

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135565.11 + phase: 0
(225 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
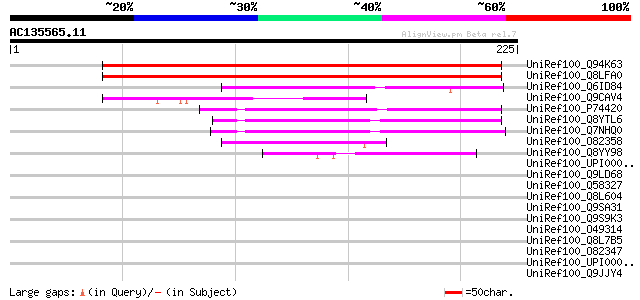
UniRef100_Q94K63 Hypothetical protein At3g04890 [Arabidopsis tha... 214 2e-54
UniRef100_Q8LFA0 Hypothetical protein [Arabidopsis thaliana] 214 2e-54
UniRef100_Q6ID84 At2g46100 [Arabidopsis thaliana] 82 1e-14
UniRef100_Q9CAV4 Hypothetical protein T9J14.16 [Arabidopsis thal... 76 7e-13
UniRef100_P74420 Sll0364 protein [Synechocystis sp.] 69 1e-10
UniRef100_Q8YTL6 All2701 protein [Anabaena sp.] 63 5e-09
UniRef100_Q7NHQ0 Glr2485 protein [Gloeobacter violaceus] 60 6e-08
UniRef100_O82358 Hypothetical protein At2g46100 [Arabidopsis tha... 53 5e-06
UniRef100_Q8YY98 Alr0953 protein [Anabaena sp.] 53 7e-06
UniRef100_UPI00002E7627 UPI00002E7627 UniRef100 entry 45 0.002
UniRef100_Q9LD68 EST AU082314(E2976) corresponds to a region of ... 43 0.005
UniRef100_Q58327 Probable inorganic polyphosphate/ATP-NAD kinase... 41 0.020
UniRef100_Q8L604 Hypothetical protein At1g65230 [Arabidopsis tha... 39 0.10
UniRef100_Q9SA31 F3O9.12 protein [Arabidopsis thaliana] 39 0.10
UniRef100_Q9S9K3 T23K8.14 protein [Arabidopsis thaliana] 39 0.10
UniRef100_O49314 Mitochondrial chaperonin [Arabidopsis thaliana] 38 0.23
UniRef100_Q8L7B5 Mitochondrial chaperonin HSP60 [Arabidopsis tha... 38 0.23
UniRef100_O82347 Expressed protein [Arabidopsis thaliana] 38 0.23
UniRef100_UPI00004669DD UPI00004669DD UniRef100 entry 37 0.29
UniRef100_Q9JJY4 Probable ATP-dependent RNA helicase DDX20 [Mus ... 36 0.65
>UniRef100_Q94K63 Hypothetical protein At3g04890 [Arabidopsis thaliana]
Length = 216
Score = 214 bits (544), Expect = 2e-54
Identities = 105/177 (59%), Positives = 137/177 (77%)
Query: 42 KNETPQVLKIAVSGVTELLRLFSPPQQTSVLSDDIEKQNNDSTVSSVEDVLIIIKSDYDN 101
+ +TP VLK AVSGVTE LRL S ++ ++ + ++ N+ T V+DV+ I++SDY N
Sbjct: 31 EKKTPAVLKWAVSGVTEFLRLISGAPSSTSIATNKDRSKNEVTAGDVDDVMGILRSDYRN 90
Query: 102 DYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIKLLKIEKEVESD 161
YFVTG TS+IY+++CIFEDPTI F G +LY RNLKLLVPF + ASI+L +EK S
Sbjct: 91 FYFVTGVLTSAIYSDDCIFEDPTISFQGTELYERNLKLLVPFLEDASIELQNMEKSESSQ 150
Query: 162 TNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVESWNVSALEAVLQIF 218
N++ A+WKLRT LKLPWRPLI+I+G+T Y+L++DFKIVRHVESWNVSALEA+ QIF
Sbjct: 151 RNYILATWKLRTYLKLPWRPLISINGNTVYDLDKDFKIVRHVESWNVSALEAIGQIF 207
>UniRef100_Q8LFA0 Hypothetical protein [Arabidopsis thaliana]
Length = 216
Score = 214 bits (544), Expect = 2e-54
Identities = 105/177 (59%), Positives = 137/177 (77%)
Query: 42 KNETPQVLKIAVSGVTELLRLFSPPQQTSVLSDDIEKQNNDSTVSSVEDVLIIIKSDYDN 101
+ +TP VLK AVSGVTE LRL S ++ ++ + ++ N+ T V+DV+ I++SDY N
Sbjct: 31 EKKTPAVLKWAVSGVTEFLRLISGAPSSTSIATNKDRSKNEVTAGDVDDVMGILRSDYRN 90
Query: 102 DYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIKLLKIEKEVESD 161
YFVTG TS+IY+++CIFEDPTI F G +LY RNLKLLVPF + ASI+L +EK S
Sbjct: 91 FYFVTGVLTSAIYSDDCIFEDPTISFQGTELYERNLKLLVPFLEDASIELQNMEKSESSQ 150
Query: 162 TNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVESWNVSALEAVLQIF 218
N++ A+WKLRT LKLPWRPLI+I+G+T Y+L++DFKIVRHVESWNVSALEA+ QIF
Sbjct: 151 RNYILATWKLRTYLKLPWRPLISINGNTVYDLDKDFKIVRHVESWNVSALEAIGQIF 207
>UniRef100_Q6ID84 At2g46100 [Arabidopsis thaliana]
Length = 240
Score = 81.6 bits (200), Expect = 1e-14
Identities = 43/126 (34%), Positives = 61/126 (48%), Gaps = 5/126 (3%)
Query: 95 IKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIKLLKI 154
IK D+ YFVTGN T +Y E C F DP F G + RN + +++KL+K
Sbjct: 109 IKQDFKRSYFVTGNLTPEVYEEKCEFADPAGSFKGLARFKRNCTNFGSLIEKSNMKLMKW 168
Query: 155 EKEVESDTNFLRASWKLRTNLKLPWRPLIAIDGSTSYELN-EDFKIVRHVESWNVSALEA 213
E + WK + PW+P+++ G T Y + E KI RHVE WNV +
Sbjct: 169 ENFEDKGI----GHWKFSCVMSFPWKPILSATGYTEYYFDTESGKICRHVEHWNVPKIAL 224
Query: 214 VLQIFK 219
Q+ +
Sbjct: 225 FKQLLR 230
>UniRef100_Q9CAV4 Hypothetical protein T9J14.16 [Arabidopsis thaliana]
Length = 154
Score = 75.9 bits (185), Expect = 7e-13
Identities = 52/129 (40%), Positives = 69/129 (53%), Gaps = 34/129 (26%)
Query: 42 KNETPQVLKIAVSGVTELLRLFS-PPQQTSVLSD--------DIE---KQNNDSTVSSVE 89
+ +TP VLK AVSGVTE LRL S P TS+ ++ D+ + N+ T V+
Sbjct: 31 EKKTPAVLKWAVSGVTEFLRLISGAPSSTSIATNKDRLVVEIDLNTGCRSKNEVTAGDVD 90
Query: 90 DVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASI 149
DV+ I++SDY N YFVTG +LY RNLKLLVPF + ASI
Sbjct: 91 DVMGILRSDYRNFYFVTGT----------------------ELYERNLKLLVPFLEDASI 128
Query: 150 KLLKIEKEV 158
+L +EK +
Sbjct: 129 ELQNMEKNL 137
>UniRef100_P74420 Sll0364 protein [Synechocystis sp.]
Length = 139
Score = 68.6 bits (166), Expect = 1e-10
Identities = 42/134 (31%), Positives = 67/134 (49%), Gaps = 7/134 (5%)
Query: 85 VSSVEDVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFF 144
++S + +L I++ DY N + T SIY N F+DP +F G Y + ++ L +F
Sbjct: 11 MTSNDRILTILQQDYAN---FPWHQTFSIYDPNVYFQDPLSQFRGLARYEKMIQFLGRWF 67
Query: 145 DCASIKLLKIEKEVESDTNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVE 204
++L +I + + T W L LPW+P IAI G + LNE I+ H++
Sbjct: 68 QAIDLQLHEITQVNQEITT----RWTLNWTSPLPWKPRIAISGCSELTLNEAGLIISHID 123
Query: 205 SWNVSALEAVLQIF 218
W+ S + V Q F
Sbjct: 124 YWDCSPWDVVKQHF 137
>UniRef100_Q8YTL6 All2701 protein [Anabaena sp.]
Length = 128
Score = 63.2 bits (152), Expect = 5e-09
Identities = 38/128 (29%), Positives = 60/128 (46%), Gaps = 7/128 (5%)
Query: 91 VLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIK 150
++ I+K DY N T SIY E+ F+DP KF G Y + + + +F +
Sbjct: 3 IIEILKQDYQR---FPVNQTYSIYAEDVYFQDPLNKFRGITRYKQMINFMQTWFLNIKMD 59
Query: 151 LLKIEKEVESDTNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVESWNVSA 210
L I+ + ++ W L N +PW+P I+I G + LN IV H++ W S
Sbjct: 60 LHDIQHLEDK----IKTEWTLSWNTPVPWKPRISISGWSELGLNSKGLIVSHIDYWQCSP 115
Query: 211 LEAVLQIF 218
+ + Q F
Sbjct: 116 FDVIKQHF 123
>UniRef100_Q7NHQ0 Glr2485 protein [Gloeobacter violaceus]
Length = 141
Score = 59.7 bits (143), Expect = 6e-08
Identities = 37/131 (28%), Positives = 61/131 (46%), Gaps = 7/131 (5%)
Query: 90 DVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASI 149
D+L I+K+DY + T +IY + F+DP F G + Y R + + +F +
Sbjct: 6 DILEILKADYQR---FPEDQTYAIYANDVYFKDPLNSFRGIERYRRMIGWMHRWFQPIRL 62
Query: 150 KLLKIEKEVESDTNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVESWNVS 209
+L IE++ + +W L LPW+P I IDG + +L+ +I H + W S
Sbjct: 63 ELHSIERQQTR----IVTTWTLSWQAPLPWKPHIVIDGWSELDLDTQGQIAAHTDHWRCS 118
Query: 210 ALEAVLQIFKF 220
+ Q F
Sbjct: 119 PNDVAWQHLGF 129
>UniRef100_O82358 Hypothetical protein At2g46100 [Arabidopsis thaliana]
Length = 286
Score = 53.1 bits (126), Expect = 5e-06
Identities = 29/74 (39%), Positives = 38/74 (51%), Gaps = 1/74 (1%)
Query: 95 IKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIKLLKI 154
IK D+ YFVTGN T +Y E C F DP F G + RN + +++KL+K
Sbjct: 109 IKQDFKRSYFVTGNLTPEVYEEKCEFADPAGSFKGLARFKRNCTNFGSLIEKSNMKLMKW 168
Query: 155 EK-EVESDTNFLRA 167
E EV S T R+
Sbjct: 169 ENFEVRSTTKRCRS 182
>UniRef100_Q8YY98 Alr0953 protein [Anabaena sp.]
Length = 139
Score = 52.8 bits (125), Expect = 7e-06
Identities = 28/104 (26%), Positives = 56/104 (52%), Gaps = 17/104 (16%)
Query: 113 IYTENCIFEDPTIKFSGRDLYAR-------NLKLLVP--FFDCASIKLLKIEKEVESDTN 163
IYT++ F+DP KF G+ Y + +L P +FD + + ++ D +
Sbjct: 30 IYTKDIYFQDPVNKFKGKFNYRIIFWTLRFHARLFFPEIYFD--------LHEVLQLDKD 81
Query: 164 FLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRHVESWN 207
+ A W +R L++PWR + +G ++Y+L ++ I +H+++W+
Sbjct: 82 TILAKWTVRGTLRVPWRSQMLFNGYSTYKLRQNNLIYQHIDTWD 125
>UniRef100_UPI00002E7627 UPI00002E7627 UniRef100 entry
Length = 122
Score = 44.7 bits (104), Expect = 0.002
Identities = 22/56 (39%), Positives = 30/56 (53%)
Query: 74 DDIEKQNNDSTVSSVEDVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSG 129
DD + + + ++S ED + I++DYD YFVTG Y E C F DP F G
Sbjct: 62 DDDDDDADAAPIASREDAVRAIRADYDAAYFVTGEGDMRAYDEACEFADPFASFRG 117
>UniRef100_Q9LD68 EST AU082314(E2976) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 238
Score = 43.1 bits (100), Expect = 0.005
Identities = 30/103 (29%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query: 113 IYTENCIFEDPTIKFSGRDLYARNLKLL----VPFFDCASIKLLKIEKEVESDTNFLRAS 168
IY ++ +F+DP KF G D Y R L FF + ++ I + E N +
Sbjct: 121 IYRDDIVFKDPLNKFEGIDNYKRIFWALRFTGRIFFKALWVDIVSIWQPAE---NLIMIR 177
Query: 169 WKLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRH-VESWNVSA 210
W ++PW DG++ Y+L+++ KI H V++ N A
Sbjct: 178 WIAHGIPRVPWEAHGRFDGASEYKLDKNGKIYEHKVQACNTDA 220
>UniRef100_Q58327 Probable inorganic polyphosphate/ATP-NAD kinase (EC 2.7.1.23)
(Poly(P)/ATP NAD kinase) [Methanococcus jannaschii]
Length = 574
Score = 41.2 bits (95), Expect = 0.020
Identities = 41/165 (24%), Positives = 77/165 (45%), Gaps = 18/165 (10%)
Query: 42 KNETPQVLKIAVSGVTELLRLFSPPQQTSVLSDDIEKQNND-STVSSVEDVLIIIKSD-- 98
K+E +++ + G E+ ++ S LS I K N T S++ ++++I K+
Sbjct: 395 KDEVFEIIDKVIYGEYEI-------EKRSKLSCKIIKDNRVIKTPSALNEMVVITKNPAK 447
Query: 99 -YDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNL--KLLVPFFDCASIKLLKIE 155
+ D +V ++ + I PT G Y+ + ++ P DC I +
Sbjct: 448 ILEFDVYVNDTLVENVRADGIIVSTPT----GSTAYSLSAGGPIVEPNVDCFIISPICPF 503
Query: 156 KEVESDTNFLRASWKLRTNLKLPWRPLIAIDGSTSYELNEDFKIV 200
K + S + AS +++ LKL L+ IDGS YE+N+D +++
Sbjct: 504 K-LSSRPLVISASNRIKLKLKLEKPALLVIDGSVEYEINKDDELI 547
>UniRef100_Q8L604 Hypothetical protein At1g65230 [Arabidopsis thaliana]
Length = 286
Score = 38.9 bits (89), Expect = 0.10
Identities = 27/121 (22%), Positives = 55/121 (45%), Gaps = 18/121 (14%)
Query: 107 GNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIK--LLKIEKEVESDTNF 164
GN + Y + ++DP F+GR+ Y R P + + ++ + +++ V T+
Sbjct: 96 GNRAFACYAVSVKYKDPVRSFTGREKYKR------PMWITSGLENPTVTVQEMVMLSTSV 149
Query: 165 LRASWKLRTNLKLPWRPLIAIDGSTSYELNEDF-------KIVRHVESWNVSALEAVLQI 217
LR W ++ P L A+ G ++ +F ++ H ESW++S+ + Q
Sbjct: 150 LRIKWTVKGK---PKSILAAVSGDLIVKVKSEFTLNQISGQVFEHEESWDLSSSSPIAQA 206
Query: 218 F 218
+
Sbjct: 207 Y 207
>UniRef100_Q9SA31 F3O9.12 protein [Arabidopsis thaliana]
Length = 273
Score = 38.9 bits (89), Expect = 0.10
Identities = 36/153 (23%), Positives = 62/153 (39%), Gaps = 13/153 (8%)
Query: 59 LLRLFSPP---QQTSVLSDDIEKQNNDSTVSSVEDVLIIIKS-DYDNDYFVTGNFTSSIY 114
LL + SPP Q +K NN ++ + +++ D T + IY
Sbjct: 51 LLSIQSPPLKDTQVQTRHSSQDKHNNHDRDEFYINLGVAVRTLREDLPLLFTRDLNYDIY 110
Query: 115 TENCIFEDPTIKFSGRDLY-----ARNLKLLVPFFDCASIKLLKIEKEVESDTNFLRASW 169
++ F DP F+G D Y A + F D + L+I + + N + W
Sbjct: 111 RDDITFVDPMNTFTGMDNYKIIFWALRFHGKILFRDIS----LEIFRVWQPSENMILIRW 166
Query: 170 KLRTNLKLPWRPLIAIDGSTSYELNEDFKIVRH 202
L+ ++PW G++ Y+L+ + KI H
Sbjct: 167 NLKGVPRVPWEAKGEFQGTSRYKLDRNGKIYEH 199
>UniRef100_Q9S9K3 T23K8.14 protein [Arabidopsis thaliana]
Length = 218
Score = 38.9 bits (89), Expect = 0.10
Identities = 27/121 (22%), Positives = 55/121 (45%), Gaps = 18/121 (14%)
Query: 107 GNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIK--LLKIEKEVESDTNF 164
GN + Y + ++DP F+GR+ Y R P + + ++ + +++ V T+
Sbjct: 57 GNRAFACYAVSVKYKDPVRSFTGREKYKR------PMWITSGLENPTVTVQEMVMLSTSV 110
Query: 165 LRASWKLRTNLKLPWRPLIAIDGSTSYELNEDF-------KIVRHVESWNVSALEAVLQI 217
LR W ++ P L A+ G ++ +F ++ H ESW++S+ + Q
Sbjct: 111 LRIKWTVKGK---PKSILAAVSGDLIVKVKSEFTLNQISGQVFEHEESWDLSSSSPIAQA 167
Query: 218 F 218
+
Sbjct: 168 Y 168
>UniRef100_O49314 Mitochondrial chaperonin [Arabidopsis thaliana]
Length = 524
Score = 37.7 bits (86), Expect = 0.23
Identities = 39/144 (27%), Positives = 63/144 (43%), Gaps = 11/144 (7%)
Query: 38 MVREKNETPQVLKIAVSGVTELLRLFSPPQQTSVLSDDIEKQNNDSTVSSVEDVL-IIIK 96
M+ E QV I+ +G E+ L + +T I Q+ + + +E V + I
Sbjct: 107 MISTSEEIAQVGTISANGDREIGELIAKAMETVGKEGVITIQDGKTLFNELEVVEGMKID 166
Query: 97 SDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIK---LLK 153
Y + YF+T T+ C EDP I + + N+ +V + A K LL
Sbjct: 167 RGYISPYFITNP-----KTQKCELEDPLILIHEKKI--SNINAMVKVLELALKKQRPLLI 219
Query: 154 IEKEVESDTNFLRASWKLRTNLKL 177
+ ++VESD KLR N+K+
Sbjct: 220 VAEDVESDALATLILNKLRANIKV 243
>UniRef100_Q8L7B5 Mitochondrial chaperonin HSP60 [Arabidopsis thaliana]
Length = 585
Score = 37.7 bits (86), Expect = 0.23
Identities = 39/144 (27%), Positives = 63/144 (43%), Gaps = 11/144 (7%)
Query: 38 MVREKNETPQVLKIAVSGVTELLRLFSPPQQTSVLSDDIEKQNNDSTVSSVEDVL-IIIK 96
M+ E QV I+ +G E+ L + +T I Q+ + + +E V + I
Sbjct: 168 MISTSEEIAQVGTISANGDREIGELIAKAMETVGKEGVITIQDGKTLFNELEVVEGMKID 227
Query: 97 SDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLYARNLKLLVPFFDCASIK---LLK 153
Y + YF+T T+ C EDP I + + N+ +V + A K LL
Sbjct: 228 RGYISPYFITNP-----KTQKCELEDPLILIHEKKI--SNINAMVKVLELALKKQRPLLI 280
Query: 154 IEKEVESDTNFLRASWKLRTNLKL 177
+ ++VESD KLR N+K+
Sbjct: 281 VAEDVESDALATLILNKLRANIKV 304
>UniRef100_O82347 Expressed protein [Arabidopsis thaliana]
Length = 241
Score = 37.7 bits (86), Expect = 0.23
Identities = 29/133 (21%), Positives = 59/133 (43%), Gaps = 9/133 (6%)
Query: 74 DDIEKQNNDSTVSSVEDVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFSGRDLY 133
++ E+++ S ++ + I+ ++ ++ NF IY ++ +F+DP F G D Y
Sbjct: 74 EEEEEEDKQSYYVNMGHAVRSIREEFPLLFYKELNF--DIYRDDIVFKDPMNTFMGIDNY 131
Query: 134 ARNLKLLV----PFFDCASIKLLKIEKEVESDTNFLRASWKLRTNLKLPWRPLIAIDGST 189
L FF + ++ + + E N L W + + PW DG++
Sbjct: 132 KSIFGALRFHGRIFFRALCVDIVSVWQPTE---NTLMIRWTVHGIPRGPWETRGRFDGTS 188
Query: 190 SYELNEDFKIVRH 202
Y+ +++ KI H
Sbjct: 189 EYKFDKNGKIYEH 201
>UniRef100_UPI00004669DD UPI00004669DD UniRef100 entry
Length = 307
Score = 37.4 bits (85), Expect = 0.29
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query: 69 TSVLSDDIEKQNNDSTVSSVEDVLIIIKSDYDNDYFVTGNFTSSIYTENCIFEDPTIKFS 128
T +L + +K+NN++ VS V+ +L IIK + N+T+ ++E T+K
Sbjct: 130 TKLLKREYQKKNNENNVSRVKHILAIIKGRKNRHMACLNNYTN--INRIPLYEQNTLKER 187
Query: 129 GRDLYARNLK 138
+ +Y R +K
Sbjct: 188 NKKIYIRKIK 197
>UniRef100_Q9JJY4 Probable ATP-dependent RNA helicase DDX20 [Mus musculus]
Length = 825
Score = 36.2 bits (82), Expect = 0.65
Identities = 27/96 (28%), Positives = 47/96 (48%), Gaps = 9/96 (9%)
Query: 2 FTESLIKTRTPPLQLRK-------QHCSNSYTPSSRNTCAIYNMVREKNETPQVLKIAVS 54
++ + T++P Q++K Q + TPS RNT A R K+ P+ L +
Sbjct: 463 YSSPTVATQSPKKQVQKLERAFQSQRTPGNQTPSPRNTSASALSARPKHSKPK-LPVKSH 521
Query: 55 GVTELLRLFSPPQQTSVLSDDIEKQNNDSTVSSVED 90
+L +PPQ++ + +E+Q +S +SVED
Sbjct: 522 SECGVLEKAAPPQESGCPA-QLEEQVKNSVQTSVED 556
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 377,526,799
Number of Sequences: 2790947
Number of extensions: 15343813
Number of successful extensions: 37172
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 37139
Number of HSP's gapped (non-prelim): 60
length of query: 225
length of database: 848,049,833
effective HSP length: 123
effective length of query: 102
effective length of database: 504,763,352
effective search space: 51485861904
effective search space used: 51485861904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC135565.11


