

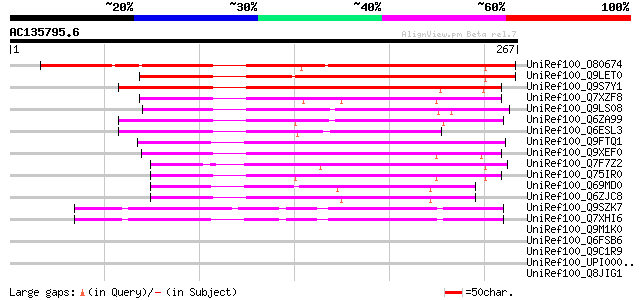
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.6 - phase: 0
(267 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80674 Hypothetical protein At2g41130 [Arabidopsis tha... 233 3e-60
UniRef100_Q9LET0 Putative HLH DNA binding protein [Arabidopsis t... 196 4e-49
UniRef100_Q9S7Y1 Putative DNA-binding protein; 36199-34606 [Arab... 154 2e-36
UniRef100_Q7XZF8 Putative DNA binding protein [Oryza sativa] 146 6e-34
UniRef100_Q9LS08 Putative HLH DNA-binding protein [Arabidopsis t... 139 6e-32
UniRef100_Q6ZA99 DNA-binding protein-like [Oryza sativa] 124 3e-27
UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa] 120 3e-26
UniRef100_Q9FTQ1 DNA binding protein-like [Oryza sativa] 110 4e-23
UniRef100_Q9XEF0 Hypothetical protein T07M07.8 [Arabidopsis thal... 106 5e-22
UniRef100_Q7F7Z2 ESTs C26093 [Oryza sativa] 98 3e-19
UniRef100_Q75IR0 Hypothetical protein OSJNBb0099P06.13 [Oryza sa... 91 4e-17
UniRef100_Q69MD0 BHLH-like protein [Oryza sativa] 81 3e-14
UniRef100_Q6ZJC8 BHLH protein family-like [Oryza sativa] 81 3e-14
UniRef100_Q9SZK7 Hypothetical protein F20D10.190 [Arabidopsis th... 75 1e-12
UniRef100_Q7XHI6 Putative bHLH131 transcription factor [Arabidop... 72 1e-11
UniRef100_Q9M1K0 Hypothetical protein F24I3.60 [Arabidopsis thal... 44 0.003
UniRef100_Q6FSB6 Tr|Q9C1R9 Candida glabrata Centromere binding p... 44 0.006
UniRef100_Q9C1R9 Centromere binding protein 1 [Candida glabrata] 44 0.006
UniRef100_UPI000036202E UPI000036202E UniRef100 entry 43 0.010
UniRef100_Q8JIG1 BHLH-PAS transcription factor [Brachydanio rerio] 43 0.010
>UniRef100_O80674 Hypothetical protein At2g41130 [Arabidopsis thaliana]
Length = 253
Score = 233 bits (595), Expect = 3e-60
Identities = 137/263 (52%), Positives = 176/263 (66%), Gaps = 33/263 (12%)
Query: 17 LFQFLVANNPSFFEYSTPSSTMM-QQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAA 75
L+ FL N Y M QS C SS + ++YY P +S I +T +Q DRALAA
Sbjct: 10 LYSFLAGNEVGGGGYCVSGDYMTTMQSLCGSSSSTSSYY-PLAISGIGETMAQ-DRALAA 67
Query: 76 LKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVE 135
L+NHKEAE+RRRERINSHL+ LR +L CNSKT K + LAKVV+
Sbjct: 68 LRNHKEAERRRRERINSHLNKLRNVLSCNSKT-----------------DKATLLAKVVQ 110
Query: 136 RVKELRQQTCQITQVET--VPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDL 193
RV+EL+QQT + + + +P E D +V+ G D S +G +IFKASLCCEDRSDL+PDL
Sbjct: 111 RVRELKQQTLETSDSDQTLLPSETDEISVLHFG-DYSNDGHIIFKASLCCEDRSDLLPDL 169
Query: 194 IEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSS---- 249
+EILKSL++KTL+AEM T+GGRTR+VLVVAA+KE + +ES+HFLQN+L+SLL+RSS
Sbjct: 170 MEILKSLNMKTLRAEMVTIGGRTRSVLVVAADKEMHGVESVHFLQNALKSLLERSSKSLM 229
Query: 250 ------GCNDRSKRRRGLDRRMM 266
G +RSKRRR LD +M
Sbjct: 230 ERSSGGGGGERSKRRRALDHIIM 252
>UniRef100_Q9LET0 Putative HLH DNA binding protein [Arabidopsis thaliana]
Length = 230
Score = 196 bits (499), Expect = 4e-49
Identities = 110/209 (52%), Positives = 145/209 (68%), Gaps = 29/209 (13%)
Query: 69 QDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGS 128
+D+ALA+L+NHKEAE++RR RINSHL+ LR LL CNSKT K +
Sbjct: 39 EDKALASLRNHKEAERKRRARINSHLNKLRKLLSCNSKT-----------------DKST 81
Query: 129 RLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGR-LIFKASLCCEDRS 187
LAKVV+RVKEL+QQT +IT ET+P E D +V++ G+ R +IFK S CCEDR
Sbjct: 82 LLAKVVQRVKELKQQTLEITD-ETIPSETDEISVLNIEDCSRGDDRRIIFKVSFCCEDRP 140
Query: 188 DLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDR 247
+L+ DL+E LKSL ++TL A+M T+GGRTRNVLVVAA+KEH+ ++S++FLQN+L+SLL+R
Sbjct: 141 ELLKDLMETLKSLQMETLFADMTTVGGRTRNVLVVAADKEHHGVQSVNFLQNALKSLLER 200
Query: 248 SS----------GCNDRSKRRRGLDRRMM 266
SS G +R KRRR LD +M
Sbjct: 201 SSKSVMVGHGGGGGEERLKRRRALDHIIM 229
>UniRef100_Q9S7Y1 Putative DNA-binding protein; 36199-34606 [Arabidopsis thaliana]
Length = 368
Score = 154 bits (389), Expect = 2e-36
Identities = 87/210 (41%), Positives = 131/210 (61%), Gaps = 25/210 (11%)
Query: 58 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS 117
E+ ++T +ALAA K+H EAE+RRRERIN+HL LR++LP +KT
Sbjct: 157 ELGKMTAQEIMDAKALAASKSHSEAERRRRERINNHLAKLRSILPNTTKT---------- 206
Query: 118 RTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIF 177
K S LA+V++ VKEL+++T I++ VP E+D TV + +G+GR +
Sbjct: 207 -------DKASLLAEVIQHVKELKRETSVISETNLVPTESDELTVAFTEEEETGDGRFVI 259
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAE----KEHNSIES 233
KASLCCEDRSDL+PD+I+ LK++ LKTLKAE+ T+GGR +NVL V E +E
Sbjct: 260 KASLCCEDRSDLLPDMIKTLKAMRLKTLKAEITTVGGRVKNVLFVTGEESSGEEVEEEYC 319
Query: 234 IHFLQNSLRSLLDRS----SGCNDRSKRRR 259
I ++ +L++++++S S + +KR+R
Sbjct: 320 IGTIEEALKAVMEKSNVEESSSSGNAKRQR 349
>UniRef100_Q7XZF8 Putative DNA binding protein [Oryza sativa]
Length = 268
Score = 146 bits (368), Expect = 6e-34
Identities = 95/214 (44%), Positives = 124/214 (57%), Gaps = 40/214 (18%)
Query: 69 QDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGS 128
QDRALAA +NH+EAEKRRRERI SHLD LR +L C+ K K S
Sbjct: 61 QDRALAASRNHREAEKRRRERIKSHLDRLRAVLACDPKI-----------------DKAS 103
Query: 129 RLAKVVERVKELRQQTCQITQVETV---PCEADGXTVI-SAGSDMSGEG--RLIFKASLC 182
LAK VERV++L+Q+ I + P E D V+ S G + G G +F+AS+C
Sbjct: 104 LLAKAVERVRDLKQRMAGIGEAAPAHLFPTEHDEIVVLASGGGGVGGAGGAAAVFEASVC 163
Query: 183 CEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVA----------------AEK 226
C+DR DL+P+LIE L++L L+TL+AEMATLGGR RNVLV+A A
Sbjct: 164 CDDRCDLLPELIETLRALRLRTLRAEMATLGGRVRNVLVLARDAGGAGEGGDGDDDRAGY 223
Query: 227 EHNSIESIHFLQNSLRSLLDR-SSGCNDRSKRRR 259
S + FL+ +LR+L++R + DR KRRR
Sbjct: 224 SAVSNDGGDFLKEALRALVERPGAAAGDRPKRRR 257
>UniRef100_Q9LS08 Putative HLH DNA-binding protein [Arabidopsis thaliana]
Length = 344
Score = 139 bits (351), Expect = 6e-32
Identities = 83/216 (38%), Positives = 126/216 (57%), Gaps = 42/216 (19%)
Query: 71 RALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRL 130
+ALAA K+H EAE+RRRERIN+HL LR++LP +KT K S L
Sbjct: 128 KALAASKSHSEAERRRRERINTHLAKLRSILPNTTKT-----------------DKASLL 170
Query: 131 AKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLI 190
A+V++ +KEL++QT QIT VP E D TV S+ +D EG L+ +AS CC+DR+DL+
Sbjct: 171 AEVIQHMKELKRQTSQITDTYQVPTECDDLTVDSSYNDE--EGNLVIRASFCCQDRTDLM 228
Query: 191 PDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAA----EKEHNSI--------------- 231
D+I LKSL L+TLKAE+AT+GGR +N+L ++ E++H+S
Sbjct: 229 HDVINALKSLRLRTLKAEIATVGGRVKNILFLSREYDDEEDHDSYRRNFDGDDVEDYDEE 288
Query: 232 ----ESIHFLQNSLRSLLDRSSGCNDRSKRRRGLDR 263
+ ++ +L++++++ ND S L++
Sbjct: 289 RMMNNRVSSIEEALKAVIEKCVHNNDESNDNNNLEK 324
>UniRef100_Q6ZA99 DNA-binding protein-like [Oryza sativa]
Length = 345
Score = 124 bits (311), Expect = 3e-27
Identities = 83/236 (35%), Positives = 121/236 (51%), Gaps = 53/236 (22%)
Query: 58 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS 117
E+ +T +ALAA ++H EAE+RRR+RIN HL LR+LLP +KT
Sbjct: 104 ELGRMTAKEIMDAKALAASRSHSEAERRRRQRINGHLARLRSLLPNTTKT---------- 153
Query: 118 RTCERFKSKGSRLAKVVERVKELRQQTCQITQ------------VETVPCEADGXTVISA 165
K S LA+V+E VKEL++QT + + V +P E D V +A
Sbjct: 154 -------DKASLLAEVIEHVKELKRQTSAMMEDGAAGGEAAAAPVVLLPTEDDELEVDAA 206
Query: 166 GSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAE 225
+ GRL+ +ASLCCEDR+DLIP + L +L L+ +AE+ATLGGR R+VL++AA
Sbjct: 207 ADE---GGRLVARASLCCEDRADLIPGIARALAALRLRARRAEIATLGGRVRSVLLIAAV 263
Query: 226 KE---------------------HNSIESIHFLQNSLRSLLDRSSGCNDRSKRRRG 260
+E H E + + +LR +++R + +D S G
Sbjct: 264 EEEDPDEAGNDDDGEHGYGVAASHRRHELVASIHEALRGVMNRKAASSDTSSSGAG 319
>UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa]
Length = 363
Score = 120 bits (302), Expect = 3e-26
Identities = 74/196 (37%), Positives = 105/196 (52%), Gaps = 46/196 (23%)
Query: 58 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS 117
E+ +T +ALAA ++H EAE+RRR+RIN HL LR+LLP +KT
Sbjct: 89 ELGRVTAREIMDAKALAASRSHSEAERRRRQRINGHLARLRSLLPNTTKT---------- 138
Query: 118 RTCERFKSKGSRLAKVVERVKELRQQTCQITQV--------------------------E 151
K S LA+V+E VKEL++QT I +
Sbjct: 139 -------DKASLLAEVIEHVKELKRQTTAIAAAAAAGDYHGNDEDDDDAVVGRRSAAAQQ 191
Query: 152 TVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMAT 211
+P EAD V +A + EG+L+ +ASLCCEDR DLIPD+ L +L L+ +AE+ T
Sbjct: 192 LLPTEADELAVDAA---VDAEGKLVVRASLCCEDRPDLIPDIARALAALRLRARRAEITT 248
Query: 212 LGGRTRNVLVVAAEKE 227
LGGR R+VL++ A+++
Sbjct: 249 LGGRVRSVLLITADEQ 264
>UniRef100_Q9FTQ1 DNA binding protein-like [Oryza sativa]
Length = 267
Score = 110 bits (275), Expect = 4e-23
Identities = 70/196 (35%), Positives = 111/196 (55%), Gaps = 19/196 (9%)
Query: 68 QQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKG 127
++++A ALK+H EAE+RRRERIN+HL LRT++PC K K
Sbjct: 71 REEKAAMALKSHSEAERRRRERINAHLATLRTMVPCTDK-----------------MDKA 113
Query: 128 SRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISA-GSDMSGEGRLIFKASLCCEDR 186
+ LA+VV VK+L+ ++ + TVP AD V A + GEG L+ +A+L C+DR
Sbjct: 114 ALLAEVVGHVKKLKSAAARVGRRATVPSGADEVAVDEASATGGGGEGPLLLRATLSCDDR 173
Query: 187 SDLIPDLIEILKSLHLKTLKAEMATLGGRTR-NVLVVAAEKEHNSIESIHFLQNSLRSLL 245
+DL D+ L+ L L+ + +E+ TLGGR R LV + + ++ ++++L+S+L
Sbjct: 174 ADLFVDVKRALQPLGLEVVGSEVTTLGGRVRLAFLVSCGSRGGAAAAAMASVRHALQSVL 233
Query: 246 DRSSGCNDRSKRRRGL 261
D++S D + R L
Sbjct: 234 DKASSGFDFAPRAASL 249
>UniRef100_Q9XEF0 Hypothetical protein T07M07.8 [Arabidopsis thaliana]
Length = 254
Score = 106 bits (265), Expect = 5e-22
Identities = 75/203 (36%), Positives = 110/203 (53%), Gaps = 30/203 (14%)
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSR 129
++A + ++H+ AEKRRR+RINSHL LR L+P + K K +
Sbjct: 58 EKAESLSRSHRLAEKRRRDRINSHLTALRKLVPNSDKL-----------------DKAAL 100
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAG-SDM-SGEGRLIFKASLCCEDRS 187
LA V+E+VKEL+Q+ + + +P EAD TV SD S +IFKAS CCED+
Sbjct: 101 LATVIEQVKELKQKAAESPIFQDLPTEADEVTVQPETISDFESNTNTIIFKASFCCEDQP 160
Query: 188 DLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVA---AEKEHNSIESIHFLQNSLRSL 244
+ I ++I +L L L+T++AE+ ++GGR R ++ + N S L+ SL S
Sbjct: 161 EAISEIIRVLTKLQLETIQAEIISVGGRMRINFILKDSNCNETTNIAASAKALKQSLCSA 220
Query: 245 LDR--------SSGCNDRSKRRR 259
L+R SS C RSKR+R
Sbjct: 221 LNRITSSSTTTSSVCRIRSKRQR 243
>UniRef100_Q7F7Z2 ESTs C26093 [Oryza sativa]
Length = 258
Score = 97.8 bits (242), Expect = 3e-19
Identities = 66/203 (32%), Positives = 113/203 (55%), Gaps = 32/203 (15%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
ALK H EAE+RRRERIN+HL LR ++P T+Q K + LA+VV
Sbjct: 65 ALKIHSEAERRRRERINAHLTTLRRMIP---DTKQ--------------MDKATLLARVV 107
Query: 135 ERVKELRQQTCQITQVETVPCEADGXTV-------ISAGSDMSGEGRLIF-KASLCCEDR 186
++VK+L+++ +ITQ +P E + ++ +A + ++G + ++ KAS+ C+DR
Sbjct: 108 DQVKDLKRKASEITQRTPLPPETNEVSIECFTGDAATAATTVAGNHKTLYIKASISCDDR 167
Query: 187 SDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLD 246
DLI + L L+T++AEM +LGGR ++V ++ E+ S+ L+ ++R L
Sbjct: 168 PDLIAGITHAFHGLRLRTVRAEMTSLGGRVQHVFILCREEGIAGGVSLKSLKEAVRQALA 227
Query: 247 RSS-------GCNDRSKRRRGLD 262
+ + + +SKR+R L+
Sbjct: 228 KVASPELVYGSSHFQSKRQRILE 250
>UniRef100_Q75IR0 Hypothetical protein OSJNBb0099P06.13 [Oryza sativa]
Length = 271
Score = 90.5 bits (223), Expect = 4e-17
Identities = 65/197 (32%), Positives = 100/197 (49%), Gaps = 29/197 (14%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A+ H EAE+RRRERIN+HL LR +LP + K + LA VV
Sbjct: 66 AMTIHSEAERRRRERINAHLATLRRILPDAKQM-----------------DKATLLASVV 108
Query: 135 ERVKELRQQTCQITQ---VETVPCEADGXTV-ISAGSDMSGEGRLIFKASLCCEDRSDLI 190
+VK L+ + + T T+P EA+ TV AG + + R +A++ C+DR L+
Sbjct: 109 NQVKHLKTRATEATTPSTAATIPPEANEVTVQCYAGGEHTAAARTYVRATVSCDDRPGLL 168
Query: 191 PDLIEILKSLHLKTLKAEMATLGGRTRNVLVVA--AEKEHNSIESIHFLQNSLRSLLDRS 248
D+ + L L+ L A+M+ LGGRTR+ V+ E+E ++ L+ ++R L +
Sbjct: 169 ADIAATFRRLRLRPLSADMSCLGGRTRHAFVLCREEEEEEDAAAEARPLKEAVRQALAKV 228
Query: 249 S------GCNDRSKRRR 259
+ G RSKR+R
Sbjct: 229 ALPETVYGGGGRSKRQR 245
>UniRef100_Q69MD0 BHLH-like protein [Oryza sativa]
Length = 215
Score = 80.9 bits (198), Expect = 3e-14
Identities = 60/185 (32%), Positives = 92/185 (49%), Gaps = 33/185 (17%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A ++H EAE++RRERIN+HLD LR L+P S+ K + L +VV
Sbjct: 28 ARRSHSEAERKRRERINAHLDTLRGLVPSASR-----------------MDKAALLGEVV 70
Query: 135 ERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSG--------EGRLIFKASLCCEDR 186
V++LR + V VP E D V ++ G + KAS+CC DR
Sbjct: 71 RYVRKLRSEAAGSAAV--VPGEGDEVVVEEEEVEVEGCSCDAGERQAARRVKASVCCADR 128
Query: 187 SDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVL------VVAAEKEHNSIESIHFLQNS 240
L+ +L + +S+ + ++AE+AT+GGRTR+ L AA N + LQ +
Sbjct: 129 PGLMSELGDAERSVSARAVRAEIATVGGRTRSDLELDVARTAAAGGGSNGASQLPALQAA 188
Query: 241 LRSLL 245
LR+++
Sbjct: 189 LRAVI 193
>UniRef100_Q6ZJC8 BHLH protein family-like [Oryza sativa]
Length = 223
Score = 80.9 bits (198), Expect = 3e-14
Identities = 62/191 (32%), Positives = 98/191 (50%), Gaps = 37/191 (19%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A+++H EAE++RR+RIN+HL LRTL+P S+ K + L +VV
Sbjct: 19 AVRSHSEAERKRRQRINAHLATLRTLVPSASRM-----------------DKAALLGEVV 61
Query: 135 ERVKELRQQTCQITQVETVPCEADGXTV-ISAGSDMSGEG--------------RLIFKA 179
V+ELR + T+ V +G V + D GE R +A
Sbjct: 62 RHVRELRCRADDATEGADVVVPGEGDEVGVEDEDDDEGERDEGCYVVGGGDRRWRRRVRA 121
Query: 180 SLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVL----VVAAEKEHNS-IESI 234
+CC DR L+ DL ++S+ + ++AE+AT+GGRTR+VL VVA++ N ++
Sbjct: 122 WVCCADRPGLMSDLGRAVRSVSARPVRAEVATVGGRTRSVLELDVVVASDAADNDRAVAL 181
Query: 235 HFLQNSLRSLL 245
L+ +LR++L
Sbjct: 182 SALRAALRTVL 192
>UniRef100_Q9SZK7 Hypothetical protein F20D10.190 [Arabidopsis thaliana]
Length = 1496
Score = 75.5 bits (184), Expect = 1e-12
Identities = 61/226 (26%), Positives = 103/226 (44%), Gaps = 17/226 (7%)
Query: 35 SSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHL 94
S+TM + F ++ ++ + F V T S+ + A K H +AE+RRR RINS
Sbjct: 1281 STTMGRSFFAGAATSSKLFSRGFSV---TKPKSKTESKEVAAKKHSDAERRRRLRINSQF 1337
Query: 95 DHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVP 154
LRT+LP K F + + + K S L + V EL++ + + T P
Sbjct: 1338 ATLRTILPNLVKRSNTFCIMFNE---TKQQDKASVLGETVRYFNELKK---MVQDIPTTP 1391
Query: 155 CEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGG 214
D + D R + + C DR L+ ++ E +K++ K ++AE+ T+GG
Sbjct: 1392 SLEDNLRL-----DHCNNNRDLARVVFSCSDREGLMSEVAESMKAVKAKAVRAEIMTVGG 1446
Query: 215 RTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSSGCNDRSKRRRG 260
RT+ L V N E + L+ SL+ +++ S ++ G
Sbjct: 1447 RTKCALFVQGV---NGNEGLVKLKKSLKLVVNGKSSSEAKNNNNGG 1489
>UniRef100_Q7XHI6 Putative bHLH131 transcription factor [Arabidopsis thaliana]
Length = 256
Score = 72.4 bits (176), Expect = 1e-11
Identities = 60/226 (26%), Positives = 100/226 (43%), Gaps = 31/226 (13%)
Query: 35 SSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHL 94
S+TM + F ++ ++ + F V T S+ + A K H +AE+RRR RINS
Sbjct: 55 STTMGRSFFAGAATSSKLFSRGFSV---TKPKSKTESKEVAAKKHSDAERRRRLRINSQF 111
Query: 95 DHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVP 154
LRT+LP K + K S L + V EL++ + + T P
Sbjct: 112 ATLRTILPNLVK-----------------QDKASVLGETVRYFNELKK---MVQDIPTTP 151
Query: 155 CEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGG 214
D + D R + + C DR L+ ++ E +K++ K ++AE+ T+GG
Sbjct: 152 SLEDNLRL-----DHCNNNRDLARVVFSCSDREGLMSEVAESMKAVKAKAVRAEIMTVGG 206
Query: 215 RTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSSGCNDRSKRRRG 260
RT+ L V N E + L+ SL+ +++ S ++ G
Sbjct: 207 RTKCALFVQGV---NGNEGLVKLKKSLKLVVNGKSSSEAKNNNNGG 249
>UniRef100_Q9M1K0 Hypothetical protein F24I3.60 [Arabidopsis thaliana]
Length = 258
Score = 44.3 bits (103), Expect = 0.003
Identities = 32/140 (22%), Positives = 62/140 (43%), Gaps = 9/140 (6%)
Query: 8 NIGMENNSELFQFLVA----NNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEIT 63
N G + E + +A NN F ++ P T + + N+ E +EI
Sbjct: 11 NFGWPSTGEYDSYYLAGDILNNGGFLDFPVPEETYGAVTAVTQHQNSFGVSVSSEGNEID 70
Query: 64 DTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERF 123
+ P + NH +E+ RR +INS LR+ LP + ++++ + SR+ +
Sbjct: 71 NNP-----VVVKKLNHNASERDRRRKINSLFSSLRSCLPASGQSKKLSIPATVSRSLKYI 125
Query: 124 KSKGSRLAKVVERVKELRQQ 143
++ K++++ +EL Q
Sbjct: 126 PELQEQVKKLIKKKEELLVQ 145
>UniRef100_Q6FSB6 Tr|Q9C1R9 Candida glabrata Centromere binding protein 1 [Candida
glabrata]
Length = 442
Score = 43.5 bits (101), Expect = 0.006
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query: 67 SQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSK 126
S QD ++HKE E+RRRE IN +D+LR LLP ++ G +S ++ + +S+
Sbjct: 273 SIQDWRKIRKESHKEVERRRRETINLAIDNLRKLLPKPEYSKAGILNSAAALIQKLKESE 332
Query: 127 GSRLAK------VVERVKELRQQT 144
S++ K V E+ K Q++
Sbjct: 333 SSQMNKWAIQRIVTEQTKTAHQKS 356
>UniRef100_Q9C1R9 Centromere binding protein 1 [Candida glabrata]
Length = 441
Score = 43.5 bits (101), Expect = 0.006
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query: 67 SQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSK 126
S QD ++HKE E+RRRE IN +D+LR LLP ++ G +S ++ + +S+
Sbjct: 272 SIQDWRKIRKESHKEVERRRRETINLAIDNLRKLLPKPEYSKAGILNSAAALIQKLKESE 331
Query: 127 GSRLAK------VVERVKELRQQT 144
S++ K V E+ K Q++
Sbjct: 332 SSQMNKWAIQRIVTEQTKTAHQKS 355
>UniRef100_UPI000036202E UPI000036202E UniRef100 entry
Length = 613
Score = 42.7 bits (99), Expect = 0.010
Identities = 26/79 (32%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 32 STPSSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERIN 91
STP ++ S++D YH + + QQ R A + H + EKRRR+++N
Sbjct: 29 STPGMDYTRKRKGSTTD-----YHDMDPDKDKLGSDQQGRIKNAREAHSQIEKRRRDKMN 83
Query: 92 SHLDHLRTLLP-CNSKTRQ 109
S +D L +L+P CN+ +R+
Sbjct: 84 SFIDELASLVPTCNAMSRK 102
>UniRef100_Q8JIG1 BHLH-PAS transcription factor [Brachydanio rerio]
Length = 622
Score = 42.7 bits (99), Expect = 0.010
Identities = 33/97 (34%), Positives = 49/97 (50%), Gaps = 17/97 (17%)
Query: 30 EYSTPSSTMMQQSFCSSSDNNNNYYHP------------FEVSEITDTPSQQDRALAALK 77
E+ +PSST + S S+ + N FE S TD RA +K
Sbjct: 14 EFMSPSSTELISSSMGSTGMDFNRKRKGSITDYEIDGFSFEDSMDTDKDKPLGRADQQMK 73
Query: 78 NHKEA----EKRRRERINSHLDHLRTLLP-CNSKTRQ 109
N +EA EKRRR+++NS +D L +L+P CN+ +R+
Sbjct: 74 NAREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRK 110
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 390,030,966
Number of Sequences: 2790947
Number of extensions: 14501888
Number of successful extensions: 52130
Number of sequences better than 10.0: 363
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 217
Number of HSP's that attempted gapping in prelim test: 51876
Number of HSP's gapped (non-prelim): 382
length of query: 267
length of database: 848,049,833
effective HSP length: 125
effective length of query: 142
effective length of database: 499,181,458
effective search space: 70883767036
effective search space used: 70883767036
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC135795.6


