

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.8 - phase: 0
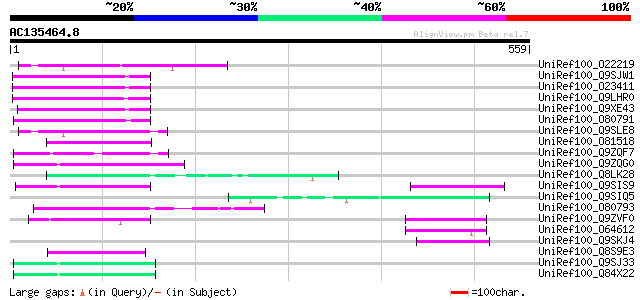
(559 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O22219 Putative Ta11-like non-LTR retroelement protein... 71 7e-11
UniRef100_Q9SJW1 Hypothetical protein At2g01050 [Arabidopsis tha... 70 1e-10
UniRef100_O23411 Hypothetical protein AT4g15600 [Arabidopsis tha... 65 5e-09
UniRef100_Q9LHR0 Non-LTR retrolelement reverse transcriptase-lik... 62 4e-08
UniRef100_Q9XE43 Putative non-LTR retrolelement reverse transcri... 61 1e-07
UniRef100_O80791 Putative Ta11-like non-LTR retroelement protein... 61 1e-07
UniRef100_Q9SLE8 Putative Ta11-like non-LTR retroelement protein... 60 2e-07
UniRef100_O81518 T24M8.9 protein [Arabidopsis thaliana] 60 2e-07
UniRef100_Q9ZQF7 Putative Ta11-like non-LTR retroelement protein... 59 3e-07
UniRef100_Q9ZQG0 Putative Ta11-like non-LTR retroelement protein... 58 6e-07
UniRef100_Q8LK28 Putative DNA/RNA binding protein [Brassica napus] 58 8e-07
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 58 8e-07
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 58 8e-07
UniRef100_O80793 Putative Ta11-like non-LTR retroelement protein... 57 1e-06
UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcrip... 57 1e-06
UniRef100_O64612 Putative reverse transcriptase [Arabidopsis tha... 56 3e-06
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 55 4e-06
UniRef100_Q8S9E3 ORF285 [Silene latifolia] 55 4e-06
UniRef100_Q9SJ33 Putative Ta11-like non-LTR retroelement protein... 55 5e-06
UniRef100_Q84X22 Hypothetical protein At2g05290/F5G3.19 [Arabido... 55 5e-06
>UniRef100_O22219 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 418
Score = 71.2 bits (173), Expect = 7e-11
Identities = 60/243 (24%), Positives = 103/243 (41%), Gaps = 26/243 (10%)
Query: 10 FEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQ-----VWVRLHEL 64
F+F+F S +D+ + LK G+ EW ++ T VW+RL +
Sbjct: 77 FQFFFKSEDDLLEI-------LKTGVWTQDEWCVVMERWIEKSTEEYLMFLPVWMRLRNI 129
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHI--FNEVMIERTG 122
P Y+ T+K+IAS +G L ++ + Y RV V +D+ + F E+ + TG
Sbjct: 130 PVNYYTQDTIKKIASCVGKVLKVELDLEKSQAQDYIRVQVIIDVRNPLRNFKEIQLP-TG 188
Query: 123 FSFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVKEPLIDKKKKTIISQK--------- 173
S+ YER+ C C + H C P K ++D +++ +K
Sbjct: 189 EIVSVTFDYERIRKRCFLCQRLTHEKGDCDSGRPGKSSVMDGFQRSFFEKKEDKEKSDLR 248
Query: 174 -PQPPKWLPRDNLDGIGSSKAFEAPVGNNNAESIPTT-HKVATKLPVQTDLTESADAQHS 231
PQ LP +LD I + A N+ + +T +V + V+ E+ ++Q
Sbjct: 249 FPQSNHPLPSSDLDRIEGNSALSTKTNLLNSRIMSSTPSEVQSTGLVEKIRKEAKESQEH 308
Query: 232 DSA 234
+ A
Sbjct: 309 EKA 311
>UniRef100_Q9SJW1 Hypothetical protein At2g01050 [Arabidopsis thaliana]
Length = 515
Score = 70.5 bits (171), Expect = 1e-10
Identities = 44/148 (29%), Positives = 67/148 (44%), Gaps = 4/148 (2%)
Query: 4 ELGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHE 63
+L +F F E+ + + G + L + +WS F VWVRL
Sbjct: 113 DLPRQFFMIRFELEEEYMAALTGGPWRVLGNYLLVQDWSSRFDPLRDDIVTTPVWVRLSN 172
Query: 64 LPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIERTGF 123
+P Y+ L EIA +G PL +D T N G +ARV ++++L+K + V+I +
Sbjct: 173 IPYNYYHRCLLMEIARGLGRPLKVDMNTINFDKGRFARVCIEVNLAKPLKGTVLINGDRY 232
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSC 151
+ YE L C+ CG GH SC
Sbjct: 233 F----VAYEGLSKICSSCGIYGHLVHSC 256
>UniRef100_O23411 Hypothetical protein AT4g15600 [Arabidopsis thaliana]
Length = 655
Score = 65.1 bits (157), Expect = 5e-09
Identities = 38/148 (25%), Positives = 68/148 (45%), Gaps = 4/148 (2%)
Query: 4 ELGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHE 63
+L +F F ED + G + +L + WS +F VWVR+
Sbjct: 128 DLPRQFFMVRFEVEEDYMMALTGGPWRVLGSILMVQAWSPEFNPLRDVIETTPVWVRVAN 187
Query: 64 LPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIERTGF 123
LP ++ + L IA+ +G P+ +D T + G +ARV V+++L + +++ +
Sbjct: 188 LPVTFYHNEILLGIAAGLGKPIKVDLTTLRKERGRFARVCVEVNLKNPLKGTLVVNGERY 247
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSC 151
++YE L C+ CG GH ++C
Sbjct: 248 F----VSYEGLQTICSLCGIYGHTVNNC 271
Score = 42.4 bits (98), Expect = 0.036
Identities = 23/80 (28%), Positives = 38/80 (46%), Gaps = 1/80 (1%)
Query: 436 LWALWSKDL-SITVLFVSDQCIALQISCHQSIVYIAAVYASTYYLKRRQLWADLTHIQEH 494
LW LW + ++ ++ + Q I +I V++ VYA+ +R LW +L
Sbjct: 575 LWLLWRSSVGNVEIVSSTSQFIHAKILTGLESVHLVVVYAAPSVSRRSGLWNELREAVSG 634
Query: 495 FVGPWLFVGDFNAVLGAHEK 514
GP + GDFN +L E+
Sbjct: 635 LDGPLIIGGDFNTILRVDER 654
>UniRef100_Q9LHR0 Non-LTR retrolelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 591
Score = 62.0 bits (149), Expect = 4e-08
Identities = 35/148 (23%), Positives = 64/148 (42%), Gaps = 4/148 (2%)
Query: 4 ELGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHE 63
+L +F F E+ + + G L + WS +F +WVR+ +
Sbjct: 113 DLPRHFFMVRFEKEEEFLTALTGGPWRAFGSCLLVKAWSPEFNPLKDEIVTTPIWVRIAD 172
Query: 64 LPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIERTGF 123
+P ++ L +A +G PL +D + G +ARV V++DL + + +++ +
Sbjct: 173 MPVSFYHKSILMSVAQGLGRPLKVDLTMLHFERGRFARVCVEVDLKRPLQGALLVNGDRY 232
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSC 151
+ YE L C+ CG GH +C
Sbjct: 233 ----YVAYEGLTNICSKCGIYGHLVHNC 256
>UniRef100_Q9XE43 Putative non-LTR retrolelement reverse transcriptase [Arabidopsis
thaliana]
Length = 855
Score = 60.8 bits (146), Expect = 1e-07
Identities = 35/143 (24%), Positives = 64/143 (44%), Gaps = 4/143 (2%)
Query: 9 YFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHELPQEY 68
+F F ++ S + G L + WS +F T +WVRL +P
Sbjct: 93 FFMIRFEREDEYLSALTGGPWRAFGSYLLVQAWSPEFDPLRDEITTTPIWVRLMNIPLSL 152
Query: 69 WMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIERTGFSFSIE 128
+ L IA ++G P+ +D T + +AR+ +++DL+K + +++ +
Sbjct: 153 YHTSILMGIAGSLGKPVKVDMTTLHVERARFARMCIEVDLAKPLKGTLLLNGERYF---- 208
Query: 129 ITYERLPAFCTHCGNIGHHNSSC 151
++YE L C+ CG GH +C
Sbjct: 209 VSYEGLANICSRCGMYGHLIHTC 231
>UniRef100_O80791 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 409
Score = 60.8 bits (146), Expect = 1e-07
Identities = 38/147 (25%), Positives = 61/147 (40%), Gaps = 4/147 (2%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
L +F F ED + + +L + WS DF VWVR+ L
Sbjct: 66 LPRSFFMVKFDLEEDYLAAAIGDPWRVLGSILMIRAWSPDFNPIRDEIVTTLVWVRISNL 125
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIERTGFS 124
P Y+ + L I +G P+ ++ + H+ARV V ++L + + V++ +
Sbjct: 126 PVNYYHETILTAIVEGLGKPIKVNLTILRKERAHFARVCVKVNLKQPLKGTVVVNGECYY 185
Query: 125 FSIEITYERLPAFCTHCGNIGHHNSSC 151
S YE L C+ CG GH +C
Sbjct: 186 AS----YEGLNVICSLCGVFGHLAGAC 208
>UniRef100_Q9SLE8 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 240
Score = 60.1 bits (144), Expect = 2e-07
Identities = 38/167 (22%), Positives = 73/167 (42%), Gaps = 21/167 (12%)
Query: 10 FEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQ-----VWVRLHEL 64
F+F++ S +D+ + LK G+ EW ++ +W+RL +
Sbjct: 77 FQFFYKSNDDLLEI-------LKTGVWTQDEWCVIMDRWVEKPPEEYLMILPIWIRLRNI 129
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIE-RTGF 123
P Y+ + T+K+IA +G + ++ + Y RV V++D+ + N ++ +G
Sbjct: 130 PVNYYTEETIKKIAGCVGQVVKVELDLEKSQAQDYVRVQVNLDVRNPLRNSKSVQVPSGE 189
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVKEPLIDKKKKTII 170
S+ YER+ C C + H + C P ++KK + I
Sbjct: 190 VVSVTFDYERIRKRCFFCQRLTHDKADC--------PYVNKKIRIFI 228
>UniRef100_O81518 T24M8.9 protein [Arabidopsis thaliana]
Length = 463
Score = 59.7 bits (143), Expect = 2e-07
Identities = 32/114 (28%), Positives = 53/114 (46%), Gaps = 1/114 (0%)
Query: 40 EWSKDFTARTQRQTHAQVWVRLHELPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHY 99
+W T T A +W+ L +P +++ D L+ IAS +G P + +T N+
Sbjct: 189 KWEPGLTPVKPELTTAPIWLELRHVPFQFFNDNGLEHIASLVGEPKFLHPSTANKTNLEV 248
Query: 100 ARVLVDMDLSKHIFNEVMIE-RTGFSFSIEITYERLPAFCTHCGNIGHHNSSCR 152
ARV +D K + V + +G IE++ +P C HC +GH C+
Sbjct: 249 ARVFTIIDPRKPLPEAVNVHFASGDIRRIEVSSPWMPPVCAHCKEVGHSLRKCK 302
>UniRef100_Q9ZQF7 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 589
Score = 59.3 bits (142), Expect = 3e-07
Identities = 39/168 (23%), Positives = 74/168 (43%), Gaps = 19/168 (11%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
LG+ F+F+F S D++ V +K + L W T WVR +
Sbjct: 85 LGDARFQFFFESEVDLQKVLNKRPCHFNKWSFALERWEPH--VGTSFPNIMTFWVRTEGI 142
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHI-FNEVMIERTGF 123
P E+W + L+ +++G +D + R+L+ + + FN+ +G
Sbjct: 143 PAEFWDEEVLRNFGNSLGLVRRVDPSK--------GRILISVTADVPLRFNKNAQLPSGT 194
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVKEPLIDKKKKTIIS 171
++++YE+L +C++C I H C PL+D ++K ++S
Sbjct: 195 VVKVKLSYEKLFRWCSYCRRICHELEQC--------PLLDAEQKAVLS 234
>UniRef100_Q9ZQG0 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 483
Score = 58.2 bits (139), Expect = 6e-07
Identities = 44/186 (23%), Positives = 78/186 (41%), Gaps = 4/186 (2%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
L N F+F F S D+ V +KG + + W ++ R + +WV++ +
Sbjct: 69 LNNERFQFIFDSEHDLEEVLAKGVHTFNDWSIVVDRWYENPPDDYLR--YMLLWVQIWNI 126
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIE-RTGF 123
P Y + + ++ IG + + + + RV V D+S+ + ++ R G
Sbjct: 127 PVNYNTAKAITKLGDLIGEVKEVVFNPELKQIREFVRVKVLFDVSRPLRRSKVVNFRNGG 186
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVKEPLIDKKKKTIISQK-PQPPKWLPR 182
S ++ YER+ C C + H C L ++ L ++K II K PP
Sbjct: 187 SATVNFHYERIQKRCYECQRLTHEKDVCPILVKARQDLATARRKGIIIPKATTPPILKES 246
Query: 183 DNLDGI 188
D L G+
Sbjct: 247 DPLFGV 252
>UniRef100_Q8LK28 Putative DNA/RNA binding protein [Brassica napus]
Length = 306
Score = 57.8 bits (138), Expect = 8e-07
Identities = 60/320 (18%), Positives = 124/320 (38%), Gaps = 31/320 (9%)
Query: 40 EWSKDFTARTQRQTHAQVWVRLHELPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHY 99
+W + VW+ H +P +++ + L+ IA +G PL + AT N
Sbjct: 13 KWKPGLQPEIPELSSVPVWLDFHNVPPQFYSEEGLEHIAGTLGHPLFLHPATANMSNLEV 72
Query: 100 ARVLVDMDLSKHIFNEVMIE-RTGFSFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVK 158
ARV +D +K + V + +G +E++ LP C HC +GH +C +
Sbjct: 73 ARVFTIIDPTKPLPEAVNVRFDSGHIERVEVSSPWLPPTCDHCKEVGHSIKNC-----LT 127
Query: 159 EPLIDKKKKTIISQKPQPPKWLPRDNLDGIGSSKAFEAPVGNNNAESIPTTHKVATKLPV 218
P+ K+ + PK + + + G KV T P
Sbjct: 128 APITCSLCKSTAHKLEDCPK-----------AKRQTDTKDGERKKRKTRKKTKV-TDPPA 175
Query: 219 QTDLTESADAQHSDSANVSAAIHHSDDEDNLSAAAHHSDDVSPQNVTKDPLDFEFIPVMP 278
T+ T + ++ + + + D+ N++A DD +++T++ + + ++
Sbjct: 176 DTEQTAAVIVENEGNIIIDIGL----DKINITA-----DDSRERSITEEVQNSDSDALVS 226
Query: 279 DDINKATLPSTSIHVLEETADTAEVPIEDTTAAAEPLFTDVMALEA----QTQTNRTDVE 334
LPS E +++ E + + + F + ++ + + Q + +E
Sbjct: 227 SGSEIEELPSAEESSQESSSEEEEGSSDSSNPEEDSGFEEALSKKTKKMQRKQVRKAALE 286
Query: 335 LIAARVASLQHHEIPISKNV 354
L R L+ + S+N+
Sbjct: 287 LRLQREKELKRSKSSKSRNL 306
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 57.8 bits (138), Expect = 8e-07
Identities = 32/102 (31%), Positives = 45/102 (43%)
Query: 432 LQPNLWALWSKDLSITVLFVSDQCIALQISCHQSIVYIAAVYASTYYLKRRQLWADLTHI 491
L L W K LSI V+ + + L + Y++ +Y +R LW L +
Sbjct: 426 LSGGLVVYWKKHLSIQVISHDVRLVDLYVEYKNFNFYLSCIYGHPIPSERHHLWEKLQRV 485
Query: 492 QEHFVGPWLFVGDFNAVLGAHEKCGRRPPPPLSCLDFLNWSN 533
H GPW+ GDFN +L +EK G R S +F N N
Sbjct: 486 SAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQNFTNMIN 527
Score = 50.8 bits (120), Expect = 1e-04
Identities = 33/146 (22%), Positives = 60/146 (40%), Gaps = 3/146 (2%)
Query: 7 NGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHELPQ 66
N F+F F S E + +V +G + +L + W+ D W+++ E+P
Sbjct: 75 NRRFQFIFPSEESLDTVLRRGPWSFADRMLVVERWTPDMDPLVLN--FIPFWIQVREIPL 132
Query: 67 EYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHI-FNEVMIERTGFSF 125
++ + IA ++G +D + R+ + D++ + F G +
Sbjct: 133 QFLNLEVIDNIAGSLGERKAVDFDPFTTTRVEFVRIQIKWDVNHPLRFQRNYQFSLGVNT 192
Query: 126 SIEITYERLPAFCTHCGNIGHHNSSC 151
+ YERL FC CG + H SC
Sbjct: 193 VLSFYYERLRGFCDVCGRMTHDAGSC 218
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 57.8 bits (138), Expect = 8e-07
Identities = 65/291 (22%), Positives = 115/291 (39%), Gaps = 27/291 (9%)
Query: 236 VSAAIHHSDDEDNLSAAAHHSD----DVSPQNVTKDPLDFEFIPVMPDDINKATLPSTSI 291
+S + SDD+D HH D D+S + + IP + ++K
Sbjct: 30 LSNPVETSDDDDADDDDDHHDDNKEHDISDTDTLQTVDPATLIPGLQSSLSKGNQRQ--- 86
Query: 292 HVLEETADTAEVP--IEDTTAAAEPLFTDVMALEAQTQTNRTDVELIAARVASLQHHEIP 349
+++ +D VP IEDT E L L A+ +TD + + +
Sbjct: 87 --VQKLSDGYSVPCVIEDTDLTVERL----RYLHAKLAGVKTDAPVEKDLL-------LE 133
Query: 350 ISKNVQHDLDLW----ARIREYDQRMAEEGFTQVLSKKQQKDAKKASLRKGFVQHPRKGY 405
S N Q++ R+ E Q++ VLS+ +K K S + H
Sbjct: 134 SSDNAQNEFVFMKRKRVRLEEMYQQVEAADEMGVLSQLCKKGRKTESAGSCSI-HDGLET 192
Query: 406 SSSLILMNILFWNIRVTKYFLNVRDNLQPNLWALWSKDLSITVLFVSDQCIALQISCHQS 465
+ + L +++ + L +W ++S++++ ++ I ++ +
Sbjct: 193 LNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALMWKNNVSLSLISQDERLIDSHVTFNNK 252
Query: 466 IVYIAAVYASTYYLKRRQLWADLTHIQEHFVGPWLFVGDFNAVLGAHEKCG 516
Y++ VY +R QLW L HI ++ WL VGDFN +L EK G
Sbjct: 253 SFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIG 303
>UniRef100_O80793 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 530
Score = 57.0 bits (136), Expect = 1e-06
Identities = 56/250 (22%), Positives = 101/250 (40%), Gaps = 27/250 (10%)
Query: 26 KGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHELPQEYWMDRTLKEIASAIGTPL 85
+G ++ L+ + WS T + VWV L +P + + IAS +G P+
Sbjct: 210 RGLWHVDDCLMFVAPWSTVNTFDLPEISTIPVWVTLKNIPNRLYSILGISHIASGLGAPM 269
Query: 86 LIDSATQNRVFGHYARVLVDMDLSKHIFNEVM-IERTGFSFSIEITYERLPAFCTHCGNI 144
+ A +LV+++LSK + +++ G + + Y +PA C CG +
Sbjct: 270 ATYKPRLDPSLMSEANILVEVELSKAFPPRIAAVDKKGNISMVNVEYAWIPAKCGKCGQL 329
Query: 145 GHHNSSCRWLHPVKEPLIDKKKKTIISQKPQPPKWLPRDNLDGIGSSKAFEAPVGNNNAE 204
GH S C +K L +K I+S++ P A V +A
Sbjct: 330 GHKASRC-----MKPHLAHEKVTEIVSEEIITP------------------AIVSLASAT 366
Query: 205 SIPTTHKVATKLPVQTDLTESADAQHSDSANVSAAIHHSDDEDNLSAAAHHSDDVSPQNV 264
++ + + TK P+ +T S Q ++ A D+DN + A +V + V
Sbjct: 367 NLVSPITLQTKTPIDVPITNS-KIQIDTVFDIEAG--PIQDKDNCTGVADCFTNVEAEVV 423
Query: 265 TKDPLDFEFI 274
T++ E +
Sbjct: 424 TEETCTVESV 433
>UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1449
Score = 57.0 bits (136), Expect = 1e-06
Identities = 38/135 (28%), Positives = 68/135 (50%), Gaps = 5/135 (3%)
Query: 21 RSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHA-QVWVRLHELPQEYWMDRTLKEIAS 79
R ++G N+ + + +WS F TQ + Q+W+ L ++P + D+ L+ +AS
Sbjct: 173 RRALNRGMWNIMDLPMVVSKWSP-FAEETQPAMKSIQLWITLTDVPPSLFTDKGLEFLAS 231
Query: 80 AIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMI---ERTGFSFSIEITYERLPA 136
A+G P+ + T++ A++LV+ DL+K + E + E + I+ +Y LP
Sbjct: 232 AVGKPIRLHPKTESCSSFDEAQLLVEADLTKELPKEFVFTGEEEGELAAVIKYSYPWLPP 291
Query: 137 FCTHCGNIGHHNSSC 151
C+ C GH SC
Sbjct: 292 RCSCCDKWGHLRDSC 306
Score = 52.0 bits (123), Expect = 5e-05
Identities = 28/92 (30%), Positives = 48/92 (51%), Gaps = 5/92 (5%)
Query: 427 NVRDNLQPNLWALWSKDLSITVLFVSDQCI--ALQISCHQSIVYIAAVYASTYYLKRRQL 484
N N + LW +W +++ T + SDQ I ++++ + + + VYAS + +R+ L
Sbjct: 475 NYEFNRRGRLWVVWRENVRFTPFYKSDQLITCSVKLESQEEEFFYSFVYASNFAEERKIL 534
Query: 485 WADL-THIQEHFV--GPWLFVGDFNAVLGAHE 513
W DL H+ + PW+ GDFN +L E
Sbjct: 535 WNDLRDHMDSPIIRDKPWIIFGDFNEILDMDE 566
>UniRef100_O64612 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 1412
Score = 55.8 bits (133), Expect = 3e-06
Identities = 30/92 (32%), Positives = 46/92 (49%), Gaps = 5/92 (5%)
Query: 427 NVRDNLQPNLWALWSKDLSITVLFVSDQCIALQISC-HQSIVYIAA-VYASTYYLKRRQL 484
N N +W +WS + + V+F S Q I + H + +I + +YAS + +R++L
Sbjct: 362 NYEFNRLGRIWVVWSSSVQLQVIFKSSQMIVCLVRVEHYDVEFICSFIYASNFVEERKKL 421
Query: 485 WADLTHIQEHFV---GPWLFVGDFNAVLGAHE 513
W DL ++Q PWL GDFN L E
Sbjct: 422 WQDLHNLQNSVAFRNKPWLLFGDFNETLKMEE 453
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 55.5 bits (132), Expect = 4e-06
Identities = 24/78 (30%), Positives = 42/78 (53%)
Query: 439 LWSKDLSITVLFVSDQCIALQISCHQSIVYIAAVYASTYYLKRRQLWADLTHIQEHFVGP 498
+W ++S++++ ++ I ++ + Y++ VY +R QLW L HI ++
Sbjct: 452 MWKNNVSLSLISQDERLIDSHVTFNNKSFYLSCVYGHPTQSERHQLWQTLEHISDNRNAE 511
Query: 499 WLFVGDFNAVLGAHEKCG 516
WL VGDFN +L EK G
Sbjct: 512 WLLVGDFNEILSNAEKIG 529
Score = 42.4 bits (98), Expect = 0.036
Identities = 33/158 (20%), Positives = 66/158 (40%), Gaps = 4/158 (2%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
+ G +F F S E M V +G + +L + W + + + WV++ +
Sbjct: 73 IDKGRVQFKFQSEEAMNLVLRRGPWSFNDWMLSIHRWYPNLSEAEMKII--PFWVQITGI 130
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHI-FNEVMIERTGF 123
P + + + + + +G +D + G + RV ++ +L + F G
Sbjct: 131 PLLFLTNAMARCVGNRLGHVADVDFDENSNHTG-FVRVRINWNLDDPLRFQRNFQFADGE 189
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPVKEPL 161
+ I+ +ERL FCT CG++ H C ++P+
Sbjct: 190 NTVIKFRFERLRNFCTKCGSLKHDIKVCTLSFDNEDPV 227
>UniRef100_Q8S9E3 ORF285 [Silene latifolia]
Length = 285
Score = 55.5 bits (132), Expect = 4e-06
Identities = 31/107 (28%), Positives = 54/107 (49%), Gaps = 2/107 (1%)
Query: 41 WSKDFTARTQRQTHAQVWVRLHELPQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYA 100
W+ + + T VWVRL LP ++W + IA +G + D AT++R Y
Sbjct: 178 WTPNESLTKAEITVVPVWVRLLNLPLKFW-GNCIPWIAGLLGDYVRCDGATEDRTRLAYT 236
Query: 101 RVLVDMDLSKHIFNEV-MIERTGFSFSIEITYERLPAFCTHCGNIGH 146
RVL+D+ K +V ++ +G ++++ ++ P CT G +GH
Sbjct: 237 RVLIDVTFGKSAPKDVKFLDESGELITLKVEFKWNPILCTDFGGVGH 283
>UniRef100_Q9SJ33 Putative Ta11-like non-LTR retroelement protein [Arabidopsis
thaliana]
Length = 233
Score = 55.1 bits (131), Expect = 5e-06
Identities = 38/154 (24%), Positives = 60/154 (38%), Gaps = 3/154 (1%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
L F+F F ED++ V G + + W +D +W+RL +
Sbjct: 63 LSRDRFQFIFKYEEDLQEVLKTGVWTQDDWGVVMERWVEDLPPNYLM--FLLIWLRLRNI 120
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIE-RTGF 123
P ++ T+K IA +G + Y R+ + D+SK + N I+ G
Sbjct: 121 PVNHYTQATIKSIAKCVGQVIEFPFDENEAQSKDYVRIRILFDVSKGLRNSKEIQLPNGS 180
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPV 157
I I YER+ C C + H C +L V
Sbjct: 181 MVKIGIDYERIRKRCFQCQRLTHEKPRCPYLPSV 214
>UniRef100_Q84X22 Hypothetical protein At2g05290/F5G3.19 [Arabidopsis thaliana]
Length = 305
Score = 55.1 bits (131), Expect = 5e-06
Identities = 38/154 (24%), Positives = 60/154 (38%), Gaps = 3/154 (1%)
Query: 5 LGNGYFEFYFSSYEDMRSVWSKGTQNLKPGLLRLFEWSKDFTARTQRQTHAQVWVRLHEL 64
L F+F F ED++ V G + + W +D +W+RL +
Sbjct: 74 LSRDRFQFIFKYEEDLQEVLKTGVWTQDDWGVVMERWVEDLPPNYLM--FLLIWLRLRNI 131
Query: 65 PQEYWMDRTLKEIASAIGTPLLIDSATQNRVFGHYARVLVDMDLSKHIFNEVMIE-RTGF 123
P ++ T+K IA +G + Y R+ + D+SK + N I+ G
Sbjct: 132 PVNHYTQATIKSIAKCVGQVIEFPFDENEAQSKDYVRIRILFDVSKGLRNSKEIQLPNGS 191
Query: 124 SFSIEITYERLPAFCTHCGNIGHHNSSCRWLHPV 157
I I YER+ C C + H C +L V
Sbjct: 192 MVKIGIDYERIRKRCFQCQRLTHEKPRCPYLPSV 225
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 967,540,620
Number of Sequences: 2790947
Number of extensions: 40413801
Number of successful extensions: 106538
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 106332
Number of HSP's gapped (non-prelim): 252
length of query: 559
length of database: 848,049,833
effective HSP length: 133
effective length of query: 426
effective length of database: 476,853,882
effective search space: 203139753732
effective search space used: 203139753732
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135464.8


