

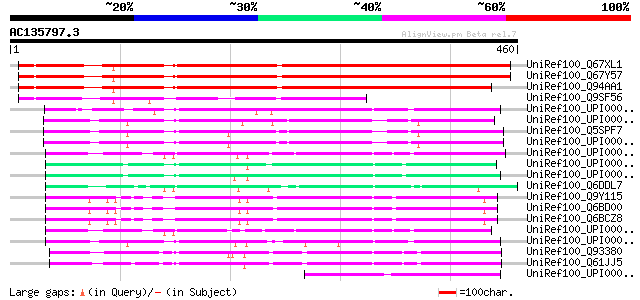
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135797.3 - phase: 0 /pseudo
(460 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q67XL1 MRNA, complete cds, clone: RAFL25-35-M14 [Arabi... 412 e-113
UniRef100_Q67Y57 MRNA, complete cds, clone: RAFL25-13-J23 [Arabi... 411 e-113
UniRef100_Q94AA1 AT3g09470/F11F8_4 [Arabidopsis thaliana] 392 e-107
UniRef100_Q9SF56 Hypothetical protein F11F8_4 [Arabidopsis thali... 218 2e-55
UniRef100_UPI0000430C57 UPI0000430C57 UniRef100 entry 114 4e-24
UniRef100_UPI0000248FBF UPI0000248FBF UniRef100 entry 114 6e-24
UniRef100_Q5SPF7 Novel protein similar to vertebrate unc-93 homo... 114 8e-24
UniRef100_UPI000024A98E UPI000024A98E UniRef100 entry 113 1e-23
UniRef100_UPI00004322B2 UPI00004322B2 UniRef100 entry 110 6e-23
UniRef100_UPI00003ACE0C UPI00003ACE0C UniRef100 entry 110 8e-23
UniRef100_UPI00003ACE0A UPI00003ACE0A UniRef100 entry 107 5e-22
UniRef100_Q6DDL7 MGC83353 protein [Xenopus laevis] 107 7e-22
UniRef100_Q9Y115 CG4928-PA, isoform A [Drosophila melanogaster] 104 6e-21
UniRef100_Q6BD00 CG4928 [Drosophila simulans] 104 6e-21
UniRef100_Q6BCZ8 CG4928 [Drosophila teissieri] 104 6e-21
UniRef100_UPI00004322B4 UPI00004322B4 UniRef100 entry 101 4e-20
UniRef100_UPI0000360999 UPI0000360999 UniRef100 entry 99 2e-19
UniRef100_Q93380 Hypothetical protein C46F11.1 [Caenorhabditis e... 97 1e-18
UniRef100_Q61JJ5 Hypothetical protein CBG09771 [Caenorhabditis b... 96 3e-18
UniRef100_UPI0000318FB5 UPI0000318FB5 UniRef100 entry 95 4e-18
>UniRef100_Q67XL1 MRNA, complete cds, clone: RAFL25-35-M14 [Arabidopsis thaliana]
Length = 437
Score = 412 bits (1059), Expect = e-113
Identities = 237/453 (52%), Positives = 298/453 (65%), Gaps = 34/453 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQLK+WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLKVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTLLCLSFCSFLWLALKVGN 454
+PYIS QA+++VML ++C+S SFL+LALKV N
Sbjct: 399 SPYISLQAMLIVMLVMVCVSLFSFLFLALKVEN 431
>UniRef100_Q67Y57 MRNA, complete cds, clone: RAFL25-13-J23 [Arabidopsis thaliana]
Length = 437
Score = 411 bits (1056), Expect = e-113
Identities = 236/453 (52%), Positives = 298/453 (65%), Gaps = 34/453 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQL++WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLRVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTLLCLSFCSFLWLALKVGN 454
+PYIS QA+++VML ++C+S SFL+LALKV N
Sbjct: 399 SPYISLQAMLIVMLVMVCVSLFSFLFLALKVEN 431
>UniRef100_Q94AA1 AT3g09470/F11F8_4 [Arabidopsis thaliana]
Length = 464
Score = 392 bits (1006), Expect = e-107
Identities = 226/436 (51%), Positives = 284/436 (64%), Gaps = 34/436 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQLK+WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLKVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTL 437
+PYIS QA+++VML +
Sbjct: 399 SPYISLQAMLIVMLVM 414
>UniRef100_Q9SF56 Hypothetical protein F11F8_4 [Arabidopsis thaliana]
Length = 285
Score = 218 bits (556), Expect = 2e-55
Identities = 143/324 (44%), Positives = 182/324 (56%), Gaps = 53/324 (16%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTI-- 62
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
+ I++V ++F V G ++ + W F +
Sbjct: 63 -------------SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFE--AQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNG 180
F +Y+G + WV QGTYLTS ARSHA D+ HEG+VIG FNG
Sbjct: 110 WFTMVPASLYLGFAASIIWV----------GQGTYLTSIARSHATDHGLHEGSVIGVFNG 159
Query: 181 EFWGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDY 239
EFW ++ H QEGST+GTTLL +VFL MT G IL F+ K G+
Sbjct: 160 EFWAMFACH---------------QEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGED 204
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
KG + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +V
Sbjct: 205 GKG---PVGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIV 261
Query: 300 TPEIGVSGVGIAMAVYGAFDGICS 323
TP IGVSGVG AMAVYGA D + S
Sbjct: 262 TPAIGVSGVGGAMAVYGALDAVVS 285
>UniRef100_UPI0000430C57 UPI0000430C57 UniRef100 entry
Length = 439
Score = 114 bits (286), Expect = 4e-24
Identities = 115/459 (25%), Positives = 187/459 (40%), Gaps = 76/459 (16%)
Query: 32 TRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYI 91
TR+V ++ +AF++ F A+ A NLQS++N A G L G F + + I
Sbjct: 7 TRNVLVIGLAFMVNFTAFMGATNLQSSIN-ADGSL--------GTFTLASIYGSLIFSNI 57
Query: 92 FFCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVY------------VGSGFCLSW 139
F + W L ++ F A Y VG G W
Sbjct: 58 FLPTLIISWLGCK-------WTISLSILAYVPFMAAQFYPKFYTMIPAGLLVGIGAAPLW 110
Query: 140 VLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFA 199
A C TYLT A ++A ++ ++ F G F+ Y + Q GNLI+ A
Sbjct: 111 C--------AKC--TYLTVVAEAYATLSDVATEILVTRFFGLFFMFYQMAQVWGNLISSA 160
Query: 200 LLSDGQEGSTNGTTLLFVVFLSV---------------------------MTFGAILTCF 232
+LS G + + TL V V + G L C
Sbjct: 161 VLSYGIDAAPTNITLNTSVVAEVCGAKFCGATSADNDGSNLERPSEKRIYLISGIYLCCM 220
Query: 233 LHKR-----GDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQH 287
+ G S Y G+ KS L L + +LI+P+I + G +
Sbjct: 221 IMASLIVAFGVDSLTRYNKNRTGSATGKSGLKLLVVTLKLLKEKNQILILPIIMFIGAEQ 280
Query: 288 AFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQ 347
AF++A++ V+ G++ +G M +G + I +L AG + LT +++F FV
Sbjct: 281 AFLFADYNASFVSCAWGINNIGYVMICFGVTNAIAALGAGSI-MKLTGRRPLMAFAFFVH 339
Query: 348 AVVLILLLLDFSMSSGFIGTLYILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQ 406
VLI LL M +I FL + L G+ D + + Q+NAL G+LF E AF+
Sbjct: 340 MGVLIFLLHWKPMPE----QNFIFFLISGLWGLCDAMWLVQINALSGLLFPGKEEAAFSN 395
Query: 407 LKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSF 445
++W+S + + +PY+ + + +++ +LC +
Sbjct: 396 FRLWESTGSVITYVYSPYLCTEIKLYILIGILCFGMIGY 434
>UniRef100_UPI0000248FBF UPI0000248FBF UniRef100 entry
Length = 411
Score = 114 bits (285), Expect = 6e-24
Identities = 112/442 (25%), Positives = 183/442 (41%), Gaps = 64/442 (14%)
Query: 31 HTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFY 90
+T++V ++S FLL+F AYG Q+LQS+LN G ++ + +A I+ +
Sbjct: 2 NTKNVLVVSFGFLLLFTAYGGLQSLQSSLNAEEGMGVISLSVI-------YAAIILSSMF 54
Query: 91 IFFCVCFFGGADSRI---QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMG 147
+ + G I C+ ++FG L + +G G W
Sbjct: 55 LPPIMIKNLGCKWTIVVSMGCYVAYSFGNLAPGWASLMSTSAILGMGGSPLWS------- 107
Query: 148 WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQEG 207
A C TYLT + +N +I + G F+ ++ GNL++ + Q
Sbjct: 108 -AKC--TYLTISGNRQGQKHNKKGQDLINQYFGIFFFIFQSSGVWGNLMSSLIFGQDQNI 164
Query: 208 ST-----NGTTLLFVVFLSVMTFGAILTCFLHKRG--------------DYSKGGYKHLD 248
T N + +F++ + + C L G + + K
Sbjct: 165 VTIKLLKNAAKQVDKLFVASYSPTLLTKCVLLHSGVGLLAIIFVAVFLDNIDRDEAKEFR 224
Query: 249 AGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGV 308
+ G +KS + L D ++LL+IPL YSG + +F+ E+TK VT +G+ V
Sbjct: 225 STKG-NKSFWDTFLATFKLLRDPRLLLLIPLTMYSGFEQSFLSGEYTKNYVTCALGIHNV 283
Query: 309 GIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTL 368
G M + A + +CS GRL + + L L ++ S F+G L
Sbjct: 284 GFVMICFAASNSLCSFAFGRL-------------AQYTGRIALFCLAAAINLGS-FLGLL 329
Query: 369 Y---------ILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMV 418
Y I F+ AL G+ D V TQ NAL G+LF + E AFA ++W+S +
Sbjct: 330 YWKPHPDQLAIFFVFPALWGMADAVWQTQTNALYGILFAKNKEAAFANYRMWESLGFVIA 389
Query: 419 FFLAPYISFQAVIMVMLTLLCL 440
F + +I I + L +L L
Sbjct: 390 FAYSTFICLSTKIYIALAVLAL 411
>UniRef100_Q5SPF7 Novel protein similar to vertebrate unc-93 homolog A [Brachydanio
rerio]
Length = 465
Score = 114 bits (284), Expect = 8e-24
Identities = 112/458 (24%), Positives = 186/458 (40%), Gaps = 74/458 (16%)
Query: 31 HTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFY 90
+T++V ++S FLL+F AYG Q+LQS+LN G ++ + +A I+ +
Sbjct: 4 NTKNVLVVSFGFLLLFTAYGGLQSLQSSLNAEEGMGVISLSVI-------YAAIILSSMF 56
Query: 91 IFFCVCFFGGADSRI---QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMG 147
+ + G I C+ ++FG L + +G G W
Sbjct: 57 LPPIMIKNLGCKWTIVVSMGCYVAYSFGNLAPGWASLMSTSAILGMGGSPLWS------- 109
Query: 148 WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---------- 197
A C TYLT + +N +I + G F+ ++ GNL++
Sbjct: 110 -AKC--TYLTISGNRQGQKHNKKGQDLINQYFGIFFFIFQSSGVWGNLMSSLIFGQDQNI 166
Query: 198 -----------------FALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGDYS 240
F ++ + S + L ++ V I D
Sbjct: 167 VPKENLEFCGVSTCLDNFTVIGNSTRPSKHLVDTLLGCYIGVGLLAIIFVAVFLDNIDRD 226
Query: 241 KGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVT 300
+ K + G +KS + L D ++LL+IPL YSG + +F+ E+TK VT
Sbjct: 227 EA--KEFRSTKG-NKSFWDTFLATFKLLRDPRLLLLIPLTMYSGFEQSFLSGEYTKNYVT 283
Query: 301 PEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSM 360
+G+ VG M + A + +CS GRL + + L L ++
Sbjct: 284 CALGIHNVGFVMICFAASNSLCSFAFGRL-------------AQYTGRIALFCLAAAINL 330
Query: 361 SSGFIGTLY---------ILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIW 410
S F+G LY I F+ AL G+ D V TQ NAL G+LF + E AFA ++W
Sbjct: 331 GS-FLGLLYWKPHPDQLAIFFVFPALWGMADAVWQTQTNALYGILFAKNKEAAFANYRMW 389
Query: 411 QSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWL 448
+S + F + +I I + L +L L+ ++L++
Sbjct: 390 ESLGFVIAFAYSTFICLSTKIYIALAVLALTMVTYLYV 427
>UniRef100_UPI000024A98E UPI000024A98E UniRef100 entry
Length = 437
Score = 113 bits (282), Expect = 1e-23
Identities = 112/460 (24%), Positives = 186/460 (40%), Gaps = 76/460 (16%)
Query: 31 HTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFY 90
+T++V ++S FLL+F AYG Q+LQS+LN G ++ + +A I+ +
Sbjct: 4 NTKNVLVVSFGFLLLFTAYGGLQSLQSSLNAEEGMGVISLSVI-------YAAIILSSMF 56
Query: 91 IFFCVCFFGGADSRI---QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMG 147
+ + G I C+ ++FG L + +G G W
Sbjct: 57 LPPIMIKNLGCKWTIVVSMGCYVAYSFGNLAPGWASLMSTSAILGMGGSPLWS------- 109
Query: 148 WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---------- 197
A C TYLT + +N +I + G F+ ++ GNL++
Sbjct: 110 -AKC--TYLTISGNRQGQKHNKKGQDLINQYFGIFFFIFQSSGVWGNLMSSLIFGQDQNI 166
Query: 198 -------------------FALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGD 238
F ++ + S + L ++ V I D
Sbjct: 167 VEVPKENLEFCGVSTCLDNFTVIGNSTRPSKHLVDTLLGCYIGVGLLAIIFVAVFLDNID 226
Query: 239 YSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYV 298
+ K + G +KS + L D ++LL+IPL YSG + +F+ E+TK
Sbjct: 227 RDEA--KEFRSTKG-NKSFWDTFLATFKLLRDPRLLLLIPLTMYSGFEQSFLSGEYTKNY 283
Query: 299 VTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDF 358
VT +G+ VG M + A + +CS GRL + + L L
Sbjct: 284 VTCALGIHNVGFVMICFAASNSLCSFAFGRL-------------AQYTGRIALFCLAAAI 330
Query: 359 SMSSGFIGTLY---------ILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
++ S F+G LY I F+ AL G+ D V TQ NAL G+LF + E AFA +
Sbjct: 331 NLGS-FLGLLYWKPHPDQLAIFFVFPALWGMADAVWQTQTNALYGILFAKNKEAAFANYR 389
Query: 409 IWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWL 448
+W+S + F + +I I + L +L L+ ++L++
Sbjct: 390 MWESLGFVIAFAYSTFICLSTKIYIALAVLALTMVTYLYV 429
>UniRef100_UPI00004322B2 UPI00004322B2 UniRef100 entry
Length = 524
Score = 110 bits (276), Expect = 6e-23
Identities = 112/462 (24%), Positives = 197/462 (42%), Gaps = 75/462 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
+++ +S+AF++ F A+ NLQS++N + G V + I+ +
Sbjct: 49 KNISTVSVAFMVQFTAFQGTANLQSSINASDGLGTVSLS-------------AIYAALVL 95
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
C+ R+ + L P Y+GS F + V ++G
Sbjct: 96 SCIFVPTFVIKRLTVKWTLCISMLCYAP---------YIGSQFYPKFYTLVPAGVLLGLG 146
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
WAA Q TYLT +A + A++ F G F+ + + GNLI+ +LS
Sbjct: 147 AAPMWAA-QATYLTQVGGVYAKLTDQPVDAIVVRFFGFFFLAWQTAELWGNLISSLVLSG 205
Query: 204 GQ--EGSTNGTTL--------------------------------LFVVFLSVMTFGAIL 229
G+ GS N TT + ++LS + I+
Sbjct: 206 GEFGSGSENSTTNSNKIKHCGANFCVLGNGGHETLERPPESEIYEISAIYLSCVIVAVII 265
Query: 230 TCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAF 289
S+ G K + + ++ L + L L+IP+ + G++ AF
Sbjct: 266 VALFVD--PLSRYGEKQRKVDSQELSGIQ-LLSATAYQLKKPYQQLLIPITIWIGMEQAF 322
Query: 290 VWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAV 349
+ A+FT+ ++ +GV VG M +G + ICSLV G L ++ GA V V
Sbjct: 323 IGADFTQAYISCALGVHRVGYVMICFGVVNAICSLVFGSL-MKFVGRQPLMVLGAIVH-V 380
Query: 350 VLILLLLDFSMSSGFIGTLYILF-LAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
LI++LL + + Y+ + ++ L G+GD V TQ+N L G LF+ + E AF+ +
Sbjct: 381 SLIVVLLHWKPNPD---NPYVFYSVSGLWGVGDAVWQTQVNGLYGTLFRRNKEAAFSNYR 437
Query: 409 IWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWLAL 450
+W+SA + + + ++ + + VMLT+L + C ++ + L
Sbjct: 438 LWESAGFVIAYAYSTHLCARMKLYVMLTVLLVGTCGYIVVEL 479
>UniRef100_UPI00003ACE0C UPI00003ACE0C UniRef100 entry
Length = 420
Score = 110 bits (275), Expect = 8e-23
Identities = 103/437 (23%), Positives = 173/437 (39%), Gaps = 50/437 (11%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
++V +++ FLL+F AYG Q+LQS+LN+ G + + + +
Sbjct: 4 KNVLVIAFGFLLLFTAYGGLQSLQSSLNSEEGLGVASLSVLYAALIVSSMFLPPILIKKL 63
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQ 152
C G+ C+ ++ G V +G G W A C
Sbjct: 64 GCKWTIAGSMC----CYIAFSLGNFYASWYTLIPTSVILGLGGAPLWS--------AKC- 110
Query: 153 GTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQEGSTNGT 212
TYLT S+A +I + G F+ ++ GNLI+ +LS ++
Sbjct: 111 -TYLTIAGNSYAEKAGKIGKDIINQYFGVFFLIFQSSGIWGNLISSLILSQSSNKDSSAK 169
Query: 213 TL----------------------------LFVVFLSVMTFGAILTCFLHKRGDYSKGGY 244
T LFV+FL ++ ++ FL D K
Sbjct: 170 TSHAELSTKTITVTSNFPSFFQHSSFFFHELFVLFLQLVVAVLLIIIFL----DQIKSDQ 225
Query: 245 KHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIG 304
+ ++ S S + L D + L+IPL YSG + F+ ++TK VT +G
Sbjct: 226 AETEKEIQKTPSFWSTFLATFQHLKDKRQCLLIPLTMYSGFEQGFLAGDYTKTYVTCALG 285
Query: 305 VSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGF 364
+ VG M + A + +CSL+ G+++ T + + +I LLL F
Sbjct: 286 IHYVGYVMICFSAVNSLCSLLFGKIS-QFTGRKFLFALATVTNTSCIIALLL---WKPHF 341
Query: 365 IGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPY 424
+ AL G+ D + TQ N L G+LF+ E AFA ++W+S + F +
Sbjct: 342 NQLVVFFIFPALWGLSDAIWQTQTNGLYGVLFEKHTEAAFANYRLWESCGFVIAFGYSTT 401
Query: 425 ISFQAVIMVMLTLLCLS 441
+ + ++L +L LS
Sbjct: 402 LRVYIKLYILLAVLTLS 418
>UniRef100_UPI00003ACE0A UPI00003ACE0A UniRef100 entry
Length = 435
Score = 107 bits (268), Expect = 5e-22
Identities = 106/439 (24%), Positives = 173/439 (39%), Gaps = 44/439 (10%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
++V +++ FLL+F AYG Q+LQS+LN+ G + + + +
Sbjct: 6 KNVLVIAFGFLLLFTAYGGLQSLQSSLNSEEGLGVASLSVLYAALIVSSMFLPPILIKKL 65
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQ 152
C G+ C+ ++ G V +G G W A C
Sbjct: 66 GCKWTIAGSMC----CYIAFSLGNFYASWYTLIPTSVILGLGGAPLWS--------AKC- 112
Query: 153 GTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLS---------- 202
TYLT S+A +I + G F+ ++ GNLI+ +LS
Sbjct: 113 -TYLTIAGNSYAEKAGKIGKDIINQYFGVFFLIFQSSGIWGNLISSLILSQSSNKVEISE 171
Query: 203 ---------DGQEGSTNGTTL------LFVVFLSVMTFGAILTCFLHKRG-DYSKGGYKH 246
D +TN T L L L V T +L L D K
Sbjct: 172 EDLECCGAYDCVSNTTNTTALAEPSKSLVYTLLGVYTASGVLAVLLIIIFLDQIKSDQAE 231
Query: 247 LDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVS 306
+ ++ S S + L D + L+IPL YSG + F+ ++TK VT +G+
Sbjct: 232 TEKEIQKTPSFWSTFLATFQHLKDKRQCLLIPLTMYSGFEQGFLAGDYTKTYVTCALGIH 291
Query: 307 GVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIG 366
VG M + A + +CSL+ G+++ T + + +I LLL F
Sbjct: 292 YVGYVMICFSAVNSLCSLLFGKIS-QFTGRKFLFALATVTNTSCIIALLL---WKPHFNQ 347
Query: 367 TLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYIS 426
+ AL G+ D + TQ N L G+LF+ E AFA ++W+S + F + +
Sbjct: 348 LVVFFIFPALWGLSDAIWQTQTNGLYGVLFEKHTEAAFANYRLWESCGFVIAFGYSTTLR 407
Query: 427 FQAVIMVMLTLLCLSFCSF 445
+ ++L +L LS ++
Sbjct: 408 VYIKLYILLAVLTLSMVTY 426
>UniRef100_Q6DDL7 MGC83353 protein [Xenopus laevis]
Length = 460
Score = 107 bits (267), Expect = 7e-22
Identities = 118/474 (24%), Positives = 193/474 (39%), Gaps = 81/474 (17%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
+++ I+S FLL+F A+G Q+LQS+LN+ G + + V Y
Sbjct: 5 KNILIVSFGFLLLFTAFGGLQSLQSSLNSDEGLGVASLS----------------VIYAA 48
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
V I++ W +V C + + Y F SW + I+G
Sbjct: 49 LIVSSVFVPPIVIKKIGCKWT---IVASMCCY---ITYSLGNFYASWYTLIPTSLILGFG 102
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
WAA + TYLT + +A ++ + G F+ ++ GNLI+ +
Sbjct: 103 GAPLWAA-KCTYLTESGNRYAEKKGKLAKDIVNQYFGLFFLIFQSSGVWGNLISSLIFGQ 161
Query: 204 GQE---------------------------GSTNGTTLLFVVFLSVMTFGAILTCFL--- 233
G+T T L L V T +L L
Sbjct: 162 NYPAGSNDSFTDYSQCGANDCPGTNFGNGTGTTKPTKSLIYTLLGVYTGSGVLAVILIAV 221
Query: 234 -----HKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ R D K G K +S S K L + L D + L+IPL YSG +
Sbjct: 222 FLDTINLRTDQLKPGTKE------ESFSKKIL--ATVRHLKDKRQCLLIPLTMYSGFEQG 273
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ ++TK VT +G+ VG M + A + +CSL+ G+L+ T + A A
Sbjct: 274 FLSGDYTKSYVTCSLGIHFVGYVMICFAATNAVCSLLFGQLS-KYTGRICLFILAAVSNA 332
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
+I LLL + F ++ +F A+ G+ D + TQ NAL G+LF E AFA +
Sbjct: 333 ACVIALLLWEPYPNDF--AVFFIF-PAIWGMADAIWQTQTNALYGVLFDEHKEAAFANYR 389
Query: 409 IWQSATIAMVFFLAPY--ISFQAVIMVMLTLLCLSFCSFLWLALKVGNASSPST 460
+W+S + + + + +S + I++ + L+ + F F+ V + + ST
Sbjct: 390 LWESLGFVIAYGYSTFLCVSVKLYILLAVLLIAIVFYGFVEYLEHVEHKKAAST 443
>UniRef100_Q9Y115 CG4928-PA, isoform A [Drosophila melanogaster]
Length = 538
Score = 104 bits (259), Expect = 6e-21
Identities = 109/457 (23%), Positives = 193/457 (41%), Gaps = 76/457 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIE--FSGGGFGYDFAWDIIF--- 87
+++ I+SIAF++ F A+ NLQS++N G V + ++ F +I
Sbjct: 44 KNISIISIAFMVQFTAFQGTANLQSSINAKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 103
Query: 88 -VFYIFFC--VCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFY 144
V + C +C+ A + F F LV G + VG G W
Sbjct: 104 TVKWTLVCSMLCY---APYIAFQLFP--RFYTLVPAG-------ILVGMGAAPMW----- 146
Query: 145 IMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDG 204
A + TYLT + +A A+I F G F+ + + GNLI+ +LS G
Sbjct: 147 -----ASKATYLTQVGQVYAKITEQAVDAIIVRFFGFFFLAWQSAELWGNLISSLVLSSG 201
Query: 205 QEG--STNGTTL---------------------------------LFVVFLS-VMTFGAI 228
G S++ TT+ + +++LS ++ I
Sbjct: 202 AHGGGSSSNTTVSEEDLQFCGANFCTTGSGGHGNLERPPEDEIFEISMIYLSCIVAAVCI 261
Query: 229 LTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ FL Y G K + + S L + + + L+IP+ + G++ A
Sbjct: 262 IAFFLDPLKRY---GEKRKGSNSAAELSGLQLLSATFRQMKKPNLQLLIPITVFIGMEQA 318
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ A+FT+ V +GV+ +G M +G + +CS++ G + T I+ GA V
Sbjct: 319 FIGADFTQAYVACALGVNKIGFVMICFGVVNALCSILFGSV-MKYIGRTPIIVLGAVVH- 376
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
L+ ++ + ++ L G+GD V TQ+N L G+LF+ + E AF+ +
Sbjct: 377 --FTLITVELFWRPNPDNPIIFYAMSGLWGVGDAVWQTQINGLYGLLFRRNKEAAFSNYR 434
Query: 409 IWQSATIAMVFFLAPYISFQA---VIMVMLTLLCLSF 442
+W+SA + + A + Q +++ +LTL C+ +
Sbjct: 435 LWESAGFVIAYAYATTLCTQMKLYILLAVLTLGCIGY 471
>UniRef100_Q6BD00 CG4928 [Drosophila simulans]
Length = 514
Score = 104 bits (259), Expect = 6e-21
Identities = 109/457 (23%), Positives = 193/457 (41%), Gaps = 76/457 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIE--FSGGGFGYDFAWDIIF--- 87
+++ I+SIAF++ F A+ NLQS++N G V + ++ F +I
Sbjct: 35 KNISIISIAFMVQFTAFQGTANLQSSINAKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 94
Query: 88 -VFYIFFC--VCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFY 144
V + C +C+ A + F F LV G + VG G W
Sbjct: 95 TVKWTLVCSMLCY---APYIAFQLFP--RFYTLVPAG-------ILVGMGAAPMW----- 137
Query: 145 IMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDG 204
A + TYLT + +A A+I F G F+ + + GNLI+ +LS G
Sbjct: 138 -----ASKATYLTQVGQVYAKITEQAVDAIIVRFFGFFFLAWQSAELWGNLISSLVLSSG 192
Query: 205 QEG--STNGTTL---------------------------------LFVVFLS-VMTFGAI 228
G S++ TT+ + +++LS ++ I
Sbjct: 193 AHGGGSSSNTTVSEEDLQFCGANFCTTGSGGHGNLERPPEDEIFEISMIYLSCIVAAVCI 252
Query: 229 LTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ FL Y G K + + S L + + + L+IP+ + G++ A
Sbjct: 253 IAFFLDPLKRY---GEKRKGSNSAAELSGLQLLSATFRQMKKPNLQLLIPITVFIGMEQA 309
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ A+FT+ V +GV+ +G M +G + +CS++ G + T I+ GA V
Sbjct: 310 FIGADFTQAYVACALGVNKIGFVMICFGVVNALCSILFGSV-MKYIGRTPIIVLGAVVH- 367
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
L+ ++ + ++ L G+GD V TQ+N L G+LF+ + E AF+ +
Sbjct: 368 --FTLITVELFWRPNPDNPIIFYAMSGLWGVGDAVWQTQINGLYGLLFRRNKEAAFSNYR 425
Query: 409 IWQSATIAMVFFLAPYISFQA---VIMVMLTLLCLSF 442
+W+SA + + A + Q +++ +LTL C+ +
Sbjct: 426 LWESAGFVIAYAYATTLCTQMKLYILLAVLTLGCIGY 462
>UniRef100_Q6BCZ8 CG4928 [Drosophila teissieri]
Length = 514
Score = 104 bits (259), Expect = 6e-21
Identities = 109/457 (23%), Positives = 193/457 (41%), Gaps = 76/457 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIE--FSGGGFGYDFAWDIIF--- 87
+++ I+SIAF++ F A+ NLQS++N G V + ++ F +I
Sbjct: 35 KNISIISIAFMVQFTAFQGTANLQSSINAKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 94
Query: 88 -VFYIFFC--VCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFY 144
V + C +C+ A + F F LV G + VG G W
Sbjct: 95 TVKWTLVCSMLCY---APYIAFQLFP--RFYTLVPAG-------ILVGMGAAPMW----- 137
Query: 145 IMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDG 204
A + TYLT + +A A+I F G F+ + + GNLI+ +LS G
Sbjct: 138 -----ASKATYLTQVGQVYAKITEQAVDAIIVRFFGFFFLAWQSAELWGNLISSLVLSSG 192
Query: 205 QEG--STNGTTL---------------------------------LFVVFLS-VMTFGAI 228
G S++ TT+ + +++LS ++ I
Sbjct: 193 AHGGSSSSNTTVSEEDLQFCGANFCTTGSGGHGNLERPPEDEIFEISMIYLSCIVAAVCI 252
Query: 229 LTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ FL Y G K + + S L + + + L+IP+ + G++ A
Sbjct: 253 IAFFLDPLKRY---GEKRKGSNSAAELSGLQLLSATFRQMKKPNLQLLIPITVFIGMEQA 309
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ A+FT+ V +GV+ +G M +G + +CS++ G + T I+ GA V
Sbjct: 310 FIGADFTQAYVACALGVNKIGFVMICFGVVNALCSILFGSV-MKYIGRTPIIVLGAVVH- 367
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
L+ ++ + ++ L G+GD V TQ+N L G+LF+ + E AF+ +
Sbjct: 368 --FTLITVELFWRPNPDNPIIFYAMSGLWGVGDAVWQTQINGLYGLLFRRNKEAAFSNYR 425
Query: 409 IWQSATIAMVFFLAPYISFQA---VIMVMLTLLCLSF 442
+W+SA + + A + Q +++ +LTL C+ +
Sbjct: 426 LWESAGFVIAYAYATTLCTQMKLYILLAVLTLGCIGY 462
>UniRef100_UPI00004322B4 UPI00004322B4 UniRef100 entry
Length = 376
Score = 101 bits (252), Expect = 4e-20
Identities = 107/415 (25%), Positives = 185/415 (43%), Gaps = 49/415 (11%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
+++ +S+AF++ F A+ NLQS++N + G V + I+ +
Sbjct: 1 KNISTVSVAFMVQFTAFQGTANLQSSINASDGLGTVSLS-------------AIYAALVL 47
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
C+ R+ + L P Y+GS F + V ++G
Sbjct: 48 SCIFVPTFVIKRLTVKWTLCISMLCYAP---------YIGSQFYPKFYTLVPAGVLLGLG 98
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
WAA Q TYLT +A + A++ F G F+ + + GNLI+ S
Sbjct: 99 AAPMWAA-QATYLTQVGGVYAKLTDQPVDAIVVRFFGFFFLAWQTAELWGNLIS----SL 153
Query: 204 GQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRS 263
G+ S L F++ L + IL FL + G K + + ++ L +
Sbjct: 154 GKSKSR----LSFLLPLFQLESNRIL--FLDSLLFFHVYGEKQRKVDSQELSGIQ-LLSA 206
Query: 264 LTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICS 323
L L+IP+ + G++ AF+ A+FT+ ++ +GV VG M +G + ICS
Sbjct: 207 TAYQLKKPYQQLLIPITIWIGMEQAFIGADFTQAYISCALGVHRVGYVMICFGVVNAICS 266
Query: 324 LVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILF-LAALLGIGDG 382
LV G L ++ GA V V LI++LL + + Y+ + ++ L G+GD
Sbjct: 267 LVFGSL-MKFVGRQPLMVLGAIVH-VSLIVVLLHWKPNPD---NPYVFYSVSGLWGVGDA 321
Query: 383 VLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTL 437
V TQ+N L G LF+ + E AF+ ++W+SA + + + ++ + + VMLT+
Sbjct: 322 VWQTQVNGLYGTLFRRNKEAAFSNYRLWESAGFVIAYAYSTHLCARMKLYVMLTV 376
>UniRef100_UPI0000360999 UPI0000360999 UniRef100 entry
Length = 440
Score = 99.4 bits (246), Expect = 2e-19
Identities = 112/454 (24%), Positives = 186/454 (40%), Gaps = 72/454 (15%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
++V ++S+ FL +F+A G QNLQS+LN G + + +A II ++
Sbjct: 7 KNVLVVSVGFLFLFMAAGGLQNLQSSLNAEEGMGVASLSII-------YASIIISSMFLP 59
Query: 93 FCVCFFGGADSRI---QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWA 149
+ G + C+ ++ G L V +G G W A
Sbjct: 60 PIMIKNLGCKWTVVAGMGCYVSYSLGNLYPGWYTLMPTSVILGLGGAPLWS--------A 111
Query: 150 ACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD------ 203
C TYLT + + A VI + G F+ ++ GNL++ +
Sbjct: 112 KC--TYLTISGNTQAAAQGKRAPDVINQYFGIFFFIFQSSGVWGNLMSSLIFGQDTKIAE 169
Query: 204 --------------GQEGSTNGTT----------LLFVVFLSVMTFGAILTCFLHKRGDY 239
G +NGTT LL + + I+ FL D
Sbjct: 170 IPEEQLRTCGVTECGLVVQSNGTTPRPAQEVVQTLLGCYIGAGVLAMLIVAVFLD---DI 226
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGA---LSDVKMLLIIPLIAYSGLQHAFVWAEFTK 296
+ D + + + C++ L D ++LL++PL YSG + +F+ E+TK
Sbjct: 227 DR------DRASRFREKREPFCKTFLATFRLLKDRRLLLVVPLTMYSGFEQSFLSGEYTK 280
Query: 297 Y----VVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLI 352
VT +G+ +G M +GA + I S + GRL T +++ A +I
Sbjct: 281 AGTLTYVTCALGIHYIGFVMMCFGATNSISSFLFGRLA-RYTGRAALMCLAAATNLSCII 339
Query: 353 LLLLDFSMSSGFIGTLYILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQ 411
LL L++ FL AAL G+ D V TQ NAL G+LF D E AFA ++W+
Sbjct: 340 TFLLWKPDPK----QLHVFFLLAALWGMADAVWQTQTNALYGILFPRDKEAAFANYRMWE 395
Query: 412 SATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSF 445
S + F + ++ + + +ML +L L+ ++
Sbjct: 396 SLGFVISFAYSSFLCLEYKLYIMLGVLVLAAVTY 429
>UniRef100_Q93380 Hypothetical protein C46F11.1 [Caenorhabditis elegans]
Length = 705
Score = 96.7 bits (239), Expect = 1e-18
Identities = 104/448 (23%), Positives = 194/448 (43%), Gaps = 66/448 (14%)
Query: 37 ILSIAFLLIFLAYGAAQNLQSTLNTASG-ELIVCIEFSGGGFGYDFAWDIIFVFYIFFCV 95
ILS+AFL +F A+ QNLQ+++N G + +V + S + + +F
Sbjct: 252 ILSVAFLFLFTAFNGLQNLQTSVNGDLGSDSLVALYLS------------LAISSLFVPS 299
Query: 96 CFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTY 155
++ + + L ++ + + S FC + I G A C Y
Sbjct: 300 FMINRLGCKLTFLIAIFVYFLYIVINLRPTYSSMIPASIFC--GIAASCIWG-AKC--AY 354
Query: 156 LTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---FAL------------ 200
+T +A N + VI F G F+ + Q +GN+++ F L
Sbjct: 355 ITEMGIRYASLNFESQTTVIVRFFGYFFMIVHCGQVVGNMVSSYIFTLSYSQALRGPEDS 414
Query: 201 ------------LSDGQEGSTNG---------TTLLFVVFLSVMTFGAILTCFLHKRGDY 239
LSD E + + + V+ G I++ FL+
Sbjct: 415 IYDSCGYQFPKNLSDLTELAESNLARPPQKVYVAVCLAYLACVIISGMIMSMFLNA---L 471
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
+K A S+ + + L ++K +L++PL ++GL+ AF+ +TK V
Sbjct: 472 AKDARNRKMAQKFNSEIFYLMLKHLI----NIKFMLLVPLTIFNGLEQAFLVGVYTKAFV 527
Query: 300 TPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFS 359
+G+ +G MA +G D +CSLV G L L + FGA V ++++ L++ +
Sbjct: 528 GCGLGIWQIGFVMACFGISDAVCSLVFGPL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WP 585
Query: 360 MSSGFIGTLYILFLAALLGIGDGVLMTQLNAL-LGMLFKHDMEGAFAQLKIWQSATIAMV 418
+++ Y+ +AA+ G+ DGV TQ+N + ++ + ++ AF + + W+S IA+
Sbjct: 586 LNAADTQIFYV--VAAMWGMADGVWNTQINGFWVALVGRQSLQFAFTKYRFWESLGIAIG 643
Query: 419 FFLAPYISFQAVIMVMLTLLCLSFCSFL 446
F L +++ + +++ +L L C FL
Sbjct: 644 FALIRHVTVEIYLLITFFMLLLGMCGFL 671
>UniRef100_Q61JJ5 Hypothetical protein CBG09771 [Caenorhabditis briggsae]
Length = 716
Score = 95.5 bits (236), Expect = 3e-18
Identities = 104/448 (23%), Positives = 192/448 (42%), Gaps = 66/448 (14%)
Query: 37 ILSIAFLLIFLAYGAAQNLQSTLNTASG-ELIVCIEFSGGGFGYDFAWDIIFVFYIFFCV 95
ILS+A L +F A+ QNLQ+++N G + +V + S I +F F +
Sbjct: 247 ILSVALLFLFTAFNGLQNLQTSVNGDLGSDSLVALYLSLA---------ISSLFVPSFMI 297
Query: 96 CFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTY 155
G + + F + L ++ + + S FC + I G A C Y
Sbjct: 298 NRLGCKMTFLVAIFV---YFLYIVINLRPTYSSMIPASIFC--GIAASCIWG-AKC--AY 349
Query: 156 LTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQEGSTNG---- 211
+T +A N + VI F G F+ + Q +GN+++ + + GS G
Sbjct: 350 ITEMGIRYASLNFESQTTVIVRFFGYFFMIVHCGQVVGNVVSSYIFTLSYSGSLRGPEDS 409
Query: 212 --------------------------------TTLLFVVFLSVMTFGAILTCFLHKRGDY 239
+ ++ G I++ FL+
Sbjct: 410 IYDSCGYQFPKNLSYLSELAESNLARPPQKVYVAVCLAYLACIIISGMIMSMFLNA---L 466
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
K A S+ + + + L ++K LL++PL ++GL+ AF+ +TK V
Sbjct: 467 VKDTRNRKMAQRFNSEIITLMVKHLI----NIKFLLLVPLTIFNGLEQAFLVGVYTKAFV 522
Query: 300 TPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFS 359
+G+ +G MA +G D ICSLV G L L + FGA V ++++ L++ +
Sbjct: 523 GCGLGIWQIGFVMACFGISDAICSLVFGPL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WP 580
Query: 360 MSSGFIGTLYILFLAALLGIGDGVLMTQLNAL-LGMLFKHDMEGAFAQLKIWQSATIAMV 418
+++ Y+ +AA+ G+ DGV TQ+N + ++ + ++ AF + + W+S IA+
Sbjct: 581 LNAADTQIFYV--VAAMWGMADGVWNTQINGFWVALVGRQSLQFAFTKYRFWESLGIAIG 638
Query: 419 FFLAPYISFQAVIMVMLTLLCLSFCSFL 446
F L +++ + +++ +L L C F+
Sbjct: 639 FALIRHVTVEMYLLITFFVLLLGMCGFV 666
>UniRef100_UPI0000318FB5 UPI0000318FB5 UniRef100 entry
Length = 540
Score = 95.1 bits (235), Expect = 4e-18
Identities = 58/179 (32%), Positives = 94/179 (52%), Gaps = 6/179 (3%)
Query: 268 LSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAG 327
L D ++L +IPL YSG + +F+ E+TK VT +G+ VG M +GA + I S + G
Sbjct: 352 LKDWRLLALIPLTMYSGFEQSFLSGEYTKNYVTCALGIHYVGFVMMCFGASNSIFSFLFG 411
Query: 328 RLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLA-ALLGIGDGVLMT 386
RL + I + AV + ++ F + LY+ FL AL G+ D V
Sbjct: 412 RLARYTGRLALIC-----LAAVTNLACIITFLLWKPDPQQLYLFFLLPALWGMADAVWQP 466
Query: 387 QLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSF 445
Q+NAL G+LF D E AFA ++W+S + F + ++ + + +ML +L L+ ++
Sbjct: 467 QVNALYGILFPRDKEAAFANYRMWESLGFVIAFAYSTFLCLEYKVYIMLGVLLLTAVTY 525
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.329 0.143 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 744,624,346
Number of Sequences: 2790947
Number of extensions: 31014735
Number of successful extensions: 96058
Number of sequences better than 10.0: 184
Number of HSP's better than 10.0 without gapping: 76
Number of HSP's successfully gapped in prelim test: 108
Number of HSP's that attempted gapping in prelim test: 95793
Number of HSP's gapped (non-prelim): 263
length of query: 460
length of database: 848,049,833
effective HSP length: 131
effective length of query: 329
effective length of database: 482,435,776
effective search space: 158721370304
effective search space used: 158721370304
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135797.3


