

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135504.2 - phase: 0 /pseudo
(2338 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
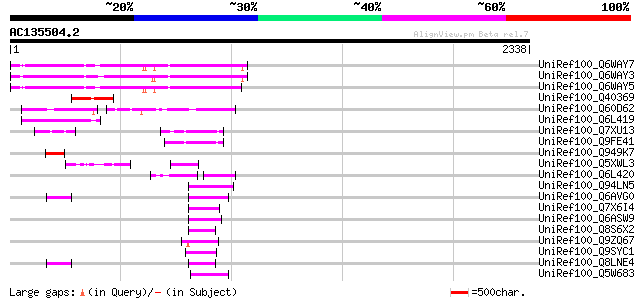
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 534 e-149
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 531 e-149
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 528 e-148
UniRef100_Q40369 RPE15 protein [Medicago sativa] 254 2e-65
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 222 9e-56
UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum] 141 2e-31
UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa] 107 6e-21
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 106 7e-21
UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum] 100 7e-19
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 99 2e-18
UniRef100_Q6L420 Putative polyprotein [Solanum demissum] 92 2e-16
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 77 5e-12
UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sat... 77 8e-12
UniRef100_Q7X6I4 OSJNBa0039G19.2 protein [Oryza sativa] 75 2e-11
UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polyme... 75 2e-11
UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa] 74 4e-11
UniRef100_Q9ZQ67 Putative retroelement pol polyprotein [Arabidop... 73 9e-11
UniRef100_Q9SYC1 F11M15.5 protein [Arabidopsis thaliana] 73 9e-11
UniRef100_Q8LNE4 Putative retroelement [Oryza sativa] 73 1e-10
UniRef100_Q5W683 Hypothetical protein OSJNBa0076A09.14 [Oryza sa... 73 1e-10
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 534 bits (1375), Expect = e-149
Identities = 390/1149 (33%), Positives = 571/1149 (48%), Gaps = 124/1149 (10%)
Query: 3 QLRTELASLREELAKANDTMTALLAAQEQSATVIPVTTSVIPTASTDARFAMPTGFPYGL 62
Q++ ELA ++ +A+ + M + QE+ ++ + IP + A P G P
Sbjct: 3 QVQAELAEMKANMAQFMNMMQGVAQGQEELRALVQRQEAAIPPVN----HAPPEGGPVN- 57
Query: 63 PPFFTPNTAAGTS------GTANSG-PIPGANLASIGITLTQATATVTEPVMITVPQGTH 115
N AA G G I G +A A P I Q
Sbjct: 58 ----GNNVAAAVPINNYAVGDELGGIRINGQPIAPDVANARAVRAPARNPAPIVDRQEDM 113
Query: 116 AGAHRGSTPITGTMEER---IEELAKELRREIKANRGSSDTIKTHDLCLVPKVDIPKKFK 172
+ G ++ER ++ LA+++R N D ++ LV + IP KFK
Sbjct: 114 FSLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLGFDVT---NMGLVEGLRIPYKFK 170
Query: 173 VPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 232
P FD+YNG +CP+ H+ Y RK+ Y+D++ + ++ FQDSL + +WY L +D +
Sbjct: 171 APSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKRDSIRC 230
Query: 233 FDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEETT 292
+ +L AF Y N + PSR L+SL QK ESF EYAQRWR AAR+ P + E E T
Sbjct: 231 WKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQPPMLERELT 290
Query: 293 QTFMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESSANASKRNGNG 352
F+ TL+ +++RM +FS +V G R E ++ G I ++ +++SK+ G
Sbjct: 291 DMFIGTLQGVFMDRMGSCPFVSFSDVVICGERTESLIKTGKIQ----DAGSSSSKKPFAG 346
Query: 353 HHKKKETEVWMVSAGAGQSMATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQP 412
+++E E V Q+ + + A +P P P Q
Sbjct: 347 APRRREGETNAVQYRRDQNRSQRCQVAAVTIPA--------------------PQPRQQQ 386
Query: 413 QVPVNAIVQQMQQQPPTQQQQQQQARPTFPPIPMLYFELLPTLLLRGHCTTRQGKPPPDP 472
Q V QQ QQQ P Q +Q+ R F +PM Y ELLP LL G R PP
Sbjct: 387 QQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT-MAPPTV 444
Query: 473 LPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHVA 532
LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P H
Sbjct: 445 LPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQHGG 503
Query: 533 --VNMIEVCE-EAPRLDVRNIATPLVPLHIKLCQASLFDHDHASCPKCFRNPLGCYVVQN 589
VN IE E E ++V ++ T L+ + +L ++ C C + GC ++
Sbjct: 504 HRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRT 563
Query: 590 DI*SLMNDNYL----------TVSDVCMI-------------VPVFHDPPVKSMPSKENV 626
I +M++ L TVS + + P + PV
Sbjct: 564 GIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTISVPVTIS 623
Query: 627 EPLVIRLPGPVPYTSEKAIPYKY-----------------NATMI*NGV--EVPLTSLAM 667
P I G + +A+P+KY N + G+ VP T
Sbjct: 624 APTTIAASGRRAVENSRAVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPA 683
Query: 668 VSNIAEGTTAALRSGRV-CPPLFQKKAATPTTPPIDKATLTDVSPITKDVSRPSQSIEDS 726
V N+ G RSGR+ P + A K + + P+ K+ P S E
Sbjct: 684 VDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA--PEGSFE-K 739
Query: 727 NLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTVDR 786
+++E ++IIK+SDYKIVDQL QTPSKIS+LSLLL SEAHRN LLK+L AYV E++V++
Sbjct: 740 DVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQ 799
Query: 787 FGDIVGNITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKS 846
++ N+++ + + F+ ++ G+ HN ALHI++ CK +LS+VLVDTG+SLNV+PK
Sbjct: 800 LEGVMANVSTRHGVGFTNLDLTPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQ 859
Query: 847 TLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCL 906
L ++ L S +V+AFDGS+++V GE+ LP+ IGPE F I F VMDI +YSCL
Sbjct: 860 ILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCL 919
Query: 907 LGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEA-GSAEGT--- 962
LGRPW H AGAV+STLHQKLK+V NG++VT+ GEE LVS LSSF +E G T
Sbjct: 920 LGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQ 979
Query: 963 AFQGLTIEG---TEPKKAGAAMASLKDAQKAVQEGQAVG*GKLIQLRGNKHKEGLGFSPT 1019
AF+ + +E E +K GA++ S K A++ V G+A G GK++ L + K G+G+ P
Sbjct: 980 AFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGYEPL 1039
Query: 1020 SGVSTG-----TFHSAGFVNAITEEATGFGP-----------RPVFVIPGGIARDWDAID 1063
G TF SAG +N ATG RP PGG +W A +
Sbjct: 1040 QAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINNWTAEE 1097
Query: 1064 IPSIMHVSE 1072
+ + ++E
Sbjct: 1098 VVQVTLLTE 1106
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 531 bits (1369), Expect = e-149
Identities = 394/1151 (34%), Positives = 572/1151 (49%), Gaps = 130/1151 (11%)
Query: 3 QLRTELASLREELAKANDTMTALLAAQEQSATVIPVTTSVIPTASTDARFAMPTGFPYGL 62
Q++TELA +R +A+ M + QE+ ++ +V P + P G
Sbjct: 3 QVQTELAEMRANMAQFMHMMQGVAQGQEELRALVQRQEAVTPPPNQ--------ALPEGN 54
Query: 63 PPFFTPNTAAGTSGTANSGPIPGANLASIGITLTQATATVTE-PVMITVP----QGTHAG 117
P TP A + A + G + I A A V P +P Q
Sbjct: 55 PVHDTPAAAIPVNNYAVGEELMGIRVDGQPIAPDAANARVIHAPARNRIPIVDRQEDLFT 114
Query: 118 AHRGSTPITGTMEER---IEELAKELRREIKANRGSSDTIKTHDLCLVPKVDIPKKFKVP 174
I G + R ++ LA+++R N D ++ LV + IP KFK P
Sbjct: 115 MFSEDEDIPGRNDARDRKVDALAEKIRAMECQNSLGFDVT---NMGLVEGLRIPYKFKAP 171
Query: 175 EFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFD 234
FD+YNG +CP+ H+ Y RK+ Y+D++ + ++ FQDSL + +WY L D + +
Sbjct: 172 SFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKSDSIRCWR 231
Query: 235 ELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEETTQT 294
+L AF Y N + PSR L+SL QK +ESF EYAQRWR AAR+ P + E E T
Sbjct: 232 DLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAARVQPPMLERELTDM 291
Query: 295 FMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESSANASKRNGNGHH 354
F+ TL+ +++RM +FS +V G R E ++ G I + +++SK+ G
Sbjct: 292 FIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKIQ----DVGSSSSKKPFAGAP 347
Query: 355 KKKETEVWMVSAGAGQSMATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQV 414
+++E E V Q+ A +P P Q Q P
Sbjct: 348 RRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQP--------------- 392
Query: 415 PVNAIVQQMQQQPPTQQQQQQQARPTFPPIPMLYFELLPTLLLRGH---CTTRQGKPPPD 471
QQ QQQ P Q +Q+ R F +PM Y ELLP LL G CT PP
Sbjct: 393 ------QQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGLVELCTMA----PPT 441
Query: 472 PLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHV 531
LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P H
Sbjct: 442 VLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINFAP-VPNVVNNPMPQHG 500
Query: 532 A--VNMIEVCEEAPRLD-VRNIATPLVPLHIKLCQASLFDHDHASCPKCFRNPLGCYVVQ 588
VN IE E +D V ++ T L+ + +L ++ C C + GC ++
Sbjct: 501 GHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAESENGCDQLR 560
Query: 589 NDI*SLMNDNYLTVS-------DVCMIVPVF-----HDPPVKSMPSKENVEPLVIRLPGP 636
I +M++ L S V I F H V S P+ N P+ I +P
Sbjct: 561 AGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGHGQRVVSAPAT-NGTPVTIPVPVT 619
Query: 637 V--PYT----------SEKAIPYKY-----------------NATMI*NGV--EVPLTSL 665
+ P T + +A+P+KY N T + G+ VP T
Sbjct: 620 ISAPTTIVASGRRAVENSRAVPWKYDNAYRSNRRVESQTKPVNQTPVTIGLANRVPATVG 679
Query: 666 AMVSNIAEGTTAALRSGRV-CPPLFQKKAATPTTPPIDKATLTDVSPITKDVSRPSQSIE 724
V N+ G RSGR+ P + A K + + P+ K+ P S E
Sbjct: 680 PAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA--PEGSFE 736
Query: 725 DSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTV 784
+++E +++IK+SDYKIVDQL QTPSKIS+LSLLL SEAHRN LLK+L AYV E++V
Sbjct: 737 -KDVEEFMKMIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISV 795
Query: 785 DRFGDIVGNITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMP 844
++ ++ N+++ + + F+ ++P G+ HN ALHI++ CK +LS+VLVDTG+SLNV+P
Sbjct: 796 NQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLP 855
Query: 845 KSTLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYS 904
K L ++ L S +V+AFD S+++V GE+ LP+ IGPE F I F VMDI +YS
Sbjct: 856 KQILKKIDVEGFVLTPSDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYS 915
Query: 905 CLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEA-GSAEGT- 962
CLLGRPW H AGAV+STLHQKLK+V NG++VT+ GEE LVS LSSF +E G T
Sbjct: 916 CLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETL 975
Query: 963 --AFQGLTIEG---TEPKKAGAAMASLKDAQKAVQEGQAVG*GKLIQLRGNKHKEGLGFS 1017
F+ + +E E +K G ++ S K A++ V G+A G GK++ L + K G+G+
Sbjct: 976 CQVFETVALEKVAYAEQRKPGVSITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYE 1035
Query: 1018 PTSGVSTG-----TFHSAGFVNAITEEATGFGP-----------RPVFVIPGGIARDWDA 1061
P G TF SAG +N ATG RP PGG +W A
Sbjct: 1036 PLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINNWTA 1093
Query: 1062 IDIPSIMHVSE 1072
++ + ++E
Sbjct: 1094 EEVVQVTLLTE 1104
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 528 bits (1360), Expect = e-148
Identities = 381/1109 (34%), Positives = 558/1109 (49%), Gaps = 113/1109 (10%)
Query: 3 QLRTELASLREELAKANDTMTALLAAQEQSATVIPVTTSVIPTASTDARFAMPTGFPYGL 62
Q++ ELA ++ +A+ + M + QE+ ++ + IP + A P G P
Sbjct: 3 QVQAELAEMKANMAQFMNMMQGVAQGQEELRAMVQRQEAAIPPVN----HAPPEGGPVN- 57
Query: 63 PPFFTPNTAAGTSGTAN---SGPIPGANLASIGITLTQATA-TVTEPVMITVP----QGT 114
N A N + G + S I A A T+ P VP Q
Sbjct: 58 -----DNNVAAAVPINNYDVGDELGGIRINSQPIVPDAANARTIRAPARNPVPIVDRQED 112
Query: 115 HAGAHRGSTPITGTMEER---IEELAKELRREIKANRGSSDTIKTHDLCLVPKVDIPKKF 171
+ G ++ER ++ LA+++R N S D ++ LV + IP KF
Sbjct: 113 MFTLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLSFDVT---NMGLVEGLRIPYKF 169
Query: 172 KVPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVH 231
K P FD+YN +CP+ H+ Y RK+ Y+D++ + ++ FQDSL + +WY L +D +
Sbjct: 170 KAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKRDSIR 229
Query: 232 TFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEET 291
+ +L AF Y N + PSR L+SL QK ESF EYAQRWR AAR+ P + E E
Sbjct: 230 CWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQPPMLEREL 289
Query: 292 TQTFMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESSANASKRNGN 351
T F+ TL+ +++RM +FS +V G R E ++ G I + +++SK+
Sbjct: 290 TDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKIQ----DVGSSSSKKPFA 345
Query: 352 GHHKKKETEVWMVSAGAGQSMATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQ 411
G +++E E +V Q+ A +P P P Q
Sbjct: 346 GAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPA--------------------PQPRQQ 385
Query: 412 PQVPVNAIVQQMQQQPPTQQQQQQQARPTFPPIPMLYFELLPTLLLRGHCTTRQGKPPPD 471
Q V QQ QQQ P Q +Q+ R F +PM Y ELLP LL G R PP
Sbjct: 386 QQQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT-MAPPT 443
Query: 472 PLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHV 531
LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P H
Sbjct: 444 VLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQHG 502
Query: 532 A--VNMIEVCE-EAPRLDVRNIATPLVPLHIKLCQASLFDHDHASCPKCFRNPLGCYVVQ 588
VN IE E E ++V ++ T L+ + +L ++ C C + GC ++
Sbjct: 503 GHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLR 562
Query: 589 NDI*SLMNDNYL----------TVSDVCMI-------------VPVFHDPPVKSMPSKEN 625
I +M++ L TVS + + P +D PV
Sbjct: 563 AGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTISVPVTI 622
Query: 626 VEPLVIRLPGPVPYTSEKAIPYKY-----------------NATMI*NGV--EVPLTSLA 666
P I G + + +P+KY N + G+ VP T
Sbjct: 623 SAPTTIVASGRRAVENSRVVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGP 682
Query: 667 MVSNIAEGTTAALRSGRV-CPPLFQKKAATPTTPPIDKATLTDVSPITKDVSRPSQSIED 725
V N+ G RSGR+ P + A K + + P+ K+ P + E
Sbjct: 683 AVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA--PEGTFE- 738
Query: 726 SNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTVD 785
+++E ++IIK+SDYKIVDQL QTPSKIS+LSLL+ SEAHRN LLK+L AYV E++V+
Sbjct: 739 KDVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLMCSEAHRNALLKMLNLAYVPQEISVN 798
Query: 786 RFGDIVGNITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPK 845
+ ++ N+++ + + F+ ++P G+ HN ALHI++ CK +LS+VLVDTG+SLNV+P
Sbjct: 799 QLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPN 858
Query: 846 STLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSC 905
L ++ L S +V+AFDGS+++V GE+ LP+ IGPE F I F VMDI +YSC
Sbjct: 859 QILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSC 918
Query: 906 LLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEA-GSAEGT-- 962
LLGRPW H AGAV+STLHQKLK+V NG++VT+ GEE LVS LSSF +E G T
Sbjct: 919 LLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLC 978
Query: 963 -AFQGLTIEG---TEPKKAGAAMASLKDAQKAVQEGQAVG*GKLIQLRGNKHKEGLGFSP 1018
AF+ + +E E +K GA++ S K A++ V G+A G GK++ L + K G+G+ P
Sbjct: 979 QAFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEP 1038
Query: 1019 TSGVSTG-----TFHSAGFVNAITEEATG 1042
G TF SAG +N ATG
Sbjct: 1039 LRAEQNGQAGPSTFTSAGLMNHGDVSATG 1067
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 254 bits (649), Expect = 2e-65
Identities = 131/192 (68%), Positives = 147/192 (76%), Gaps = 11/192 (5%)
Query: 278 AAARITPALDEEETTQTFMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFE 337
AAAR++P LDEEE TQTF+KTLKK++ ERMI++APNNFS MVTMGTRLEEAVREGIIV E
Sbjct: 13 AAARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLE 72
Query: 338 KVESSANASKRNGNGHHKKKET-EVWMVSAGAGQSMATVAPINAAQLPPLYPYAQYSQHP 396
K E SA+A K+ GNGHHK+KE+ +V MVS P+NA QLP Y Y Q SQHP
Sbjct: 73 KGECSASAPKKYGNGHHKRKESKKVGMVSGNHD------TPVNATQLPLPYQYMQISQHP 126
Query: 397 FFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQQQQQQARPT----FPPIPMLYFELL 452
FFPPFY QYPLP GQP VPVNA+ QQ+Q QPP QQQQQQQ P FPPIPM Y ELL
Sbjct: 127 FFPPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQQQGPRPKTYFPPIPMRYAELL 186
Query: 453 PTLLLRGHCTTR 464
PTLL +GHC TR
Sbjct: 187 PTLLAKGHCVTR 198
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 222 bits (566), Expect = 9e-56
Identities = 179/614 (29%), Positives = 278/614 (45%), Gaps = 98/614 (15%)
Query: 437 ARPTFPPIPMLY---FELLPTLLLRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 493
A+ F PI Y F+ L TL + + PPP L C + A GH+
Sbjct: 888 AKDEFTPIGESYASLFQKLRTLNVLSPIERKMSNPPPRNLD----YSQHCAYCSDAPGHN 943
Query: 494 VEGCYALKYIVKKLIDQGKLTFEN-NVPHVLDNPLPNHVAVNMIEVCEEAPRLDVRNIAT 552
+E C+ LK ++ LID ++ E+ N P++ NPLP H NM+E+ +A
Sbjct: 944 IERCWYLKKAIQDLIDTRRIIVESPNGPNINQNPLPRHTETNMLEMVNNH-----EEVAV 998
Query: 553 PLVPLHIKLCQASLFDHDHASCPKCFRNPLGCYVVQND----------I*SLMNDNYLTV 602
P P+ +K+ + K P G I + D + +
Sbjct: 999 PYKPI-LKVETGMESSTNVIDLTKIM--PSGAESTSEKLTPSSAPILTIKGALEDVWASQ 1055
Query: 603 SDVCMIVPVFHDPPVKSMPSKENVEPLVIRLPGPVPYTSEKAIPYKYNATMI*-NGVEVP 661
+ ++VP D P+ + + P++IR +P T+ KA+P+ Y T++ G E+
Sbjct: 1056 REARLVVPRGPDKPILIVQGAY-IPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGKEI- 1113
Query: 662 LTSLAMVSNIAEGTTAALRSGRVCPPLFQKKAATPTTPPIDKATLTDVSPITKDVSRPSQ 721
+ + RSGR LT++ D +
Sbjct: 1114 -------NEEVDEIGGMTRSGRCY-------------------ALTELRKNKNDQMQVKS 1147
Query: 722 SIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHE 781
+ + +E LR +K SDY IV+QL +TP++IS+LSLL+ S+AHR ++K+L +A+V +E
Sbjct: 1148 PVTEGEAEEFLRKMKLSDYSIVEQLRKTPAQISLLSLLIHSDAHRKAVIKILNEAHVPNE 1207
Query: 782 VTVDRFGDIVGNITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLN 841
VTV + I G I N + FS+DE+P G+ N
Sbjct: 1208 VTVSQLEKIAGRIFEVNRITFSDDELPV--------------------------EGSGAN 1241
Query: 842 VMPKSTLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINA 901
+ P STL +L +R + V+AFDG++ + +GEI+L + IG +F + FQV++INA
Sbjct: 1242 ICPLSTLQKLNVNVERVRPNNVCVRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLNINA 1301
Query: 902 SYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEAGSA-E 960
SY+ LLGRPW H AGAV STLHQ +KF + + V +H E V + SS I+A + E
Sbjct: 1302 SYNLLLGRPWVHRAGAVPSTLHQMVKFEYDRQEVIVHSEGDLSVYKDSSLPFIKANNENE 1361
Query: 961 GTAFQGLTI-------EGTEPKKAGAAMASLKDAQKAVQEGQAVG*GKLIQLRGNKHKE- 1012
+Q + EG K MAS+ + ++ G G G I L+G +
Sbjct: 1362 ALVYQAFEVVVVEHILEGNLISKPQLPMASVMMVNEMLKHGFEPGKGLGIFLQGRAYPVS 1421
Query: 1013 --------GLGFSP 1018
GLG+ P
Sbjct: 1422 PRKSLGTFGLGYKP 1435
Score = 104 bits (260), Expect = 3e-20
Identities = 94/368 (25%), Positives = 156/368 (41%), Gaps = 60/368 (16%)
Query: 55 PTGFPYGLPPFFTPNTAAGTSGTANSGPIPGANLASIGITLTQATATVTEPVMITVPQGT 114
P P P F P A T+ T+N +P +N L T E + I +
Sbjct: 598 PLNTPMMSNPLFVPT--APTNSTSNPTVVPKSNNDPSFQVLHDHGYTPEEALKIP---SS 652
Query: 115 HAGAHRGSTP--ITGTME-ERIEELAKELRREIKANR-----GSSDTIKTHDLCLVPKVD 166
+ H+ S+P I T++ E EE+AK+++ ++ R G I DLC+ P V
Sbjct: 653 YPQTHQYSSPFKIEKTVKNEEHEEMAKKMKSLEQSIRDMQGLGGHKGISFSDLCMFPHVH 712
Query: 167 IPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLS 226
+P + L++ F +SL+ A+EW+
Sbjct: 713 LPT----------------------------GAEGKEELLMAYFGESLVGIASEWFIDQD 744
Query: 227 KDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPAL 286
+ HT+D LA F + +N + P R L ++ +K E+F EYA RWR AAR+ ++
Sbjct: 745 IANWHTWDNLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWREQAARVKLSM 804
Query: 287 DEEETTQTFMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESSANAS 346
E E F++ + DY ++ A F +++ +G +E ++ G IV + + +
Sbjct: 805 KESEMIDVFLQAQEPDYFHYLLSAVGKTFVEVIKVGEMVENGIKSGKIVSQAALKATTQA 864
Query: 347 KRNGNGH--HKKKETEVWMVSAGA-------GQSMAT----------VAPINAAQLPPLY 387
+NG+G+ KK+ +V V GA G+S A+ ++PI P
Sbjct: 865 LQNGSGNIGGKKRREDVATVGNGAKDEFTPIGESYASLFQKLRTLNVLSPIERKMSNPPP 924
Query: 388 PYAQYSQH 395
YSQH
Sbjct: 925 RNLDYSQH 932
>UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum]
Length = 421
Score = 141 bits (356), Expect = 2e-31
Identities = 101/365 (27%), Positives = 171/365 (46%), Gaps = 34/365 (9%)
Query: 55 PTGFPYGLPPFFTPNTAAGTSGTANSGPIPGANLASIGITLTQATATVTEPVMITVPQGT 114
P P P F P A T+ T+N +P +N L T E + I +
Sbjct: 78 PLNTPMMSNPLFVPT--APTNSTSNPTVVPKSNNDPSFQVLHDHGYTPEEALKIP---SS 132
Query: 115 HAGAHRGSTP--ITGTME-ERIEELAKELRREIKANR-----GSSDTIKTHDLCLVPKVD 166
+ H+ S+P I T++ E EE+ K+++ + R G I DLC+ P V
Sbjct: 133 YPQTHQYSSPFKIEKTVKNEEHEEMTKKMKSLEHSIRDMQGLGGHKGISFSDLCMFPHVH 192
Query: 167 IPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLS 226
+P FK P F++Y+G P H+ +Y ++ + L++ F +SL+ A++W+
Sbjct: 193 LPTGFKTPNFEKYDGHGDPIAHLKRYYNQLRGAEGKEELLMAYFGESLVGIASKWFIDQD 252
Query: 227 KDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPAL 286
+ HT+D+LA F + +N + P R L ++ +K E+F EYA RWR AAR+ P +
Sbjct: 253 IANWHTWDDLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWREQAARVKPPM 312
Query: 287 DEEETTQTFMKTLKKDYVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESSANAS 346
E E F++ + DY ++ A F++++ +G +E ++ G IV + + +
Sbjct: 313 KELEMIDVFLQAQEPDYFHYLLSAVGKTFTEVIKVGEMVENGIKSGKIVSQAALKATTQA 372
Query: 347 KRNGNGH---HKKKETEVWMVSAGAGQSMATVAPINAAQLPPLYPYAQYSQHPFFPPFYH 403
NG+G+ K++E +VSA P Y ++QH +FPP
Sbjct: 373 LPNGSGNIGGKKRREDVATVVSA-----------------PRTYAQDNHTQH-YFPPQIP 414
Query: 404 QYPLP 408
QY +P
Sbjct: 415 QYLVP 419
>UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa]
Length = 2657
Score = 107 bits (266), Expect = 6e-21
Identities = 81/282 (28%), Positives = 135/282 (47%), Gaps = 24/282 (8%)
Query: 679 LRSGRVCPPLFQKKAATPTTPPIDKATLTDVSPITKDVSRPSQSIEDSNLDEILRIIKRS 738
LR G+ P + K + PI +A+ +P TK S+ I
Sbjct: 1343 LRGGKTLPDPHKAKPSK-VDKPIKEASPPGEAPETKARSKEKPDI--------------- 1386
Query: 739 DYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTVDRFGDIVGNITSCN 798
DY ++ L + P+ +SV L+ R L+K L+ V +EV + + + N N
Sbjct: 1387 DYNVLAHLKRIPALLSVYDALILVPDLREALIKALQTPEV-YEVDMAKHR-LFNNPLFVN 1444
Query: 799 NLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPL 858
+ F++++ G HN L+I N L +L+D G+++N++P +L + + L
Sbjct: 1445 EITFADEDNIIEGSDHNRPLYIEGNIGPAHLRRILIDPGSAVNILPVRSLTRAGFTTKDL 1504
Query: 859 RRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAV 918
+ ++ FD K+ LG I + + + +F + F V++ N SYS LLGRPW H V
Sbjct: 1505 EPTDMVICGFDNQGKSTLGAITVKIQMSTFSFKVHFFVIEANTSYSALLGRPWIHKYRVV 1564
Query: 919 TSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEAGSAE 960
STLHQ LKF+ +GK G + +V SS++ E+ A+
Sbjct: 1565 PSTLHQCLKFL-DGK-----GAQQRIVGNSSSYTIQESYHAD 1600
Score = 54.3 bits (129), Expect = 4e-05
Identities = 46/196 (23%), Positives = 82/196 (41%), Gaps = 23/196 (11%)
Query: 111 PQGTHAGAHRGSTPIT-----GTMEERIEELAKELRREIKANRGSSDTIKT----HDLCL 161
P TH STP T +E+ + K++ E S+ K HDL
Sbjct: 784 PTQTHDARRSTSTPTTLKDYHDIVEDLVNRRLKQISIEQTPRPMESELEKPYEAWHDLVA 843
Query: 162 VPKVDIPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEW 221
P P KF+ +FDR + H+ + G+ ++N SL++ F SL A W
Sbjct: 844 FPARWHPPKFR--QFDRTGDA---REHLAYFEAACGDTANNSSLLLRQFSGSLTGPAFHW 898
Query: 222 YTSLSKDDVHTFDELAAAFKNHYGFNTRLKPSREF----LRSLSQKKDESFCEYAQRWRG 277
Y+ L + ++ + K H+ + ++F L + Q++DE+ +Y R+R
Sbjct: 899 YSRLPTGSIRSWASMKEVLKKHF-----VAMKKDFSIVELSQVRQRRDEAIDDYVIRFRN 953
Query: 278 AAARITPALDEEETTQ 293
+ R+ + E+ +
Sbjct: 954 SFVRLAREMHLEDAIE 969
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 106 bits (265), Expect = 7e-21
Identities = 75/264 (28%), Positives = 126/264 (47%), Gaps = 8/264 (3%)
Query: 697 TTPPIDKATLTDVSPITKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVL 756
T P K+ + +V K S P ++ E + DYK++ L + P+ +SV
Sbjct: 1263 TLPDPHKSKVPNVDKPAKKASPPGEAPEAPETKTGSKEKPAVDYKVLAHLKRIPALLSVY 1322
Query: 757 SLLLSSEAHRNTLLKVLEQAYVDHEVTVDRFGDIVGNITSCNNLWFSEDEMPEAGKYHNL 816
L+ R L+K L+ V +EV + + + N N + F++++ G HN
Sbjct: 1323 DALMMVPDLREALIKALQAPEV-YEVDMAKHR-LYDNPLFVNEITFADEDNIIKGGDHNR 1380
Query: 817 ALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRKNVL 876
L+I N S L +L+D G+++N++P +L + + L ++ FD K L
Sbjct: 1381 PLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRAGFTTKDLEPIDVVICGFDNQGKPTL 1440
Query: 877 GEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVT 936
G I + + + +F + F V++ N SYS LLGRPW H V STLHQ LKF+
Sbjct: 1441 GAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFLDG----- 1495
Query: 937 IHGEETYLVSQLSSFSCIEAGSAE 960
+G + + S S ++ E+ A+
Sbjct: 1496 -NGVQQRITSNFSPYTIQESYHAD 1518
>UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum]
Length = 88
Score = 100 bits (248), Expect = 7e-19
Identities = 43/88 (48%), Positives = 61/88 (68%)
Query: 159 LCLVPKVDIPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDA 218
+CLVP V IP+KFKV EF++Y G TCP+NH+ Y RKM +++ ND L+IH FQDSL +
Sbjct: 1 MCLVPDVTIPRKFKVLEFEKYKGATCPKNHLTMYGRKMASHAHNDKLLIHFFQDSLSGAS 60
Query: 219 AEWYTSLSKDDVHTFDELAAAFKNHYGF 246
WY L ++ +H++ +LA AF Y +
Sbjct: 61 LSWYMHLERNRIHSWKDLADAFLRQYKY 88
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 98.6 bits (244), Expect = 2e-18
Identities = 48/130 (36%), Positives = 77/130 (58%)
Query: 722 SIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHE 781
SI + +E R ++ DY IV L +TP++ISV +LL+SS+ HR L+K L+ YV
Sbjct: 610 SINKAKAEEFWRRMQPKDYSIVKYLEKTPAQISVWALLMSSQLHRQALMKALDDTYVPVG 669
Query: 782 VTVDRFGDIVGNITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLN 841
D ++ + + + F ++E+P G+ HN ALH++ C+ +++ VLVD G+ LN
Sbjct: 670 TNSDNLAAMINQVIRGHRISFCDEELPFEGRMHNKALHVTTMCRDKIINRVLVDDGSGLN 729
Query: 842 VMPKSTLDQL 851
+ P STL L
Sbjct: 730 ICPLSTLRHL 739
Score = 91.7 bits (226), Expect = 2e-16
Identities = 76/292 (26%), Positives = 125/292 (42%), Gaps = 68/292 (23%)
Query: 252 PSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEETTQTFMKTLKKDYVERMIIAA 311
P R L + QK ES+ E+A RWR AAR+ P + E+E + F++ K
Sbjct: 299 PDRYSLEKMKQKSTESYREFAYRWRKEAARVRPPMSEKEIVEVFVRVAK----------- 347
Query: 312 PNNFSKMVTMGTRLEEAVREGIIVFEKVESSANASKRNGNGHHKKKETEVWMVSAGAGQS 371
F+++V +G +E+ +R G IV+ V +S +S KKK +V +S +
Sbjct: 348 ---FAEIVKIGETIEDGLRTGKIVY--VAASPESSIL-----LKKKRDDVSSISY---EG 394
Query: 372 MATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQ 431
T ++ Q + +P
Sbjct: 395 KKTAKKSSSCQ----------------------------------------GRSRPSHSS 414
Query: 432 QQQQQARPTFPPIPMLYFELLPTLLLRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALG 491
+++ AR F P+ +L L + G+ G P D +R D +C +H ++G
Sbjct: 415 FEKKPAR-NFTPLIKSRTKLFERLTVAGYIHP-VGPKPVDTSSKFYRPDQRCAYHSNSVG 472
Query: 492 HDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNH--VAVNMIEVCEE 541
HD E C LK+ ++ LI+Q ++ + P+V NPLPNH + +NMIE E+
Sbjct: 473 HDTEDCINLKHKIQDLINQNVVSLQTVAPNVNSNPLPNHGGITINMIETDED 524
>UniRef100_Q6L420 Putative polyprotein [Solanum demissum]
Length = 1483
Score = 92.0 bits (227), Expect = 2e-16
Identities = 60/163 (36%), Positives = 87/163 (52%), Gaps = 17/163 (10%)
Query: 873 KNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNG 932
++ +GEI+L + IGP +F + FQV++INASY+ LLGRPW H AGAV STLHQ +KF +
Sbjct: 213 EDAIGEIELILKIGPVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDR 272
Query: 933 KLVTIHGEETYLVSQLSSFSCIEAGSA-EGTAFQGL-------TIEGTEPKKAGAAMASL 984
+ V +H E V + SS I+A + E +Q +EG K MAS+
Sbjct: 273 QEVIVHCERDLSVYKDSSLPFIKANNENEALVYQAFEVVVVEHNLEGNLISKPQLPMASV 332
Query: 985 KDAQKAVQEGQAVG*GKLIQLRGNKHKE---------GLGFSP 1018
+ ++ G +G G I L+G + GLG+ P
Sbjct: 333 MMVNEMLKHGFKLGKGLGIFLQGRAYPVSPRKSLGTFGLGYKP 375
Score = 87.4 bits (215), Expect = 5e-15
Identities = 63/209 (30%), Positives = 104/209 (49%), Gaps = 28/209 (13%)
Query: 636 PVPYTSEKAIPYKYNATMI*-NGVEVPLTSLAMVSNIAEGTTAALRSGRVCPPLFQKKAA 694
P+P T+ KA+P+KY T++ G E+ V I EG T RS R P +K
Sbjct: 65 PLPMTNPKAVPWKYEPTVVTYKGKEIN----EKVDEI-EGMT---RSRRCYAPAELRKN- 115
Query: 695 TPTTPPIDKATLTDVSPITKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKIS 754
D + + + +E LR +K SDY IV+QL +TP++IS
Sbjct: 116 ------------------NNDQMQVKSPVTEREAEEFLRKMKLSDYSIVEQLRKTPAQIS 157
Query: 755 VLSLLLSSEAHRNTLLKVLEQAYVDHEVTVDRFGDIVGNITSCNNLWFSEDEMPEAGKYH 814
+LSLL+ S+ HR ++K+L +A+V +E T+ + I G I N + FS+DE+P
Sbjct: 158 LLSLLIHSDEHRKVVMKILNEAHVPNEGTMSQLEKIAGRIFEVNCITFSDDELPVEDAIG 217
Query: 815 NLALHISVNCKSDMLSNVLVDTGASLNVM 843
+ L + + ++ +++ AS N++
Sbjct: 218 EIELILKIGPVDFTVNFQVLNINASYNLL 246
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 77.4 bits (189), Expect = 5e-12
Identities = 57/203 (28%), Positives = 101/203 (49%), Gaps = 7/203 (3%)
Query: 806 EMPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFL 864
E PE + +L L+I+ +S ++VD GA++N+MP +T +L L ++ +
Sbjct: 351 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMV 410
Query: 865 VKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQ 924
+K F G+ +G +++ +T+G + TF V+D SYS LLGR W H + ST+HQ
Sbjct: 411 LKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTIHQ 470
Query: 925 KLKFVKNGKLVTIHGEETYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTEPKKAGAAMASL 984
L G + I ++ L + S+ C E + T++ + K+ M++
Sbjct: 471 CL-IQWQGDKIEIVPADSQLKMENPSY-CFEGVMEGSNVYTKDTVDDLDDKQGQGLMSA- 527
Query: 985 KDAQKAVQEGQAVG*GK--LIQL 1005
D + + A+G G+ LI+L
Sbjct: 528 -DDLEEIDISPAIGQGRHLLIEL 549
>UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sativa]
Length = 1251
Score = 76.6 bits (187), Expect = 8e-12
Identities = 52/183 (28%), Positives = 92/183 (49%), Gaps = 5/183 (2%)
Query: 803 SEDEMPEA-GKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRS 861
+E E PE G H L+I+ +S ++VD GA++N+MP +T +L L ++
Sbjct: 685 AEFEKPEGTGNRHLKPLYINGYVNGKSMSKMMVDGGAAVNLMPYATFKKLGRNAEDLIKT 744
Query: 862 TFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTST 921
++K F G+ G +++ +T+G + TF V+D SYS LLGR W H + ST
Sbjct: 745 NMVLKDFGGNPSETRGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPST 804
Query: 922 LHQKLKFVKNGKLVTIHGEETYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGTEPKKAGAA 980
+HQ L + K+ + + S++ + S G EG+ + T++ + K+
Sbjct: 805 MHQCLIQWQGDKIEIVPADSQ---SKMENPSYYFEGVVEGSNVYAKDTVDDLDDKQGQGF 861
Query: 981 MAS 983
M++
Sbjct: 862 MSA 864
Score = 57.4 bits (137), Expect = 5e-06
Identities = 35/116 (30%), Positives = 63/116 (54%), Gaps = 5/116 (4%)
Query: 164 KVDIPKKFKVPEFDRYNGL--TCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEW 221
+V +P +FKVP+F +++G HI +++ + G S D+L F+ SL A W
Sbjct: 235 RVPLPNRFKVPDFSKFSGQEGVSTYEHINRFLAQCGEASAVDALRFRLFRLSLSGSAFTW 294
Query: 222 YTSLSKDDVHTFDELAAAFKNH-YGFNTRLKPSREFLRSLSQKKDESFCEYAQRWR 276
++SL ++++ +L F ++ Y + +K S L ++ Q+ DE EY QR+R
Sbjct: 295 FSSLPCGSINSWADLEKQFHSYFYSWVHEMKLSD--LTAIKQRHDEPVHEYIQRFR 348
>UniRef100_Q7X6I4 OSJNBa0039G19.2 protein [Oryza sativa]
Length = 1136
Score = 75.5 bits (184), Expect = 2e-11
Identities = 45/142 (31%), Positives = 77/142 (53%), Gaps = 1/142 (0%)
Query: 806 EMPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFL 864
E PE K +L L+I+ +S ++VD GA++N+MP +T +L L +++ +
Sbjct: 364 EKPEGTKNRHLKPLYINGYVNEKPMSKMMVDGGAAVNLMPYATFRKLGRNTEDLIKTSMV 423
Query: 865 VKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQ 924
+K F G+ G ++L +T+G + TF V+D SYS LLGR W H + ST+HQ
Sbjct: 424 LKDFGGNPSETKGVLNLELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 483
Query: 925 KLKFVKNGKLVTIHGEETYLVS 946
L + K+ + +++ V+
Sbjct: 484 CLIQWQGNKIEIVLADKSVNVA 505
>UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polymerase) domain
containing protein [Oryza sativa]
Length = 1372
Score = 75.5 bits (184), Expect = 2e-11
Identities = 45/147 (30%), Positives = 76/147 (51%), Gaps = 1/147 (0%)
Query: 806 EMPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFL 864
E PE + +L L+I+ +S ++VD GA++N+MP +T +L L ++ +
Sbjct: 424 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNVEDLIKTNMV 483
Query: 865 VKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQ 924
+K F G+ G +++ +T+G + TF V+D SYS LLGR W H + ST+HQ
Sbjct: 484 LKDFGGNPSETKGVLNVELTVGNKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 543
Query: 925 KLKFVKNGKLVTIHGEETYLVSQLSSF 951
L + K+ + + L+ L F
Sbjct: 544 CLIQWQGDKIQIVPADRAKLIELLKEF 570
>UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa]
Length = 505
Score = 74.3 bits (181), Expect = 4e-11
Identities = 38/121 (31%), Positives = 64/121 (52%)
Query: 806 EMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLV 865
E P+ H L++ + +S +LVD GA++N+MP S +L + L+++ ++
Sbjct: 310 EKPDESNRHMKPLYLKGHIDGKPVSRMLVDRGAAVNLMPYSLFKRLGRGDDELKKTNMIL 369
Query: 866 KAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQK 925
F+G G + +T+G + TF ++D+ SY+ +LGR W H V STLHQ
Sbjct: 370 NGFNGEPTEAKGIFSVELTMGNKTLPTTFFIVDVQGSYNVILGRCWIHANCCVPSTLHQC 429
Query: 926 L 926
L
Sbjct: 430 L 430
>UniRef100_Q9ZQ67 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 764
Score = 73.2 bits (178), Expect = 9e-11
Identities = 51/195 (26%), Positives = 88/195 (44%), Gaps = 27/195 (13%)
Query: 774 EQAYVDHEVTVDRFGDIVGNITSCNN---------------LW------------FSEDE 806
+Q + D V R I+G +T+C + LW F+ ++
Sbjct: 277 DQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSED 336
Query: 807 MPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVK 866
+ HN L I ++ ++ +L+DTG+S+NV+ K L ++ + ++ S +
Sbjct: 337 LFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLT 396
Query: 867 AFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKL 926
FDG+ G I LP+ +G F V+D Y+ +LG PW HD A+ S+ HQ +
Sbjct: 397 GFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCI 456
Query: 927 KFVKNGKLVTIHGEE 941
K + + TI G +
Sbjct: 457 KIPTSIGIETIRGNQ 471
>UniRef100_Q9SYC1 F11M15.5 protein [Arabidopsis thaliana]
Length = 1295
Score = 73.2 bits (178), Expect = 9e-11
Identities = 44/136 (32%), Positives = 75/136 (54%)
Query: 793 NITSCNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLC 852
N C + FSE+E + K H+ AL IS++ + + VL+DTG+S++++ TL ++
Sbjct: 441 NQIDCPEISFSEEETSDLDKPHDDALVISLDVGNCEVQRVLIDTGSSVDLIFLDTLVRMG 500
Query: 853 YRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWT 912
+ ++ + + +F +L I LP+T +I F V D A+Y+ +LG PW
Sbjct: 501 ISKKDIKGALSPLVSFTSETSMLLRTITLPVTAQGIVKMIEFTVFDRPAAYNIILGTPWL 560
Query: 913 HDAGAVTSTLHQKLKF 928
++ AV ST HQ +KF
Sbjct: 561 YEMKAVPSTYHQCVKF 576
Score = 39.3 bits (90), Expect = 1.5
Identities = 29/114 (25%), Positives = 52/114 (45%), Gaps = 11/114 (9%)
Query: 137 AKELRREIKANRGSSDTIKTHDLCLVPKVDIPKKFKVPEFDRYNGLTCPQNHIIKYVRKM 196
A E+ + I+A R + T + L ++ + FK+P YNG P+ ++ +
Sbjct: 129 APEVEKVIEATRRTPFTSRISKL----RIRDFRDFKLPG---YNGKGDPKEYLTFFQVIA 181
Query: 197 GNYS----DNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGF 246
G + D+ + F +SL A W+T L + + F +L+ AF YG+
Sbjct: 182 GRVPLEPHEEDAGLCKLFSESLSGPALTWFTQLEEGSIDNFKQLSTAFIKQYGY 235
>UniRef100_Q8LNE4 Putative retroelement [Oryza sativa]
Length = 1170
Score = 72.8 bits (177), Expect = 1e-10
Identities = 41/122 (33%), Positives = 68/122 (55%), Gaps = 1/122 (0%)
Query: 806 EMPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFL 864
E PE + +L L+I+ +S ++VD GA++N+MP +T +L L ++ +
Sbjct: 437 EKPEGTENRHLKPLYINGYVNGKTMSKMMVDGGAAVNLMPYATFRKLGRNTEALIKTNMV 496
Query: 865 VKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQ 924
+K F G+ G +++ +T+G + TF V++ N SYS LLGR W H + ST+HQ
Sbjct: 497 LKDFGGNPSETRGVLNVELTVGSKTIPTTFFVINGNGSYSLLLGRDWIHANCCIPSTMHQ 556
Query: 925 KL 926
L
Sbjct: 557 CL 558
Score = 55.1 bits (131), Expect = 3e-05
Identities = 31/115 (26%), Positives = 62/115 (52%), Gaps = 3/115 (2%)
Query: 164 KVDIPKKFKVPEFDRYNGL--TCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEW 221
+V +P +FKVP+F +++G HI +++ + S D+L + F+ SL A W
Sbjct: 210 RVPLPNRFKVPDFSKFSGQEGVSTYEHISRFLAQCHEASAVDALKVRLFRLSLSGSAFTW 269
Query: 222 YTSLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWR 276
++SL ++++ +L F +++ ++ + L ++ Q+ DE EY QR+R
Sbjct: 270 FSSLPYGSINSWADLEKQFHSYF-YSGVHEMKLSDLTAIKQRHDEPVHEYIQRFR 323
>UniRef100_Q5W683 Hypothetical protein OSJNBa0076A09.14 [Oryza sativa]
Length = 1681
Score = 72.8 bits (177), Expect = 1e-10
Identities = 46/171 (26%), Positives = 87/171 (49%), Gaps = 4/171 (2%)
Query: 814 HNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRK 873
H+ L+++ +S ++VD GA++N+MP +T +L L ++ ++K F G+
Sbjct: 752 HHKPLYVNGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKHFGGNPS 811
Query: 874 NVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGK 933
G +++ +T+G + TF V+D SYS LLGR W H + ST+HQ L + K
Sbjct: 812 ETKGVLNVELTVGSKTIPTTFFVIDGKRSYSLLLGRDWIHTNCCIPSTMHQCLIQWQGHK 871
Query: 934 LVTIHGEETYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGTEPKKAGAAMAS 983
+ + + S++ + S G EG+ + T++ + K+ M++
Sbjct: 872 IEVVPADSQ---SKMENPSYYFEGVVEGSDVYAKDTVDDLDDKQGQGFMSA 919
Score = 37.0 bits (84), Expect = 7.3
Identities = 26/87 (29%), Positives = 45/87 (50%), Gaps = 3/87 (3%)
Query: 191 KYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTRL 250
++ + G S D+L + F+ SL A W++SL V+++ +L F H F + +
Sbjct: 348 EWFERCGEASAVDALRVRMFRLSLSGLAFTWFSSLPYGSVNSWADLEKQF--HSYFYSGV 405
Query: 251 KPSREF-LRSLSQKKDESFCEYAQRWR 276
+ F L ++ Q+ DE EY QR+R
Sbjct: 406 HEMKLFDLTAIKQRYDEPVHEYIQRFR 432
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.340 0.147 0.493
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,542,978,006
Number of Sequences: 2790947
Number of extensions: 143778793
Number of successful extensions: 628555
Number of sequences better than 10.0: 1040
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 934
Number of HSP's that attempted gapping in prelim test: 616420
Number of HSP's gapped (non-prelim): 5433
length of query: 2338
length of database: 848,049,833
effective HSP length: 144
effective length of query: 2194
effective length of database: 446,153,465
effective search space: 978860702210
effective search space used: 978860702210
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 83 (36.6 bits)
Medicago: description of AC135504.2


