAESE 553
Query: 361 LGYCVVQNDIQSLMNDNYLTVS 382
G ++ IQ +M++ L S
Sbjct: 554 NGCDQLRAGIQGMMDEGCLQFS 575
Score = 46.6 bits (109), Expect = 0.002
Identities = 24/55 (43%), Positives = 34/55 (61%)
Query: 106 LDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPETFLITFQVMDINAA 160
L ++ G L S L+V+AF ++++V GE+ LP+ I PE F I F VMDI A
Sbjct: 859 LKKIDVEGFVLTPSDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPA 913
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 69.3 bits (168), Expect = 2e-10
Identities = 38/116 (32%), Positives = 58/116 (49%), Gaps = 3/116 (2%)
Query: 207 PAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQ 266
P+ ++ F P+ +L L + G Y G P D +R D +C +H
Sbjct: 410 PSHSSFEKKPARNFTPLIKSRTKLFERLTVAG-YIHPVGPKPVDTSSKFYRPDQRCAYHS 468
Query: 267 GALGHDVEGCYALKYIVKKLINQGKLTFENNIPHVLDNPLPNHA--AVNMIEVYEE 320
++GHD E C LK+ ++ LINQ ++ + P+V NPLPNH +NMIE E+
Sbjct: 469 NSVGHDTEDCINLKHKIQDLINQNVVSLQTVAPNVNSNPLPNHGGITINMIETDED 524
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 67.8 bits (164), Expect = 7e-10
Identities = 62/243 (25%), Positives = 107/243 (43%), Gaps = 17/243 (6%)
Query: 216 ARPTFPPIPMLYAEL---LPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 272
A+ F PI YA L L TL + + PPP L C + A GH+
Sbjct: 888 AKDEFTPIGESYASLFQKLRTLNVLSPIERKMSNPPPRNLD----YSQHCAYCSDAPGHN 943
Query: 273 VEGCYALKYIVKKLINQGKLTFEN-NIPHVLDNPLPNHAAVNMIEV---YEE--APRLDV 326
+E C+ LK ++ LI+ ++ E+ N P++ NPLP H NM+E+ +EE P +
Sbjct: 944 IERCWYLKKAIQDLIDTRRIIVESPNGPNINQNPLPRHTETNMLEMVNNHEEVAVPYKPI 1003
Query: 327 RNVTTPLVPLHIKLCQASLFDHDHASCPECFCNPLGYCVVQNDIQSLMNDNYLTVSDVCV 386
V T + + + S E P ++ I+ + D + + + +
Sbjct: 1004 LKVETGMESSTNVIDLTKIMPSGAESTSEKL-TPSSAPIL--TIKGALEDVWASQREARL 1060
Query: 387 IVSVFHDPPVKSMLLKENVEPLVIRLPGPVPYTSAKAIPYKYNATGIFYGAEFVNAITEE 446
+V D P+ ++ + P++IR +P T+ KA+P+ Y T + Y + +N +E
Sbjct: 1061 VVPRGPDKPI-LIVQGAYIPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGKEINEEVDE 1119
Query: 447 ATG 449
G
Sbjct: 1120 IGG 1122
Score = 40.8 bits (94), Expect = 0.087
Identities = 20/57 (35%), Positives = 35/57 (61%)
Query: 104 ATLDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPETFLITFQVMDINAA 160
+TL +L + + + V+AF GT+ + +GEI+L + I F + FQV++INA+
Sbjct: 1246 STLQKLNVNVERVRPNNVCVRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLNINAS 1302
>UniRef100_Q6T6V4 Hypothetical protein [Vitis cinerea x Vitis rupestris]
Length = 248
Score = 57.0 bits (136), Expect = 1e-06
Identities = 32/122 (26%), Positives = 57/122 (46%), Gaps = 14/122 (11%)
Query: 182 HQYPLPSGQPQVPVNAVVQQMQQQPPAQQQ-------------QHQQARPTFPPIPMLYA 228
H P P PQ +++ M +PP++ + H++ R F + + +
Sbjct: 92 HPRPRPHSVPQQHSSSISLAMSTRPPSRVRGPPVVTVPQGRPHPHRRCRRHFSDLSIPLS 151
Query: 229 ELLPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIN 288
T G + PDP+PP+FR DL C +HQ ++GH + C AL++ ++ +I+
Sbjct: 152 RAFETFQAMGFLAPLAPRALPDPVPPQFRLDLYCMYHQ-SVGHHTDRCTALRHAIQDIID 210
Query: 289 QG 290
G
Sbjct: 211 SG 212
>UniRef100_UPI000024B6BB UPI000024B6BB UniRef100 entry
Length = 519
Score = 46.2 bits (108), Expect = 0.002
Identities = 36/126 (28%), Positives = 45/126 (35%), Gaps = 29/126 (23%)
Query: 153 QVMDINAAQLPPSYPYAQYSQHPFFPP----FYHQYPLPSGQPQVPVNA-------VVQQ 201
Q+M + + LPP P Q P PP + PLP P + N +
Sbjct: 394 QIMPMQSHPLPPPPPLVQMQSQPMPPPPPTVSFQSQPLPPPPPPLQANGLPPPPPLITNG 453
Query: 202 MQQQ-------------PPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPP 248
++Q PPA Q P PP P L PTL H T+ PP
Sbjct: 454 LKQAFAHKFPSRPQDVLPPANQNNFPPPPPPPPPPP-----LAPTLPPYQHVTSTSNPPP 508
Query: 249 PDPLPP 254
P P PP
Sbjct: 509 PPPPPP 514
Score = 36.6 bits (83), Expect = 1.6
Identities = 32/103 (31%), Positives = 39/103 (37%), Gaps = 15/103 (14%)
Query: 162 LPPSYPYAQYSQHPFFPPFY----HQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQAR 217
LPP P P PP +PLP P V + Q Q PP Q++
Sbjct: 375 LPPPPPIMPMQSEPLPPPAQIMPMQSHPLPPPPPLVQM-----QSQPMPPPPPTVSFQSQ 429
Query: 218 PTFPPIPMLYAELLP------TLLLRGHYTTRQGKPPPDPLPP 254
P PP P L A LP T L+ + + P D LPP
Sbjct: 430 PLPPPPPPLQANGLPPPPPLITNGLKQAFAHKFPSRPQDVLPP 472
>UniRef100_Q7RP11 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 1115
Score = 46.2 bits (108), Expect = 0.002
Identities = 31/74 (41%), Positives = 36/74 (47%), Gaps = 10/74 (13%)
Query: 161 QLPPSYPYAQYS--QHPFFPPFYHQYPLPS--GQP------QVPVNAVVQQMQQQPPAQQ 210
Q PPS+ + Q S QHP PP H P P QP Q PV+ Q QQQP +
Sbjct: 359 QQPPSHQHPQPSSHQHPQPPPHQHSQPPPHQHSQPPPHQHQQQPVSXSPQHHQQQPVSPS 418
Query: 211 QQHQQARPTFPPIP 224
QH Q +PT P P
Sbjct: 419 PQHHQHQPTPIPTP 432
>UniRef100_Q7S0G4 Hypothetical protein [Neurospora crassa]
Length = 1059
Score = 45.8 bits (107), Expect = 0.003
Identities = 27/58 (46%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query: 167 PYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIP 224
P+ QYSQHP PP Q+P P Q Q + QQ Q Q QQQQH A P P P
Sbjct: 42 PHPQYSQHP--PPPPPQHPQPLLQRQQSYHLQQQQHQHQQQQQQQQHHPAYPQPPTTP 97
Score = 34.7 bits (78), Expect = 6.2
Identities = 29/117 (24%), Positives = 40/117 (33%), Gaps = 24/117 (20%)
Query: 159 AAQLPPSYPYAQYSQH----------PFFPPFYHQ-----------YPLPSGQPQVPVNA 197
A + PP P Q S H P P+ H YP P P +
Sbjct: 161 ANRAPPPPPLQQVSAHDPRSPSLASGPGPSPYRHTPTSSISAQSGGYPFPPSAQHTPTSP 220
Query: 198 VVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
V + Q PP+ +R +PP P + + +Q +PPP P PP
Sbjct: 221 VQRPYNQYPPSSTGAAYPSREGYPPTP---GGSVGVAGPQQQQQQQQQQPPPPPPPP 274
>UniRef100_Q41632 Alpha/beta-type gliadin [Triticum urartu]
Length = 296
Score = 45.4 bits (106), Expect = 0.004
Identities = 30/75 (40%), Positives = 35/75 (46%), Gaps = 13/75 (17%)
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQV-----PVNAVVQQMQQQPPAQQQQHQQAR 217
PP PY Q PPF Q P P QPQ P++ Q QQQ QQQQ QQ +
Sbjct: 88 PPQLPYPQP------PPFSPQQPYPQPQPQYPQPQQPISQQQAQQQQQQQQQQQQQQQQQ 141
Query: 218 PTFPPIPMLYAELLP 232
P I L +L+P
Sbjct: 142 QILPQI--LQQQLIP 154
Score = 34.7 bits (78), Expect = 6.2
Identities = 25/72 (34%), Positives = 32/72 (43%), Gaps = 6/72 (8%)
Query: 185 PLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP-IPMLYAELLPTLLLRGHYTTR 243
P+P QPQ P Q +Q P QQQQ + FPP P + P+ + Y
Sbjct: 24 PVPQPQPQNPSQP--QPQRQVPLVQQQQFPGQQQQFPPQQPYPQPQPFPS---QQPYLQL 78
Query: 244 QGKPPPDPLPPR 255
Q P P P PP+
Sbjct: 79 QPFPQPQPFPPQ 90
>UniRef100_Q6C2H8 Similar to tr|Q95ST9 Drosophila melanogaster CG15780 [Yarrowia
lipolytica]
Length = 595
Score = 44.7 bits (104), Expect = 0.006
Identities = 34/104 (32%), Positives = 41/104 (38%), Gaps = 15/104 (14%)
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP Q+ HP P + Q+P G P P QQ PP Q P PP
Sbjct: 476 PPHMGQQQHPGHPQHHPGHPQHPA-GGSPPGPGQPPGAYSQQMPP-------QYYPPPPP 527
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQ 266
M Y +G+Y R+ + PP PLPP R L HQ
Sbjct: 528 PHMGYPP-------QGYYPPREHQGPPPPLPPHMRPGLPQHQHQ 564
Score = 34.3 bits (77), Expect = 8.1
Identities = 27/84 (32%), Positives = 34/84 (40%), Gaps = 21/84 (25%)
Query: 161 QLPPSY------PYAQYSQHPFFPPFYHQYP-----------LPSGQPQVPVNAVVQQMQ 203
Q+PP Y P+ Y ++PP HQ P LP Q QV N Q+
Sbjct: 516 QMPPQYYPPPPPPHMGYPPQGYYPPREHQGPPPPLPPHMRPGLPQHQHQVYYNG-SPQLY 574
Query: 204 QQPPAQQQQHQQARPTFPPIPMLY 227
Q PP Q Q+ + PP P Y
Sbjct: 575 QVPPLQPPQYGR---QLPPPPQWY 595
>UniRef100_Q7S0C3 Hypothetical protein [Neurospora crassa]
Length = 643
Score = 44.7 bits (104), Expect = 0.006
Identities = 38/112 (33%), Positives = 44/112 (38%), Gaps = 21/112 (18%)
Query: 161 QLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTF 220
QLP P QH P YP P QP P + Q QQQPP QQHQQ P +
Sbjct: 299 QLPQPQPQPYPHQHAVQAP----YPPPLSQPPPPAYHYLHQRQQQPPPPPQQHQQ--PPY 352
Query: 221 ----PPIPMLYAELLPTLLLRGHYTTRQGKPPPDP--LPPRFRSDLKCDFHQ 266
PP P + + QG PPP P+ SD + HQ
Sbjct: 353 HHQGPPHPSQQQPM---------HHHHQGPPPPHHHFPSPQLPSDAQHPEHQ 395
Score = 36.6 bits (83), Expect = 1.6
Identities = 28/101 (27%), Positives = 36/101 (34%), Gaps = 15/101 (14%)
Query: 160 AQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAV---------VQQMQQQPPAQQ 210
A PP PY P PP Y +P PQ + ++ +Q Q QP
Sbjct: 250 APYPPGQPYHPDGLPPHAPPHYAHHPQNFSPPQHHMASLPPPPAGKPGIQLPQPQPQPYP 309
Query: 211 QQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDP 251
QH P PP+ P + RQ +PPP P
Sbjct: 310 HQHAVQAPYPPPLSQ------PPPPAYHYLHQRQQQPPPPP 344
>UniRef100_UPI00002F95FC UPI00002F95FC UniRef100 entry
Length = 1449
Score = 44.3 bits (103), Expect = 0.008
Identities = 40/120 (33%), Positives = 50/120 (41%), Gaps = 33/120 (27%)
Query: 160 AQLPPSYPYAQYSQHPFFPPFYHQYPL--PSGQPQVPVNAVVQQMQQQPPAQQQQHQQAR 217
AQ PP P AQ PP P+ P QPQ PV +QQ Q P Q Q+H +
Sbjct: 654 AQQPPPQPQAQQP-----PPQPQSQPVQQPPPQPQPPV---IQQQPLQAPPQLQRHHPPQ 705
Query: 218 PTFPP-------IPM------LYAELLPTLLLRGHYTT----------RQGKPPPDPLPP 254
P+ PP P+ L+A P LL + T+ +Q PPP P PP
Sbjct: 706 PSQPPPAPQPEQYPLPPQPEPLHAPPAPHLLQQQQLTSSGSNHIPPQQQQTSPPPPPPPP 765
Score = 37.4 bits (85), Expect = 0.96
Identities = 24/69 (34%), Positives = 32/69 (45%), Gaps = 3/69 (4%)
Query: 190 QPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELL--PTLLLRGHYTTRQGKP 247
QPQ Q QQ PP Q Q Q P P P++ + L P L R H+ + +P
Sbjct: 651 QPQAQQPPPQPQAQQPPPQPQSQPVQQPPPQPQPPVIQQQPLQAPPQLQR-HHPPQPSQP 709
Query: 248 PPDPLPPRF 256
PP P P ++
Sbjct: 710 PPAPQPEQY 718
>UniRef100_Q89KP2 Bll4862 protein [Bradyrhizobium japonicum]
Length = 887
Score = 43.9 bits (102), Expect = 0.010
Identities = 68/249 (27%), Positives = 86/249 (34%), Gaps = 43/249 (17%)
Query: 36 SATAIPVATSVIP--TASTNARFAMPTGFPYGLP---PFFTPSTAAGTSGTVN------- 83
SAT P A IP T + A A P+ P GLP P P T + T+
Sbjct: 589 SATVQPTARPGIPPGTPANVAPNARPSALPGGLPGTAPNVAPQTRLPAANTLPVPGSHGG 648
Query: 84 --------NGQIPGTNPVFVNTTLPQTTATLDQLRYRGTPLGRSTLLVKAFH-GTRKNVL 134
G +PG P+ T P TT + G P + H G
Sbjct: 649 PPAPPAGAAGTLPGGKPLPSTATAPGTTLPT----HPGAPTTAAPGTTPPPHLGAPTAAA 704
Query: 135 GEIDLPITIDPETFLITFQVMDINAAQLPPSYPYAQYSQ----HPFFPPFYHQYPLPSGQ 190
P T +P ++ Q AQ PP A ++ P PP P +G+
Sbjct: 705 PTTGAPTTTNPA--VVPGQPPKPPVAQTPPGAGPADRAKSATREPAGPP-----PAAAGK 757
Query: 191 PQVPVNAVVQQ---MQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLR--GHYTTRQG 245
P VP + V + Q PPA Q + A P PP P A P R
Sbjct: 758 PVVPPPSAVHEPIRPQAHPPAPPQAAKPAAP--PPRPQAVARPTPPPPPRVSAPPPRMAA 815
Query: 246 KPPPDPLPP 254
PPP P PP
Sbjct: 816 PPPPRPAPP 824
>UniRef100_Q9GYL4 Hypothetical protein R04E5.8 [Caenorhabditis elegans]
Length = 997
Score = 43.9 bits (102), Expect = 0.010
Identities = 31/93 (33%), Positives = 35/93 (37%), Gaps = 17/93 (18%)
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP YPY Q P P + Q+P P P PV Q Q PA Q +A P PP
Sbjct: 70 PPPYPYQHPHQPPPAHPHHLQHPHPYAYPGYPVPG---QEGHQVPAPQHGDHEASPPPPP 126
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPR 255
P + G PP P PPR
Sbjct: 127 PP--------------RKSRAGGSSPPPPPPPR 145
>UniRef100_Q7SC37 Predicted protein [Neurospora crassa]
Length = 1578
Score = 43.9 bits (102), Expect = 0.010
Identities = 39/110 (35%), Positives = 41/110 (36%), Gaps = 10/110 (9%)
Query: 145 PETFLITFQVMDINAAQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQ 204
P IT IN A PP Q P PP P Q Q P QQ QQ
Sbjct: 175 PSASSITSGPGPINRAMPPPPTSPPQQGPPPQAPP-----PQAHPQQQHPQQHAQQQQQQ 229
Query: 205 QP-PAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLP 253
P P QQQQ QQ +P PP P + G Y G PPP P
Sbjct: 230 HPGPHQQQQQQQQQPPQPPHPSQQGPPPNAHQMGGSY----GIPPPRAPP 275
>UniRef100_P03186 Large tegument protein [Epstein-Barr virus]
Length = 3149
Score = 43.9 bits (102), Expect = 0.010
Identities = 58/229 (25%), Positives = 79/229 (34%), Gaps = 28/229 (12%)
Query: 31 AAQEQSATAIPVATSVIPTASTNARFAMPTGFPYGLPPFFTPSTAAGTSGTVNNGQIPGT 90
AA +A A S P ++ A A + P PP F P G + V P T
Sbjct: 343 AAPASAAPASAAPASAAPASAAPASAAPASAAPASSPPLFIPIPGLGHTPGV---PAPST 399
Query: 91 NPVFVNTTLPQTTATLDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPETFLI 150
P + PQT R LG+ + K G R LP++ +T
Sbjct: 400 PPRASSGAAPQTPK-------RKKGLGKDSPHKKPTSGRR--------LPLSSTTDT--- 441
Query: 151 TFQVMDINAAQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQ-VPVNAVVQQMQQQPPAQ 209
+ + +PP P + P P HQ P S P PV+ + + P
Sbjct: 442 --EDDQLPRTHVPPHRPPSAARLPPPVIPIPHQSPPASPTPHPAPVSTIAPSVTPSPRLP 499
Query: 210 QQ---QHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPR 255
Q QA P+ P IP+ PT TT PP PP+
Sbjct: 500 LQIPIPLPQAAPSNPKIPLTTPSPSPT-AAAAPTTTTLSPPPTQQQPPQ 547
>UniRef100_UPI000042EE1F UPI000042EE1F UniRef100 entry
Length = 1009
Score = 43.5 bits (101), Expect = 0.013
Identities = 31/73 (42%), Positives = 40/73 (54%), Gaps = 8/73 (10%)
Query: 145 PETFLITFQVMDINAAQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQ 204
PETF ++ + INAAQ+ Q Q PFF P H PLP + Q P+ ++ Q QQ
Sbjct: 914 PETFDLS-AISAINAAQMQQR----QQQQPPFFTPPPHP-PLPEQRDQQPL--LLSQYQQ 965
Query: 205 QPPAQQQQHQQAR 217
QP Q +Q QQ R
Sbjct: 966 QPEQQPEQQQQPR 978
>UniRef100_UPI00003189DF UPI00003189DF UniRef100 entry
Length = 751
Score = 43.1 bits (100), Expect = 0.017
Identities = 25/54 (46%), Positives = 26/54 (47%), Gaps = 2/54 (3%)
Query: 171 YSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIP 224
Y QH PP + P QPQ P QQ QQQPP QQQ Q P PP P
Sbjct: 492 YGQHG--PPPHPGQPQQPQQPQQPQPQQQQQQQQQPPPPQQQQPQPPPQQPPPP 543
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 864,392,712
Number of Sequences: 2790947
Number of extensions: 40148392
Number of successful extensions: 192322
Number of sequences better than 10.0: 847
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 853
Number of HSP's that attempted gapping in prelim test: 182637
Number of HSP's gapped (non-prelim): 4755
length of query: 479
length of database: 848,049,833
effective HSP length: 131
effective length of query: 348
effective length of database: 482,435,776
effective search space: 167887650048
effective search space used: 167887650048
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135959.2


