

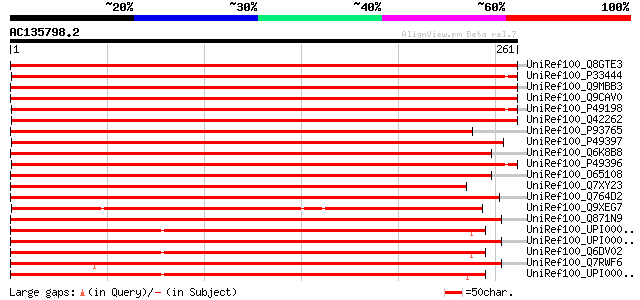
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135798.2 + phase: 0
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GTE3 Ribosomal protein S3a [Cicer arietinum] 507 e-142
UniRef100_P33444 40S ribosomal protein S3a [Catharanthus roseus] 470 e-131
UniRef100_Q9MBB3 Cyc07 [Daucus carota] 469 e-131
UniRef100_Q9CAV0 Putative 40S ribosomal protein S3A [Arabidopsis... 456 e-127
UniRef100_P49198 40S ribosomal protein S3a [Helianthus annuus] 454 e-127
UniRef100_Q42262 40S ribosomal protein S3a [Arabidopsis thaliana] 453 e-126
UniRef100_P93765 Cyc07 protein [Catharanthus roseus] 447 e-124
UniRef100_P49397 40S ribosomal protein S3a [Oryza sativa] 443 e-123
UniRef100_Q6K8B8 Putative ribosomal protein S3a, cytosolic [Oryz... 441 e-122
UniRef100_P49396 40S ribosomal protein S3a [Brassica rapa] 434 e-120
UniRef100_O65108 S-phase-specific ribosomal protein [Oryza sativa] 426 e-118
UniRef100_Q7XY23 Cyc07 [Triticum aestivum] 387 e-106
UniRef100_Q764D2 Putative S-phase specific ribosomal protein cyc... 365 e-100
UniRef100_Q9XEG7 40S ribosomal protein S3a [Tortula ruralis] 352 7e-96
UniRef100_Q871N9 Probable ribosomal protein 10, cytosolic [Neuro... 347 1e-94
UniRef100_UPI000013EBBE UPI000013EBBE UniRef100 entry 345 9e-94
UniRef100_UPI000023D8F3 UPI000023D8F3 UniRef100 entry 342 6e-93
UniRef100_Q6DV02 GekBS027P [Gecko japonicus] 342 6e-93
UniRef100_Q7RWF6 Hypothetical protein [Neurospora crassa] 342 6e-93
UniRef100_UPI000035F12A UPI000035F12A UniRef100 entry 342 7e-93
>UniRef100_Q8GTE3 Ribosomal protein S3a [Cicer arietinum]
Length = 261
Score = 507 bits (1305), Expect = e-142
Identities = 252/261 (96%), Positives = 260/261 (99%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKAADPF+KKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGKKGGKKKAADPFSKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFE+SLADLQGDEDNAFRKIRLRAEDVQGKN+LTNF+GMDFTTDKLRSLVRKWQTL
Sbjct: 61 LKHRVFEISLADLQGDEDNAFRKIRLRAEDVQGKNVLTNFYGMDFTTDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIM+NQATSCD
Sbjct: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMVNQATSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILK+PKFDLGKLMEVHGDYSEDVG
Sbjct: 181 LKELVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKSPKFDLGKLMEVHGDYSEDVG 240
Query: 241 TKVERPADETMVEGTPEIVGA 261
TKVE PADET+VEGTPEIVGA
Sbjct: 241 TKVESPADETVVEGTPEIVGA 261
>UniRef100_P33444 40S ribosomal protein S3a [Catharanthus roseus]
Length = 259
Score = 470 bits (1209), Expect = e-131
Identities = 229/260 (88%), Positives = 252/260 (96%), Gaps = 1/260 (0%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVF V+NVGKTLV+RTQGTKIASEGL
Sbjct: 1 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFSVRNVGKTLVTRTQGTKIASEGL 60
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
+HRVFE+SLADLQGDED++FRKIRLRAED+QGKN+LTNFWGMDFTTDKLRSLVRKWQ+LI
Sbjct: 61 EHRVFEISLADLQGDEDHSFRKIRLRAEDIQGKNVLTNFWGMDFTTDKLRSLVRKWQSLI 120
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDL 181
EAHVDVKTTD+YTLRMFCIGFTK+R+NQ KRTCYAQSSQIRQIRRKMREIM+NQA SCDL
Sbjct: 121 EAHVDVKTTDSYTLRMFCIGFTKKRANQQKRTCYAQSSQIRQIRRKMREIMVNQAQSCDL 180
Query: 182 KDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGT 241
KDLV+KFIPE IG+EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY+ED+GT
Sbjct: 181 KDLVQKFIPESIGREIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYNEDIGT 240
Query: 242 KVERPADETMVEGTPEIVGA 261
K++RPA+E + E T E++GA
Sbjct: 241 KLDRPAEEAVAEPT-EVIGA 259
>UniRef100_Q9MBB3 Cyc07 [Daucus carota]
Length = 261
Score = 469 bits (1206), Expect = e-131
Identities = 233/261 (89%), Positives = 248/261 (94%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKA DPFAKKDWYDIKAP+VFQ KNVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGKKGGKKKATDPFAKKDWYDIKAPNVFQNKNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEV LADLQGDED A+RKIRLRAEDVQGKN+LTNF+GMDFTTDKLRSLVRKWQTL
Sbjct: 61 LKHRVFEVCLADLQGDEDQAYRKIRLRAEDVQGKNVLTNFYGMDFTTDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD+YTLRMFCIGFTK+R+NQ KRTCYAQSSQIRQIRRKM EIM NQA+SCD
Sbjct: 121 IEAHVDVKTTDSYTLRMFCIGFTKKRANQQKRTCYAQSSQIRQIRRKMVEIMRNQASSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LV KFIPE IG+EIEKATSSI+PLQNVFIRKVKILKAPKFD+GKLMEVHGDYSEDVG
Sbjct: 181 LKELVAKFIPESIGREIEKATSSIFPLQNVFIRKVKILKAPKFDIGKLMEVHGDYSEDVG 240
Query: 241 TKVERPADETMVEGTPEIVGA 261
K+ERP +ETMVEG E+VGA
Sbjct: 241 VKLERPIEETMVEGETEVVGA 261
>UniRef100_Q9CAV0 Putative 40S ribosomal protein S3A [Arabidopsis thaliana]
Length = 262
Score = 456 bits (1172), Expect = e-127
Identities = 224/262 (85%), Positives = 244/262 (92%), Gaps = 1/262 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KGGKKKA DPF+KKDWYD+KAPS+F +NVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGGKKKAVDPFSKKDWYDVKAPSIFTHRNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDEDNA+RKIRLRAEDVQG+N+L FWGMDFTTDKLRSLV+KWQTL
Sbjct: 61 LKHRVFEVSLADLQGDEDNAYRKIRLRAEDVQGRNVLCQFWGMDFTTDKLRSLVKKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD+YTLR+FCI FTKRR+NQVKRTCYAQSSQIRQIRRKMR+IM+ +A+SCD
Sbjct: 121 IEAHVDVKTTDSYTLRLFCIAFTKRRANQVKRTCYAQSSQIRQIRRKMRDIMVREASSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYS-EDV 239
LKDLV KFIPE IG+EIEKAT IYPLQNVFIRKVKILKAPKFDLGKLM+VHGDYS EDV
Sbjct: 181 LKDLVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKAPKFDLGKLMDVHGDYSAEDV 240
Query: 240 GTKVERPADETMVEGTPEIVGA 261
G KV+RPADE VE EI+GA
Sbjct: 241 GVKVDRPADEMAVEEPTEIIGA 262
>UniRef100_P49198 40S ribosomal protein S3a [Helianthus annuus]
Length = 259
Score = 454 bits (1169), Expect = e-127
Identities = 222/260 (85%), Positives = 247/260 (94%), Gaps = 1/260 (0%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGKNKRISKGKKGGKKKAADPF+KKDWYDIKAPS+F +NVGKTLV+RTQGTKIASEGL
Sbjct: 1 AVGKNKRISKGKKGGKKKAADPFSKKDWYDIKAPSLFITRNVGKTLVTRTQGTKIASEGL 60
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
KHRVFEVSLADLQ DED+++RKIRLRAEDVQGKN+LTNFWGMD TTDKLRSLV+KWQ+LI
Sbjct: 61 KHRVFEVSLADLQNDEDHSYRKIRLRAEDVQGKNVLTNFWGMDVTTDKLRSLVKKWQSLI 120
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDL 181
EAHVDVKTTDN+TLRMFCIGFTK+R+NQVKRTCYAQSSQI+QIRRKMREIM+ QA +CDL
Sbjct: 121 EAHVDVKTTDNFTLRMFCIGFTKKRANQVKRTCYAQSSQIKQIRRKMREIMVTQAQTCDL 180
Query: 182 KDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGT 241
K+LV+KFIPE IG+EIEKATSSIYPLQNVFIRKVKILKAPKFD+GKLMEVHGDYSEDVG
Sbjct: 181 KELVQKFIPESIGREIEKATSSIYPLQNVFIRKVKILKAPKFDIGKLMEVHGDYSEDVGV 240
Query: 242 KVERPADETMVEGTPEIVGA 261
K++RPA+E E T E++GA
Sbjct: 241 KLDRPAEEAAPEAT-EVIGA 259
>UniRef100_Q42262 40S ribosomal protein S3a [Arabidopsis thaliana]
Length = 261
Score = 453 bits (1165), Expect = e-126
Identities = 224/261 (85%), Positives = 241/261 (91%), Gaps = 1/261 (0%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGKNKRISKG+KGGKKKA DPF+KKDWYD+KAP F +NVGKTLVSRTQGTKIASEGL
Sbjct: 1 AVGKNKRISKGRKGGKKKAVDPFSKKDWYDVKAPGSFTNRNVGKTLVSRTQGTKIASEGL 60
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
KHRVFEVSLADLQ DEDNA+RKIRLRAEDVQG+N+LT FWGMDFTTDKLRSLV+KWQTLI
Sbjct: 61 KHRVFEVSLADLQNDEDNAYRKIRLRAEDVQGRNVLTQFWGMDFTTDKLRSLVKKWQTLI 120
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDL 181
EAHVDVKTTD YTLRMFCI FTKRR+NQVKRTCYAQSSQIRQIRRKM EIM+ +A+SCDL
Sbjct: 121 EAHVDVKTTDGYTLRMFCIAFTKRRANQVKRTCYAQSSQIRQIRRKMSEIMVKEASSCDL 180
Query: 182 KDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDVG 240
K+LV KFIPE IG+EIEKAT IYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY +EDVG
Sbjct: 181 KELVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYTAEDVG 240
Query: 241 TKVERPADETMVEGTPEIVGA 261
KV+RPADETMVE EI+GA
Sbjct: 241 VKVDRPADETMVEEPTEIIGA 261
>UniRef100_P93765 Cyc07 protein [Catharanthus roseus]
Length = 245
Score = 447 bits (1150), Expect = e-124
Identities = 218/238 (91%), Positives = 233/238 (97%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVF V+NVGKTLV+RTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFSVRNVGKTLVTRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFE+SLADLQGDED++FRKIRLRAED+QGKN+LTNFWGMDFTTDKLRSLVRKWQ+L
Sbjct: 61 LKHRVFEISLADLQGDEDHSFRKIRLRAEDIQGKNVLTNFWGMDFTTDKLRSLVRKWQSL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD+YTLRMFCIGFTK+R+NQ KRTCYAQSSQIRQIRRKMREIM+NQA SCD
Sbjct: 121 IEAHVDVKTTDSYTLRMFCIGFTKKRANQQKRTCYAQSSQIRQIRRKMREIMVNQAPSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSED 238
LKDLV+KFIPE IG+EIEKAT SIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY+ED
Sbjct: 181 LKDLVQKFIPESIGREIEKATPSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYNED 238
>UniRef100_P49397 40S ribosomal protein S3a [Oryza sativa]
Length = 261
Score = 443 bits (1140), Expect = e-123
Identities = 218/254 (85%), Positives = 237/254 (92%), Gaps = 1/254 (0%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGKNKRISKGKKG KKK DPFAKKDWYDIKAPSVF V+N+GKTLVSRTQGTKIASEGL
Sbjct: 1 AVGKNKRISKGKKGSKKKTVDPFAKKDWYDIKAPSVFNVRNIGKTLVSRTQGTKIASEGL 60
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
KHRVFEVSLADLQ DED A+RKIRLRAEDVQGKN+LTNFWGM FTTDKLRSLV+KWQTLI
Sbjct: 61 KHRVFEVSLADLQNDEDQAYRKIRLRAEDVQGKNVLTNFWGMSFTTDKLRSLVKKWQTLI 120
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDL 181
EAHVDVKTTD Y LR+FCIGFTKRR NQVKRTCYAQ+SQIRQIRRKM EIM NQA+SCDL
Sbjct: 121 EAHVDVKTTDGYMLRLFCIGFTKRRPNQVKRTCYAQASQIRQIRRKMVEIMANQASSCDL 180
Query: 182 KDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYS-EDVG 240
K+LV KFIPE+IGKEIEKATSSI+PLQNVF+RKVKILKAPKFDLGKLMEVHGDY+ ED+G
Sbjct: 181 KELVSKFIPEVIGKEIEKATSSIFPLQNVFVRKVKILKAPKFDLGKLMEVHGDYAKEDIG 240
Query: 241 TKVERPADETMVEG 254
TK++RPA++ + G
Sbjct: 241 TKLDRPAEDEAMAG 254
>UniRef100_Q6K8B8 Putative ribosomal protein S3a, cytosolic [Oryza sativa]
Length = 261
Score = 441 bits (1133), Expect = e-122
Identities = 213/248 (85%), Positives = 234/248 (93%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KG KKK DPF+KKDWYDIKAP+VF V+N+GKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGSKKKTVDPFSKKDWYDIKAPTVFSVRNIGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQ DED A+RK+RLRAEDVQG+N+LTNFWGM FTTDKLRSLV+KWQTL
Sbjct: 61 LKHRVFEVSLADLQNDEDQAYRKVRLRAEDVQGRNVLTNFWGMSFTTDKLRSLVKKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNY LR+FCIGFTKRR NQVKRTCYAQ+SQIRQIRRKM EIM NQA++CD
Sbjct: 121 IEAHVDVKTTDNYMLRLFCIGFTKRRPNQVKRTCYAQASQIRQIRRKMVEIMANQASTCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LV KFIPE+IGKEIEK+TSSI+PLQNVFIRKVKILKAPKFDLGKLMEVHGDY EDVG
Sbjct: 181 LKELVSKFIPEVIGKEIEKSTSSIFPLQNVFIRKVKILKAPKFDLGKLMEVHGDYKEDVG 240
Query: 241 TKVERPAD 248
K++RPA+
Sbjct: 241 MKLDRPAE 248
>UniRef100_P49396 40S ribosomal protein S3a [Brassica rapa]
Length = 261
Score = 434 bits (1116), Expect = e-120
Identities = 221/262 (84%), Positives = 238/262 (90%), Gaps = 3/262 (1%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGKNKRISKG+KGGKKK DPFAKKDWYDIKAPS F +NVGKTLVSRTQGTKIASEGL
Sbjct: 1 AVGKNKRISKGRKGGKKKIVDPFAKKDWYDIKAPSSFTHRNVGKTLVSRTQGTKIASEGL 60
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
KHRVFEVSLADL DED A+RKIRLRAEDVQG+N+LT FWGMDFTTDKLRSLV+KWQTLI
Sbjct: 61 KHRVFEVSLADLNKDEDQAYRKIRLRAEDVQGRNVLTQFWGMDFTTDKLRSLVKKWQTLI 120
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMI-NQATSCD 180
E+HVDVKTTDNYTLR+FCI FTKRR+NQVKRTCYAQSSQIRQIR KMREIMI +A+SCD
Sbjct: 121 ESHVDVKTTDNYTLRLFCIAFTKRRANQVKRTCYAQSSQIRQIRSKMREIMIKEEASSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDV 239
LK+LV KFIPE IGK+IEKAT IYPLQNVFIRKVKILKAPKFDL KLMEVHGDY +EDV
Sbjct: 181 LKELVAKFIPESIGKDIEKATQGIYPLQNVFIRKVKILKAPKFDLRKLMEVHGDYTAEDV 240
Query: 240 GTKVERPADETMVEGTPEIVGA 261
G KV+RPADE + E T EI+GA
Sbjct: 241 GVKVDRPADEVVEEPT-EIIGA 261
>UniRef100_O65108 S-phase-specific ribosomal protein [Oryza sativa]
Length = 261
Score = 426 bits (1096), Expect = e-118
Identities = 206/248 (83%), Positives = 230/248 (92%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KG KKK DPF+KKDWYDIKAP+VF V+N+GKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGSKKKTVDPFSKKDWYDIKAPTVFSVRNIGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADL+ DED A+RK+RLRAEDVQG+N+LTNFWGM FTTDKLRSLV+KWQTL
Sbjct: 61 LKHRVFEVSLADLRNDEDQAYRKVRLRAEDVQGRNVLTNFWGMSFTTDKLRSLVKKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNY LR+FCIGFTKRR NQVKRTCYAQ ++ + RRKM EIM NQA++CD
Sbjct: 121 IEAHVDVKTTDNYMLRLFCIGFTKRRPNQVKRTCYAQVARSGRSRRKMVEIMANQASTCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LV KFIPE+IGKEIEK+TSSI+PLQNVFIRKVKILKAPKFDLGKLMEVHGDY EDVG
Sbjct: 181 LKELVSKFIPEVIGKEIEKSTSSIFPLQNVFIRKVKILKAPKFDLGKLMEVHGDYKEDVG 240
Query: 241 TKVERPAD 248
K++RPA+
Sbjct: 241 MKLDRPAE 248
>UniRef100_Q7XY23 Cyc07 [Triticum aestivum]
Length = 242
Score = 387 bits (995), Expect = e-106
Identities = 194/236 (82%), Positives = 207/236 (87%), Gaps = 1/236 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KG KKKA DPF KK WYDIKAP +F +NVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGSKKKAVDPFTKKQWYDIKAPLLFTSRNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQ DED A+RKIRLRAEDVQG N+LTNFWGMDFTTDKLRSLVRKWQTL
Sbjct: 61 LKHRVFEVSLADLQNDEDQAYRKIRLRAEDVQGMNVLTNFWGMDFTTDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNY LRMF IGFTKRR NQVKRTCYAQ+SQIRQIRRKM EIM+NQA SCD
Sbjct: 121 IEAHVDVKTTDNYMLRMFAIGFTKRRPNQVKRTCYAQASQIRQIRRKMVEIMVNQAASCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDL-GKLMEVHGDY 235
L +LV KFIPE+IGKEIE ATS I PLQNV++RKVKI + P+ G EVHGDY
Sbjct: 181 LXELVNKFIPEVIGKEIEXATSGIXPLQNVYVRKVKIPEGPQVSTWGSFXEVHGDY 236
>UniRef100_Q764D2 Putative S-phase specific ribosomal protein cyc07 [Lentinula
edodes]
Length = 256
Score = 365 bits (938), Expect = e-100
Identities = 175/252 (69%), Positives = 215/252 (84%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKR+SKGKKG KKK DPF++KDWYDIKAPS+F V+NVGKTLV+R+QG K A++
Sbjct: 1 MAVGKNKRLSKGKKGIKKKVVDPFSRKDWYDIKAPSIFDVRNVGKTLVNRSQGLKNANDS 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LK R+ EVSL DL +E+ +FRKI+LR ++VQGKN LTNF GMDFT+DKLRSLVRKWQTL
Sbjct: 61 LKGRIIEVSLGDLNKEEEQSFRKIKLRVDEVQGKNCLTNFHGMDFTSDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD Y LR+F IGFTKRR QV++T YAQSSQ+R+IR+KM EIM +AT+CD
Sbjct: 121 IEAHVDVKTTDGYLLRLFAIGFTKRRPTQVRKTTYAQSSQVREIRKKMFEIMTREATNCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LV+KF+PE IG+EIEKA+ SIYPLQNV++RK KILKAPKFD+ KLME+HG+ +++ G
Sbjct: 181 LKELVQKFVPEAIGREIEKASRSIYPLQNVYVRKAKILKAPKFDVSKLMELHGESTDETG 240
Query: 241 TKVERPADETMV 252
TK+ + E V
Sbjct: 241 TKIAKDFKEPEV 252
>UniRef100_Q9XEG7 40S ribosomal protein S3a [Tortula ruralis]
Length = 247
Score = 352 bits (902), Expect = 7e-96
Identities = 181/243 (74%), Positives = 205/243 (83%), Gaps = 4/243 (1%)
Query: 2 AVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGL 61
AVGK+KRISKGKKGGKKK DPF+KKDWYDIKAP + G+ RT GTKIAS+GL
Sbjct: 1 AVGKDKRISKGKKGGKKKIVDPFSKKDWYDIKAPPRSRRARSGRRC-HRTAGTKIASDGL 59
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLI 121
RVFEVSLADLQ DED AFRKI+LRAEDVQGKN+LTNFWGMDFTTDKLRSLVRKWQ+LI
Sbjct: 60 NFRVFEVSLADLQNDEDQAFRKIKLRAEDVQGKNVLTNFWGMDFTTDKLRSLVRKWQSLI 119
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDL 181
+AHVDVKTTDNY +R+FCI FTK+R N R YAQSSQI IRRKMREIM+ +A +CDL
Sbjct: 120 QAHVDVKTTDNYQIRIFCIAFTKKRPNHT-RNFYAQSSQI-AIRRKMREIMVKEAQTCDL 177
Query: 182 KDLVRKFIPEMIGKEIEK-ATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
K+LV KFIPE+IGKEI++ + IYPLQ+ FIRKVKILKAPKFDL KLMEVHGDY+E+VG
Sbjct: 178 KELVAKFIPEVIGKEIDEWRSQGIYPLQSTFIRKVKILKAPKFDLMKLMEVHGDYNEEVG 237
Query: 241 TKV 243
K+
Sbjct: 238 AKI 240
>UniRef100_Q871N9 Probable ribosomal protein 10, cytosolic [Neurospora crassa]
Length = 256
Score = 347 bits (891), Expect = 1e-94
Identities = 174/254 (68%), Positives = 209/254 (81%), Gaps = 1/254 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKR+SKGKKG KKK DPFA+KDWY IKAP+ F V++VGKTLV+RT G K A++
Sbjct: 1 MAVGKNKRLSKGKKGLKKKTQDPFARKDWYGIKAPAPFNVRDVGKTLVNRTTGLKNANDA 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LK R+FEVSLADLQ DED++FRK++LR +++QGKN LTNF G+DFT+DKLRSLVRKWQTL
Sbjct: 61 LKGRIFEVSLADLQKDEDHSFRKVKLRVDEIQGKNCLTNFHGLDFTSDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEA++ VKTTD+Y LR+F I FTKRR NQVK+T YA SSQIR IRRKM EI+ +A+SC
Sbjct: 121 IEANITVKTTDDYLLRLFAIAFTKRRPNQVKKTTYAASSQIRAIRRKMTEIIQREASSCT 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDV 239
L L K IPE+IG+EIEKAT IYPLQNV IRKVK+LK PKFDLG LM +HG+ S++
Sbjct: 181 LTQLTSKLIPEVIGREIEKATQGIYPLQNVHIRKVKLLKQPKFDLGALMSLHGESGSDEA 240
Query: 240 GTKVERPADETMVE 253
G KVER ET++E
Sbjct: 241 GQKVEREFKETVLE 254
>UniRef100_UPI000013EBBE UPI000013EBBE UniRef100 entry
Length = 264
Score = 345 bits (884), Expect = 9e-94
Identities = 170/253 (67%), Positives = 208/253 (82%), Gaps = 9/253 (3%)
Query: 1 MAVGKNKRISKG-KKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASE 59
MAVGKNKR++KG KKG KKK DPF+KKDWYD+KAP++F ++N+GKTLV+RTQGTKIAS+
Sbjct: 1 MAVGKNKRLTKGGKKGAKKKVVDPFSKKDWYDVKAPAMFNIRNIGKTLVTRTQGTKIASD 60
Query: 60 GLKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQT 119
GLK RVFEVSLADLQ DE AFRK +L EDVQGKN LTNF GMD T DK+ S+V+KWQT
Sbjct: 61 GLKGRVFEVSLADLQNDEV-AFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMVKKWQT 119
Query: 120 LIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSC 179
+IEAHVDVKTTD Y LR+FC+GFTK+R NQ+++T YAQ Q+RQIR+KM EIMI + +
Sbjct: 120 MIEAHVDVKTTDGYLLRLFCVGFTKKRKNQIRKTSYAQHQQVRQIRKKMMEIMIREVQTN 179
Query: 180 DLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYS--- 236
DLK++V K IP+ IGK+IEKA SIYPL +VF+RKVK+LK PKF+LGKLME+HG+ S
Sbjct: 180 DLKEVVNKLIPDSIGKDIEKACQSIYPLHDVFVRKVKMLKKPKFELGKLMELHGEGSSSG 239
Query: 237 ----EDVGTKVER 245
++ G KVER
Sbjct: 240 KATGDETGAKVER 252
>UniRef100_UPI000023D8F3 UPI000023D8F3 UniRef100 entry
Length = 256
Score = 342 bits (877), Expect = 6e-93
Identities = 172/254 (67%), Positives = 208/254 (81%), Gaps = 1/254 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKR+SKGKKG KKK DPF++KDWY IKAP+ F V++VGKTLV+RT G K A++
Sbjct: 1 MAVGKNKRLSKGKKGLKKKTVDPFSRKDWYSIKAPNPFNVRDVGKTLVNRTTGLKNANDA 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LK R+ EVSLADLQ DED++FRK+RLR ++VQGKN LT F G+DFT+DKLRSLVRKWQTL
Sbjct: 61 LKGRILEVSLADLQKDEDHSFRKVRLRVDEVQGKNCLTAFHGLDFTSDKLRSLVRKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEA+V VKTTD+Y +R+F IGFTKRR NQVK+T YA SSQIR IRRKM +I+ +A+SC
Sbjct: 121 IEANVTVKTTDDYLIRLFAIGFTKRRGNQVKKTTYAASSQIRAIRRKMTDIIQREASSCT 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDV 239
L L K IPE+IG+EIEK+T IYPLQNV IRKVK+LKAPKFDLG LM +HG+ ++D
Sbjct: 181 LTQLTSKLIPEVIGREIEKSTQGIYPLQNVHIRKVKLLKAPKFDLGALMALHGESGTDDQ 240
Query: 240 GTKVERPADETMVE 253
G KVER E ++E
Sbjct: 241 GQKVEREFKERVLE 254
>UniRef100_Q6DV02 GekBS027P [Gecko japonicus]
Length = 264
Score = 342 bits (877), Expect = 6e-93
Identities = 168/253 (66%), Positives = 208/253 (81%), Gaps = 9/253 (3%)
Query: 1 MAVGKNKRISKG-KKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASE 59
MAVGKNKR++KG KKG KKK DPF+KKDWYD+KAP++F ++N+GKTLV+RTQGTKIAS+
Sbjct: 1 MAVGKNKRLTKGGKKGAKKKVVDPFSKKDWYDVKAPAMFNIRNIGKTLVTRTQGTKIASD 60
Query: 60 GLKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQT 119
GLK RVFEVSLADLQ DE AFRK +L EDVQGKN LTNF GMD T DK+ S+V+KWQT
Sbjct: 61 GLKGRVFEVSLADLQNDEV-AFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMVKKWQT 119
Query: 120 LIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSC 179
+IEAHVDVKTTD Y LR+FC+GFTK+R+NQ+++T YAQ Q+RQIR+KM EIM + +
Sbjct: 120 MIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYAQHQQVRQIRKKMMEIMTREVQTN 179
Query: 180 DLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYS--- 236
DLK++V K IP+ IGK++EKA SIYPL +VF+RKVK+LK PKF+LGKLME+HG+ S
Sbjct: 180 DLKEVVNKLIPDSIGKDMEKACQSIYPLHDVFVRKVKMLKKPKFELGKLMELHGEGSSSG 239
Query: 237 ----EDVGTKVER 245
++ G KVER
Sbjct: 240 KATGDETGAKVER 252
>UniRef100_Q7RWF6 Hypothetical protein [Neurospora crassa]
Length = 337
Score = 342 bits (877), Expect = 6e-93
Identities = 174/257 (67%), Positives = 209/257 (80%), Gaps = 4/257 (1%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKN---VGKTLVSRTQGTKIA 57
MAVGKNKR+SKGKKG KKK DPFA+KDWY IKAP+ F V++ VGKTLV+RT G K A
Sbjct: 79 MAVGKNKRLSKGKKGLKKKTQDPFARKDWYGIKAPAPFNVRDLFSVGKTLVNRTTGLKNA 138
Query: 58 SEGLKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKW 117
++ LK R+FEVSLADLQ DED++FRK++LR +++QGKN LTNF G+DFT+DKLRSLVRKW
Sbjct: 139 NDALKGRIFEVSLADLQKDEDHSFRKVKLRVDEIQGKNCLTNFHGLDFTSDKLRSLVRKW 198
Query: 118 QTLIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQAT 177
QTLIEA++ VKTTD+Y LR+F I FTKRR NQVK+T YA SSQIR IRRKM EI+ +A+
Sbjct: 199 QTLIEANITVKTTDDYLLRLFAIAFTKRRPNQVKKTTYAASSQIRAIRRKMTEIIQREAS 258
Query: 178 SCDLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-S 236
SC L L K IPE+IG+EIEKAT IYPLQNV IRKVK+LK PKFDLG LM +HG+ S
Sbjct: 259 SCTLTQLTSKLIPEVIGREIEKATQGIYPLQNVHIRKVKLLKQPKFDLGALMSLHGESGS 318
Query: 237 EDVGTKVERPADETMVE 253
++ G KVER ET++E
Sbjct: 319 DEAGQKVEREFKETVLE 335
>UniRef100_UPI000035F12A UPI000035F12A UniRef100 entry
Length = 266
Score = 342 bits (876), Expect = 7e-93
Identities = 168/255 (65%), Positives = 209/255 (81%), Gaps = 11/255 (4%)
Query: 1 MAVGKNKRISKG-KKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASE 59
MAVGKNKR++KG KKG KKK DPF+KKDWYD+KAP++F ++N+GKTLV+RTQGT+IAS+
Sbjct: 1 MAVGKNKRLTKGGKKGAKKKIVDPFSKKDWYDVKAPAMFNIRNLGKTLVTRTQGTRIASD 60
Query: 60 GLKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQT 119
GLK RV+EVSLADLQ DE AFRK +L AEDVQGKN LTNF GMD T DK+ S+V+KWQT
Sbjct: 61 GLKGRVYEVSLADLQNDEV-AFRKFKLIAEDVQGKNCLTNFHGMDLTRDKMCSMVKKWQT 119
Query: 120 LIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSC 179
+IEAHVDVKTTD Y LR+FC+GFTKRR+NQ+++T YAQ Q+RQIR+KM EIM + +
Sbjct: 120 MIEAHVDVKTTDGYLLRLFCVGFTKRRNNQIRKTSYAQHQQVRQIRKKMMEIMTREVQTN 179
Query: 180 DLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGD----- 234
DLK++V K IP+ IGK+IEKA SIYPL +V++RKVK+LK PKF+LGKLME+HG+
Sbjct: 180 DLKEVVNKLIPDSIGKDIEKACQSIYPLHDVYVRKVKMLKKPKFELGKLMELHGEGGAGS 239
Query: 235 ----YSEDVGTKVER 245
+D+G KVER
Sbjct: 240 AAKAPGDDIGAKVER 254
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 404,150,135
Number of Sequences: 2790947
Number of extensions: 15583949
Number of successful extensions: 42552
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 167
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 42167
Number of HSP's gapped (non-prelim): 190
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC135798.2


