

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
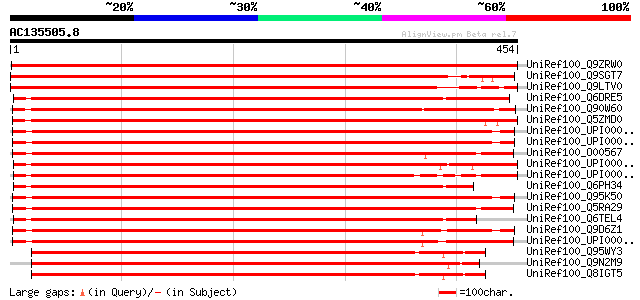
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZRW0 Nucleolar protein [Cicer arietinum] 782 0.0
UniRef100_Q9SGT7 Nucleolar protein [Arabidopsis thaliana] 647 0.0
UniRef100_Q9LTV0 Nucleolar protein [Arabidopsis thaliana] 639 0.0
UniRef100_Q6DRE5 NOP56 [Brachydanio rerio] 500 e-140
UniRef100_Q90W60 XNop56 protein [Xenopus laevis] 500 e-140
UniRef100_Q5ZMD0 Hypothetical protein [Gallus gallus] 491 e-137
UniRef100_UPI000036C2C4 UPI000036C2C4 UniRef100 entry 482 e-134
UniRef100_UPI000036C2C5 UPI000036C2C5 UniRef100 entry 481 e-134
UniRef100_O00567 Nucleolar protein Nop56 [Homo sapiens] 481 e-134
UniRef100_UPI0000363324 UPI0000363324 UniRef100 entry 481 e-134
UniRef100_UPI0000363323 UPI0000363323 UniRef100 entry 480 e-134
UniRef100_Q6PH34 Nol5a protein [Brachydanio rerio] 479 e-134
UniRef100_Q95K50 Hypothetical protein [Macaca fascicularis] 479 e-134
UniRef100_Q5RA29 Hypothetical protein DKFZp459D1517 [Pongo pygma... 479 e-134
UniRef100_Q6TEL4 Nucleolar protein 5A [Brachydanio rerio] 479 e-134
UniRef100_Q9D6Z1 Nucleolar protein Nop56 [Mus musculus] 474 e-132
UniRef100_UPI00001CF330 UPI00001CF330 UniRef100 entry 471 e-131
UniRef100_Q95WY3 Nucleolar KKE/D repeat protein [Drosophila mela... 470 e-131
UniRef100_Q9N2M9 Nucleolar protein [Drosophila subobscura] 468 e-130
UniRef100_Q8IGT5 RE33426p [Drosophila melanogaster] 468 e-130
>UniRef100_Q9ZRW0 Nucleolar protein [Cicer arietinum]
Length = 454
Score = 782 bits (2019), Expect = 0.0
Identities = 397/454 (87%), Positives = 419/454 (91%), Gaps = 1/454 (0%)
Query: 2 TDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQH 61
TDELRTVLETNLPKVKEGKK KFSLGVAESK+GS IH+ TKIP Q NEFVGEL+RGVRQH
Sbjct: 1 TDELRTVLETNLPKVKEGKKPKFSLGVAESKLGSQIHDVTKIPCQCNEFVGELLRGVRQH 60
Query: 62 FDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMR 121
F VGDLK GDL KAQLGL HSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKD+NSF+MR
Sbjct: 61 FVDSVGDLKPGDLIKAQLGLSHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDINSFSMR 120
Query: 122 VREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAA 181
VREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKL+EDK+E LTD VGDEDKAKEI+EAA
Sbjct: 121 VREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLSEDKLEGLTDQVGDEDKAKEIIEAA 180
Query: 182 KASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARL 241
KASMGQDLSPVDLINVHMFAQRVMDLSDYRRRL DYL+TKMNDIAPNL SL+G+ VGARL
Sbjct: 181 KASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLYDYLTTKMNDIAPNLASLIGEVVGARL 240
Query: 242 ISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRM 301
ISHAGSLTNLAKCPSSTLQIL AEKALFRA KTR NTPKYGLIFHSSFIGRASA+NKGRM
Sbjct: 241 ISHAGSLTNLAKCPSSTLQILCAEKALFRAFKTRRNTPKYGLIFHSSFIGRASAKNKGRM 300
Query: 302 ARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESV 361
ARYLANKCSIASRIDCFSE G++ FGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIE
Sbjct: 301 ARYLANKCSIASRIDCFSENGSTIFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIEIA 360
Query: 362 DNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDA-EDHKSEKKKKKKEKRK 420
DNKDTEMETEE SAKK+KKKKQKA D DDMAVD AAE+TNGDA EDHKSEKKKKKKEKRK
Sbjct: 361 DNKDTEMETEEASAKKSKKKKQKATDEDDMAVDNAAEVTNGDATEDHKSEKKKKKKEKRK 420
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+D EVE +D V++GAN DSSKKKKKKSK+ D +
Sbjct: 421 VDLEVETQDPSVDEGANGDSSKKKKKKSKRMDVD 454
>UniRef100_Q9SGT7 Nucleolar protein [Arabidopsis thaliana]
Length = 522
Score = 647 bits (1669), Expect = 0.0
Identities = 333/463 (71%), Positives = 385/463 (82%), Gaps = 22/463 (4%)
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
MTDELR+ LE NLPKVKEGKK KFSLG+AE K+GSHI EATKIP QSNEFV EL+RGVRQ
Sbjct: 71 MTDELRSFLELNLPKVKEGKKPKFSLGLAEPKLGSHIFEATKIPCQSNEFVLELLRGVRQ 130
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFD+F+ DLK GDLEK+QLGL HSYSRAKVKFNVNRVDNMVIQAIF+LDTLDKD+NSFAM
Sbjct: 131 HFDRFIKDLKPGDLEKSQLGLAHSYSRAKVKFNVNRVDNMVIQAIFMLDTLDKDINSFAM 190
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLY +V+K I+DKSKL ED I LT+++GDEDKAKE++EA
Sbjct: 191 RVREWYSWHFPELVKIVNDNYLYARVSKMIDDKSKLTEDHIPMLTEVLGDEDKAKEVIEA 250
Query: 181 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 240
KASMG DLSP+DLINV FAQ+VMDL+DYR++L DYL TKM+DIAPNL +L+G+ VGAR
Sbjct: 251 GKASMGSDLSPLDLINVQTFAQKVMDLADYRKKLYDYLVTKMSDIAPNLAALIGEMVGAR 310
Query: 241 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 300
LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGR
Sbjct: 311 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGR 370
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 360
+ARYLANKCSIASRIDCF++ T+AFGEKLREQVEERL+FYDKGVAPRKN+DVMK IE+
Sbjct: 371 IARYLANKCSIASRIDCFADGATTAFGEKLREQVEERLEFYDKGVAPRKNVDVMKEVIEN 430
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
+ ++ E + S KK+KKKK K + +++ E+ KSE KKKKKEKRK
Sbjct: 431 LKQEEEGKEPVDASVKKSKKKKAKGEEEEEVVA----------MEEDKSE-KKKKKEKRK 479
Query: 421 LD--QEVEVEDKV---------VEDGANADSSKKKKKKSKKKD 452
++ +E E +K E+ + S+KKKKKKSK +
Sbjct: 480 METAEENEKSEKKKTKKSKAGGEEETDDGHSTKKKKKKSKSAE 522
>UniRef100_Q9LTV0 Nucleolar protein [Arabidopsis thaliana]
Length = 499
Score = 639 bits (1648), Expect = 0.0
Identities = 328/454 (72%), Positives = 376/454 (82%), Gaps = 25/454 (5%)
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
M+DELR+ LE NLPKVKEGKK KFSLGV+E KIGS I EATKIP QSNEFV EL+RGVRQ
Sbjct: 71 MSDELRSFLELNLPKVKEGKKPKFSLGVSEPKIGSCIFEATKIPCQSNEFVHELLRGVRQ 130
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFD+F+ DLK GDLEKAQLGL HSYSRAKVKFNVNRVDNMVIQAIF+LDTLDKD+NSFAM
Sbjct: 131 HFDRFIKDLKPGDLEKAQLGLAHSYSRAKVKFNVNRVDNMVIQAIFMLDTLDKDINSFAM 190
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLY KV+K I DKSKL+E+ I LT+ +GDEDKA+E++EA
Sbjct: 191 RVREWYSWHFPELVKIVNDNYLYAKVSKIIVDKSKLSEEHIPMLTEALGDEDKAREVIEA 250
Query: 181 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 240
KASMGQDLSPVDLINV FAQRVMDL+DYR++L DYL TKM+DIAPNL +L+G+ VGAR
Sbjct: 251 GKASMGQDLSPVDLINVQTFAQRVMDLADYRKKLYDYLVTKMSDIAPNLAALIGEMVGAR 310
Query: 241 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 300
LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGR
Sbjct: 311 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGR 370
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 360
+AR+LANKCSIASRIDCFS+ T+AFGEKLREQVEERLDFYDKGVAPRKN+DVMK +E+
Sbjct: 371 IARFLANKCSIASRIDCFSDNSTTAFGEKLREQVEERLDFYDKGVAPRKNVDVMKEVLEN 430
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
++ KD +T + S KK K+K + + E+ K E+K KKK+K+
Sbjct: 431 LEKKDEGEKTVDASEKKKKRKTE-------------------EKEEEKEEEKSKKKKKK- 470
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ VE E+ D ++ KKKKK+K +D E
Sbjct: 471 -SKAVEGEELTATDNGHS----KKKKKTKSQDDE 499
>UniRef100_Q6DRE5 NOP56 [Brachydanio rerio]
Length = 548
Score = 500 bits (1288), Expect = e-140
Identities = 250/444 (56%), Positives = 334/444 (74%), Gaps = 5/444 (1%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFD 63
+L+ LETNLP G K K LGV+++K+G+ + E + +Q+ V E+IRGVR HF
Sbjct: 75 DLKLFLETNLPS---GGKKKPMLGVSDAKLGAALQEELNLSIQTGGVVAEIIRGVRLHFH 131
Query: 64 KFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVR 123
V L KAQLGL HSYSRAKVKFNVNR DNM+IQ+I LLD LDKD+N+F+MRVR
Sbjct: 132 SLVKGLTAQAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVR 191
Query: 124 EWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKA 183
EWY +HFPEL+KIV+DN YCK+AK I ++ +L+E+ +E++ ++ D KA+ I++A+++
Sbjct: 192 EWYGYHFPELIKIVSDNSTYCKLAKLIGNRKELSEEMLEAMEEITMDSAKAQAILDASRS 251
Query: 184 SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLIS 243
SMG D+SP+DLIN+ F+ RV+ L+DYR+ L +YL +KM +APNL +L+G+ VGARLIS
Sbjct: 252 SMGMDISPIDLINIECFSSRVVSLTDYRQELQEYLRSKMGQVAPNLAALIGEVVGARLIS 311
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++R
Sbjct: 312 HAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRISR 371
Query: 304 YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN 363
YLANKC+IASRIDCFSE TS FG+KLR+QVEERL FY+ G APRKN+DVMK A+
Sbjct: 372 YLANKCTIASRIDCFSEVPTSVFGDKLRDQVEERLAFYETGEAPRKNLDVMKEAVAQASE 431
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
E++ + +K KKK+QK + ++ + E TNGD E+++ KKKKK++ +
Sbjct: 432 VAAEIKRKLEKKEKKKKKRQKKLEA--LSTAEGDEETNGDVEENEVATKKKKKKQAAEEM 489
Query: 424 EVEVEDKVVEDGANADSSKKKKKK 447
EV+ +++V +G KKKKK+
Sbjct: 490 EVKEDEEVTTNGVEETPGKKKKKR 513
>UniRef100_Q90W60 XNop56 protein [Xenopus laevis]
Length = 532
Score = 500 bits (1287), Expect = e-140
Identities = 251/451 (55%), Positives = 345/451 (75%), Gaps = 8/451 (1%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++L+ +LET++P KK K LGVA++KIG+ I E KIP Q+ V E++RG+R HF
Sbjct: 74 EDLKLLLETHMP----AKKKKALLGVADAKIGAAIQEELKIPCQTGGAVVEILRGIRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HSLVKGLTAQSASKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPEL+KIV+DNY YC++AKFI ++ +L+E+K+E + ++V D KA+ +++A++
Sbjct: 190 REWYGYHFPELIKIVSDNYTYCRMAKFIGNRKELSEEKLEEMEEIVMDSAKAQAVLDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+SP+DLIN+ F+ RV+ LS+YR+ L +YL +KM+ +AP+L +L+G+ VGARLI
Sbjct: 250 SSMGMDISPIDLINIESFSSRVISLSEYRKELQEYLRSKMSQVAPSLSALIGEVVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
SHAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+ +NKGR++
Sbjct: 310 SHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAMKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKC+IASRIDCFSE TS FG+KLREQVEERL FY+ G PRKN+DVMK A +
Sbjct: 370 RYLANKCTIASRIDCFSEIPTSVFGDKLREQVEERLAFYETGEVPRKNLDVMKEAQQEAT 429
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLD 422
+E++ ++ K+ K+KK++ + +A ++A E + +++ E ++ KK+K+K
Sbjct: 430 EVVSEVK-RKLEKKEKKRKKREKRQLEALAAEEAEEPSQKKTKENGEEDEEPKKKKKKRH 488
Query: 423 QEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
E EV + +E+ SSKKKKK ++ ++A
Sbjct: 489 SEAEVSENGMEE---ETSSKKKKKNTEPEEA 516
>UniRef100_Q5ZMD0 Hypothetical protein [Gallus gallus]
Length = 535
Score = 491 bits (1265), Expect = e-137
Identities = 255/464 (54%), Positives = 341/464 (72%), Gaps = 16/464 (3%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET++P KK K LGV ++KIG+ I E Q+ V E++RG+R HF
Sbjct: 74 EDLRLLLETSMP----AKKKKALLGVGDAKIGAAILEELGYQCQTGGVVAEILRGIRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HALVKGLTAQSASKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPEL+KIV++NY YC++AKFI ++ +L+E+ +E L ++V D KA+ I+EA++
Sbjct: 190 REWYGYHFPELIKIVSENYTYCRLAKFIGNRKELSEESLEGLEEIVMDGAKAQAILEASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+SP+DLIN+ F+ RV+ LS+YR+ L +YL +KM+ +AP+L +L+G+ VGARLI
Sbjct: 250 SSMGMDISPIDLINIESFSSRVISLSEYRKGLQEYLRSKMSQVAPSLSALIGEVVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
SHAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 SHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKC+IASRIDCFSE TS FG+KLREQVEERL FY+ G PRKN++VMK A+ +
Sbjct: 370 RYLANKCTIASRIDCFSEVPTSVFGDKLREQVEERLAFYETGEPPRKNLEVMKEAMVEAN 429
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLD 422
E++ ++ +K +KK++K A + E + +AE++ E KKKKK+K++ +
Sbjct: 430 EVVAEVKRKQEKKEKKRKKREKRRLEAQAAAAEEMENSVQEAEENNIEPKKKKKKKKEQE 489
Query: 423 QE--VEVEDKVVEDG----------ANADSSKKKKKKSKKKDAE 454
QE E E+ +E+ A +KKKKK K KD E
Sbjct: 490 QEAATEPEENGLEEEVLPKKKRKVMGEATPLEKKKKKKKGKDLE 533
>UniRef100_UPI000036C2C4 UPI000036C2C4 UniRef100 entry
Length = 605
Score = 482 bits (1240), Expect = e-134
Identities = 257/452 (56%), Positives = 330/452 (72%), Gaps = 12/452 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 85 EDLRLLLETHLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 140
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 141 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 200
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L EDK+E L +L D KAK I++A++
Sbjct: 201 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEDKLEKLEELTMDGAKAKAILDASR 260
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 261 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 320
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 321 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 380
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ V
Sbjct: 381 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQVK 440
Query: 363 NKDTEMETE-EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK-RK 420
E+ + E KK KK++K +A + + T + E+ + KKKKK+K ++
Sbjct: 441 EAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEETSEKPKKKKKQKPQE 500
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ QE +ED + SK KKKKS K+
Sbjct: 501 VPQENGMEDPPI------SFSKPKKKKSFSKE 526
>UniRef100_UPI000036C2C5 UPI000036C2C5 UniRef100 entry
Length = 594
Score = 481 bits (1239), Expect = e-134
Identities = 256/452 (56%), Positives = 330/452 (72%), Gaps = 12/452 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETHLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L EDK+E L +L D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEDKLEKLEELTMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
Query: 363 NKDTEMETE-EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK-RK 420
E+ + E KK KK++K +A + + T + E+ + KKKKK+K ++
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEETSEKPKKKKKQKPQE 489
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ QE +ED + SK KKKKS K+
Sbjct: 490 VPQENGMEDPPI------SFSKPKKKKSFSKE 515
>UniRef100_O00567 Nucleolar protein Nop56 [Homo sapiens]
Length = 596
Score = 481 bits (1238), Expect = e-134
Identities = 254/452 (56%), Positives = 328/452 (72%), Gaps = 10/452 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETHLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L EDK+E L +L D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEDKLEKLEELTMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
Query: 363 NKDTEMETE---EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKR 419
++ E E KK KK++K +A + + T + E+ + KKKKK+K
Sbjct: 430 AEEAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEEMSEKPKKKKKQK- 488
Query: 420 KLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
QEV E+ + + + KKKK SK++
Sbjct: 489 --PQEVPQENGMEDPSISFSKPKKKKSFSKEE 518
>UniRef100_UPI0000363324 UPI0000363324 UniRef100 entry
Length = 563
Score = 481 bits (1237), Expect = e-134
Identities = 254/474 (53%), Positives = 335/474 (70%), Gaps = 27/474 (5%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFD 63
+L+ LETNLP K K +LGV+++KIG+ + E + +Q+ V E+ RG+R HF
Sbjct: 75 DLKLFLETNLPI---STKKKAALGVSDAKIGAALQEEFNLSIQTGGVVAEISRGIRLHFH 131
Query: 64 KFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVR 123
V L KAQLGL HSYSRAKVKFNVNR DNM+IQ+I LLD LDKD+N+F+MRVR
Sbjct: 132 SLVKGLTGLAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVR 191
Query: 124 EWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKA 183
EWY +HFPEL+KIV+DN +YC++A+ I ++ +L E+ +E L ++V D KA+ I++A+++
Sbjct: 192 EWYGYHFPELIKIVSDNSMYCRLARLIGNRKELTEESLEGLEEVVMDAAKARTILDASRS 251
Query: 184 SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLIS 243
SMG D+SP+DLIN+ F+ RV+ L+ YR L +YL +KM+ +APNL +L+G+ VGARLIS
Sbjct: 252 SMGMDISPIDLINIERFSDRVVSLAGYRLELQEYLRSKMSQVAPNLAALIGEVVGARLIS 311
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA A+NKGR++R
Sbjct: 312 HAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAGAKNKGRISR 371
Query: 304 YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN 363
YLANKC+IASRIDCFSE TS FG+KLREQVEERL FY+ G PRKN+DVMK A++ +
Sbjct: 372 YLANKCTIASRIDCFSELPTSVFGDKLREQVEERLSFYETGAMPRKNVDVMKEAVQEATD 431
Query: 364 KDTEMETEEVSAKKTKKKKQK---AADGDDMAVDKAAEITNGDAEDHKSEKKK------- 413
E++ + +K ++K++K A+GD D + GD K +KKK
Sbjct: 432 VAAEIKRKLDKKEKKRRKREKLQQQANGDTNG-DAEVCMDAGDTPVVKKKKKKQAEEAPV 490
Query: 414 ------------KKKEKRKLD-QEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KKK+KRK + +EV+VE+ V D D+ KKKKK K E
Sbjct: 491 AEEAPVAEETPGKKKKKRKSEVEEVQVEEAPVTDNGAEDTPAKKKKKRKSDATE 544
>UniRef100_UPI0000363323 UPI0000363323 UniRef100 entry
Length = 522
Score = 480 bits (1236), Expect = e-134
Identities = 251/451 (55%), Positives = 328/451 (72%), Gaps = 19/451 (4%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFD 63
+L+ LETNLP K K +LGV+++KIG+ + E + +Q+ V E+ RG+R HF
Sbjct: 72 DLKLFLETNLPI---STKKKAALGVSDAKIGAALQEEFNLSIQTGGVVAEISRGIRLHFH 128
Query: 64 KFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVR 123
V L KAQLGL HSYSRAKVKFNVNR DNM+IQ+I LLD LDKD+N+F+MRVR
Sbjct: 129 SLVKGLTGLAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVR 188
Query: 124 EWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKA 183
EWY +HFPEL+KIV+DN +YC++A+ I ++ +L E+ +E L ++V D KA+ I++A+++
Sbjct: 189 EWYGYHFPELIKIVSDNSMYCRLARLIGNRKELTEESLEGLEEVVMDAAKARTILDASRS 248
Query: 184 SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLIS 243
SMG D+SP+DLIN+ F+ RV+ L+ YR L +YL +KM+ +APNL +L+G+ VGARLIS
Sbjct: 249 SMGMDISPIDLINIERFSDRVVSLAGYRLELQEYLRSKMSQVAPNLAALIGEVVGARLIS 308
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA A+NKGR++R
Sbjct: 309 HAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAGAKNKGRISR 368
Query: 304 YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN 363
YLANKC+IASRIDCFSE TS FG+KLREQVEERL FY+ G PRKN+DVMK A++ V
Sbjct: 369 YLANKCTIASRIDCFSELPTSVFGDKLREQVEERLSFYETGAMPRKNVDVMKEAVQEVC- 427
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
M+ + K KKKKQ ++ V + A + AE+ +KKKK+K + +
Sbjct: 428 ----MDAGDTPVVKKKKKKQ----AEEAPVAEEAPV----AEETPGKKKKKRKSE---VE 472
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
EV+VE+ V D D+ KKKKK K E
Sbjct: 473 EVQVEEAPVTDNGAEDTPAKKKKKRKSDATE 503
>UniRef100_Q6PH34 Nol5a protein [Brachydanio rerio]
Length = 481
Score = 479 bits (1234), Expect = e-134
Identities = 240/412 (58%), Positives = 314/412 (75%), Gaps = 5/412 (1%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFD 63
+L+ LETNLP G K K LGV+++K+G+ + E + +Q+ V E+IRGVR HF
Sbjct: 75 DLKLFLETNLPS---GGKKKPMLGVSDAKLGAALQEELNLSIQTGGVVAEIIRGVRLHFH 131
Query: 64 KFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVR 123
V L KAQLGL HSYSRAKVKFNVNR DNM+IQ+I LLD LDKD+N+F+MRVR
Sbjct: 132 SLVKGLTAQAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVR 191
Query: 124 EWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKA 183
EWY +HFPEL+KIV+DN YCK+AK I ++ +L+E+ +E++ ++ D KA+ I++A+++
Sbjct: 192 EWYGYHFPELIKIVSDNSTYCKLAKLIGNRKELSEEMLEAMEEITMDSAKAQAILDASRS 251
Query: 184 SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLIS 243
SMG D+SP+DLIN+ F+ RV+ L+DYR+ L +YL +KM +APNL +L+G+ VGARLIS
Sbjct: 252 SMGMDISPIDLINIECFSSRVVSLTDYRQELQEYLRSKMGQVAPNLAALIGEVVGARLIS 311
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++R
Sbjct: 312 HAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRISR 371
Query: 304 YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN 363
YLANKC+IASRIDCFSE TS FG+KLR+QVEERL FY+ G APRKN+DVMK A+
Sbjct: 372 YLANKCTIASRIDCFSEVPTSVFGDKLRDQVEERLAFYETGEAPRKNLDVMKEAVAQASE 431
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKK 415
E++ + +K KKK+QK + ++ + E TNGD E++ KKKK
Sbjct: 432 VAAEIKRKLEKKEKKKKKRQKKLEA--LSTAEGDEETNGDVEENGVATKKKK 481
>UniRef100_Q95K50 Hypothetical protein [Macaca fascicularis]
Length = 594
Score = 479 bits (1234), Expect = e-134
Identities = 255/452 (56%), Positives = 330/452 (72%), Gaps = 12/452 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETHLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L E+K+E L +L D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEEKLEKLEELTMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
Query: 363 NKDTEMETE-EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK-RK 420
E+ + E KK KK++K +A + + T + E+ + KKKKK+K ++
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEETSEKPKKKKKQKPQE 489
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ QE +ED + SK KKKKS K+
Sbjct: 490 VPQENGMEDPSI------PFSKPKKKKSFSKE 515
>UniRef100_Q5RA29 Hypothetical protein DKFZp459D1517 [Pongo pygmaeus]
Length = 594
Score = 479 bits (1233), Expect = e-134
Identities = 252/450 (56%), Positives = 327/450 (72%), Gaps = 8/450 (1%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET+LP KK + LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETHLPS----KKKRVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSR KVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRTKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L EDK+E L +L D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEDKLEKLEELTMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
Query: 363 NKDTEMETE-EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKL 421
E+ + E KK KK++K +A + + T + E+ + KKKKK+K
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKRLAALALASSENSSSTPEECEETSEKPKKKKKQK--- 486
Query: 422 DQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
QEV E+ + + + KKKK SK++
Sbjct: 487 PQEVPQENGMEDPSISFSKPKKKKSFSKEE 516
>UniRef100_Q6TEL4 Nucleolar protein 5A [Brachydanio rerio]
Length = 494
Score = 479 bits (1232), Expect = e-134
Identities = 240/415 (57%), Positives = 315/415 (75%), Gaps = 5/415 (1%)
Query: 4 ELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFD 63
+L+ LETNLP G K K LGV+++K+G+ + E + +Q+ V E+IRGVR HF
Sbjct: 75 DLKLFLETNLPS---GGKKKPMLGVSDAKLGAALQEELNLSIQTGGVVAEIIRGVRLHFH 131
Query: 64 KFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVR 123
V L KAQLGL HSYSRAKVKFNVNR DNM+IQ+I LLD LDKD+N+F+MRVR
Sbjct: 132 SLVKGLTAQAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVR 191
Query: 124 EWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKA 183
EWY +HFPEL+KIV+DN YCK+AK I ++ +L+E+ +E++ ++ D KA+ I++A+++
Sbjct: 192 EWYGYHFPELIKIVSDNSTYCKLAKLIGNRKELSEEMLEAMEEITMDSAKAQAILDASRS 251
Query: 184 SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLIS 243
SMG D+SP+DLIN+ F+ RV+ L+DYR+ L +YL +KM +APNL +L+G+ VGARLIS
Sbjct: 252 SMGMDISPIDLINIECFSSRVVSLTDYRQELQEYLRSKMGQVAPNLAALIGEVVGARLIS 311
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++R
Sbjct: 312 HAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRISR 371
Query: 304 YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN 363
YLANKC+IASRIDCFSE TS FG+KL QVEERL FY+ G APRKN+DVMK A+
Sbjct: 372 YLANKCTIASRIDCFSEVPTSVFGDKLVTQVEERLAFYETGEAPRKNLDVMKEAVAQASE 431
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E++ + +K KKK+QK + ++ + E TNGD E++ KKKKK++
Sbjct: 432 VAAEIKRKLEKKEKKKKKRQKKLEA--LSTAEGVEETNGDVEENGVATKKKKKKQ 484
>UniRef100_Q9D6Z1 Nucleolar protein Nop56 [Mus musculus]
Length = 580
Score = 474 bits (1221), Expect = e-132
Identities = 252/452 (55%), Positives = 328/452 (71%), Gaps = 17/452 (3%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETYLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKIVNDN YC++A+FI ++ +L E+K+E L ++ D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIVNDNATYCRLAQFIGNRRELNEEKLEKLEEITMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHTYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAVVQAE 429
Query: 363 NKDTEM--ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
E+ + E+ K+ KK+K++ A A+ A+ + E+ + +K KK+K+
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKRLA-----ALALASSENSSTPEECEEVNEKSKKKKKL 484
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
QE +ED V K KKKK+ K++
Sbjct: 485 KPQENGMEDPPV------SLPKSKKKKAPKEE 510
>UniRef100_UPI00001CF330 UPI00001CF330 UniRef100 entry
Length = 588
Score = 471 bits (1213), Expect = e-131
Identities = 247/451 (54%), Positives = 325/451 (71%), Gaps = 12/451 (2%)
Query: 3 DELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHF 62
++LR +LET LP KK K LGV + KIG+ I E Q+ + E++RGVR HF
Sbjct: 74 EDLRLLLETYLPS----KKKKVLLGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHF 129
Query: 63 DKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRV 122
V L KAQLGL HSYSRAKVKFNVNRVDNM+IQ+I LLD LDKD+N+F+MRV
Sbjct: 130 HNLVKGLTDLSACKAQLGLGHSYSRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRV 189
Query: 123 REWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAK 182
REWY +HFPELVKI+NDN YC++A+FI ++ +L E+K+E L ++ D KAK I++A++
Sbjct: 190 REWYGYHFPELVKIINDNATYCRLAQFIGNRRELNEEKLEKLEEITMDGAKAKAILDASR 249
Query: 183 ASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLI 242
+SMG D+S +DLIN+ F+ RV+ LS+YR+ L +YL +KM+ +AP+L +L+G++VGARLI
Sbjct: 250 SSMGMDISAIDLINIESFSSRVVSLSEYRQSLHNYLRSKMSQVAPSLSALIGEAVGARLI 309
Query: 243 SHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMA 302
+HAGSLTNLAK P+ST+QILGAEKALFRALKTRGNTPKYGLIFHS+FIGRA+A+NKGR++
Sbjct: 310 AHAGSLTNLAKYPASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRIS 369
Query: 303 RYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD 362
RYLANKCSIASRIDCFSE TS FGEKLREQVEERL FY+ G PRKN+DVMK A+ +
Sbjct: 370 RYLANKCSIASRIDCFSEVPTSVFGEKLREQVEERLSFYETGEIPRKNLDVMKEAVVQAE 429
Query: 363 NKDTEM--ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
E+ + E+ K+ KK+K++ +A A N + E +K K+K+
Sbjct: 430 EAAAEITRKLEKQEKKRLKKEKKR------LAALALASSENSSTPEECEEVNEKPKKKKL 483
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
QE E+ + + + +KKKK K++
Sbjct: 484 KPQETLQENGMEDPPVSLPKTKKKKAFPKEE 514
>UniRef100_Q95WY3 Nucleolar KKE/D repeat protein [Drosophila melanogaster]
Length = 496
Score = 470 bits (1209), Expect = e-131
Identities = 249/412 (60%), Positives = 305/412 (73%), Gaps = 9/412 (2%)
Query: 20 KKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQL 79
KK K +LG+A++K+G+ I EA + V E++RGVR HF K V AQL
Sbjct: 88 KKKKCTLGIADAKLGAAITEAIGVQCSHFGVVPEILRGVRFHFAKLVKGFTDKSAGVAQL 147
Query: 80 GLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVND 139
GL HSYSRAKVKFNV+R DNM+IQ+I LLD LDKDVN+F+MR+REWYS+HFPELVKIV D
Sbjct: 148 GLGHSYSRAKVKFNVHRSDNMIIQSIALLDQLDKDVNTFSMRIREWYSYHFPELVKIVPD 207
Query: 140 NYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHM 199
NY++ K AKFI+D+ L +D++E L +V D KA+ I++AAK SMG D+S VDL+N+ +
Sbjct: 208 NYMFAKAAKFIKDRKNLTQDQLEDLEKIVMDSAKAQAIIDAAKMSMGMDISIVDLMNIEL 267
Query: 200 FAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTL 259
FA+RV+ LS+YR++LS YL KMN +APNLQSL+GD VGARLISHAGSLTNLAK P+ST+
Sbjct: 268 FAERVVKLSEYRKKLSTYLHNKMNLVAPNLQSLIGDQVGARLISHAGSLTNLAKYPASTV 327
Query: 260 QILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFS 319
QILGAEKALFRALKTR NTPKYGLI+HSSFIGRA +NKGR++R+LANKCSIASRIDCF
Sbjct: 328 QILGAEKALFRALKTRSNTPKYGLIYHSSFIGRAGLKNKGRISRFLANKCSIASRIDCFL 387
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
E+ TS FGE L++QVE+RL FY+ G PRKNI+VMK AIE+ E S KK K
Sbjct: 388 EQPTSVFGETLKQQVEDRLKFYESGDVPRKNIEVMKEAIEAAGQ---EAAITVQSEKKKK 444
Query: 380 KKKQKAAD-----GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVE 426
KKK+ A D G AAE NG+ D E KKKKK+K K + VE
Sbjct: 445 KKKRAAEDEGETNGASNGNGAAAEAENGNGND-DGEPKKKKKKKNKNKEAVE 495
>UniRef100_Q9N2M9 Nucleolar protein [Drosophila subobscura]
Length = 503
Score = 468 bits (1205), Expect = e-130
Identities = 244/412 (59%), Positives = 308/412 (74%), Gaps = 12/412 (2%)
Query: 20 KKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQL 79
KK K +LG+A++K+G+ I EA + V E++RGVR HF K V AQL
Sbjct: 88 KKKKCTLGIADAKLGAAITEAIGVQCSQVGVVPEILRGVRFHFAKLVKGFTDQSAGVAQL 147
Query: 80 GLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVND 139
GL HSYSRAKVKFNV+R DNM+IQ+I LLD LDKDVN+F+MR+REWYS+HFPELVKIV D
Sbjct: 148 GLGHSYSRAKVKFNVHRSDNMIIQSIALLDQLDKDVNTFSMRIREWYSYHFPELVKIVPD 207
Query: 140 NYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHM 199
NY++ K AKFI+D+ L++D +E L +V D KA+ I++AAK SMG D+S VDL+N+ +
Sbjct: 208 NYMFAKAAKFIKDRKNLSQDLLEDLEKIVMDSAKAQAIIDAAKMSMGMDISIVDLMNIEL 267
Query: 200 FAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTL 259
FA+RV+ LS+YR++LS YL KMN +APNLQSL+GD VGARLISHAGSLTNLAK P+ST+
Sbjct: 268 FAERVVKLSEYRKKLSTYLHNKMNLVAPNLQSLIGDQVGARLISHAGSLTNLAKYPASTV 327
Query: 260 QILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFS 319
QILGAEKALFRALKTR NTPKYGLI+HSSFIGRA +NKGR++R+LANKCSIASRIDCF
Sbjct: 328 QILGAEKALFRALKTRSNTPKYGLIYHSSFIGRAGLKNKGRISRFLANKCSIASRIDCFL 387
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
++ TS FGE L++QVE+RL FY+ G PRKNI+VMK AIE+ + T ++K K
Sbjct: 388 DQPTSVFGESLKQQVEDRLKFYESGDVPRKNIEVMKEAIEAAGQEAGANGTAAPQSEKKK 447
Query: 380 KKKQKAADGDDM-----------AVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
KK+K A G+D A +AA+ NG+ +D + +KKKKKK K K
Sbjct: 448 NKKKKRAAGEDETEANGSSNGNGAAAEAADNGNGN-DDGEPKKKKKKKNKNK 498
>UniRef100_Q8IGT5 RE33426p [Drosophila melanogaster]
Length = 496
Score = 468 bits (1205), Expect = e-130
Identities = 249/412 (60%), Positives = 304/412 (73%), Gaps = 9/412 (2%)
Query: 20 KKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQL 79
KK K +LG+A++K+G+ I EA + V E++RGVR HF K V AQL
Sbjct: 88 KKKKCTLGIADAKLGAAITEAIGVQCSHFGVVPEILRGVRFHFAKLVKGFTDKSAGVAQL 147
Query: 80 GLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVND 139
GL HSYSRAKVKFNV+R DNM+IQ+I LLD LDKDVN+F+MR+REWYS+HFPELVKIV D
Sbjct: 148 GLGHSYSRAKVKFNVHRSDNMIIQSIALLDQLDKDVNTFSMRIREWYSYHFPELVKIVPD 207
Query: 140 NYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHM 199
NY++ K AKFI+D+ L +D++E L +V D KA+ I+ AAK SMG D+S VDL+N+ +
Sbjct: 208 NYMFAKAAKFIKDRKNLTQDQLEDLEKIVMDSAKAQAIIGAAKMSMGMDISIVDLMNIEL 267
Query: 200 FAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTL 259
FA+RV+ LS+YR++LS YL KMN +APNLQSL+GD VGARLISHAGSLTNLAK P+ST+
Sbjct: 268 FAERVVKLSEYRKKLSTYLHNKMNLVAPNLQSLIGDQVGARLISHAGSLTNLAKYPASTV 327
Query: 260 QILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFS 319
QILGAEKALFRALKTR NTPKYGLI+HSSFIGRA +NKGR++R+LANKCSIASRIDCF
Sbjct: 328 QILGAEKALFRALKTRSNTPKYGLIYHSSFIGRAGLKNKGRISRFLANKCSIASRIDCFL 387
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
E+ TS FGE L++QVE+RL FY+ G PRKNI+VMK AIE+ E S KK K
Sbjct: 388 EQPTSVFGETLKQQVEDRLKFYESGDVPRKNIEVMKEAIEAAGQ---EAAITVQSEKKKK 444
Query: 380 KKKQKAAD-----GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVE 426
KKK+ A D G AAE NG+ D E KKKKK+K K + VE
Sbjct: 445 KKKRAAEDEGETNGASNGNGAAAEAENGNGND-DGEPKKKKKKKNKNKEAVE 495
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 702,532,827
Number of Sequences: 2790947
Number of extensions: 30274616
Number of successful extensions: 305022
Number of sequences better than 10.0: 6288
Number of HSP's better than 10.0 without gapping: 1901
Number of HSP's successfully gapped in prelim test: 4698
Number of HSP's that attempted gapping in prelim test: 211832
Number of HSP's gapped (non-prelim): 40050
length of query: 454
length of database: 848,049,833
effective HSP length: 131
effective length of query: 323
effective length of database: 482,435,776
effective search space: 155826755648
effective search space used: 155826755648
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC135505.8


