

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.6 + phase: 0
(255 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
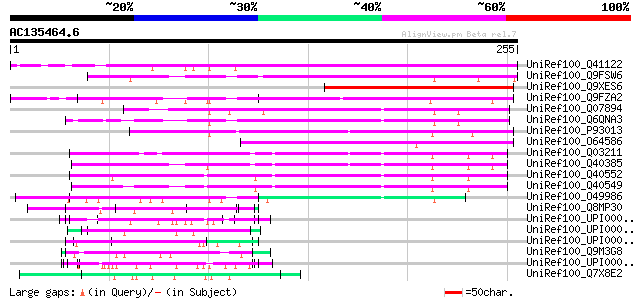
UniRef100_Q41122 Proline-rich protein precursor [Phaseolus vulga... 226 3e-58
UniRef100_Q9FSW6 Arabinogalactan protein precursor [Daucus carota] 152 6e-36
UniRef100_Q9XES6 Putative hybrid proline-rich protein PRP1 [Trif... 124 2e-27
UniRef100_Q9FZA2 F3H9.6 protein [Arabidopsis thaliana] 117 2e-25
UniRef100_Q07894 Proline-rich protein [Nicotiana alata] 114 2e-24
UniRef100_Q6QNA3 Proline-rich protein 1 [Capsicum annuum] 107 4e-22
UniRef100_P93013 Putative proline-rich protein [Arabidopsis thal... 101 2e-20
UniRef100_O64586 Putative proline-rich glycoprotein [Arabidopsis... 91 2e-17
UniRef100_Q03211 Pistil-specific extensin-like protein precursor... 87 5e-16
UniRef100_Q40385 Pistil extensin-like protein [Nicotiana alata] 85 2e-15
UniRef100_Q40552 Pistil extensin like protein [Nicotiana tabacum] 84 5e-15
UniRef100_Q40549 Pistil extensin like protein, partial CDS [Nico... 83 8e-15
UniRef100_O49986 120 kDa style glycoprotein [Nicotiana alata] 72 2e-11
UniRef100_Q8MP30 Hypothetical protein [Dictyostelium discoideum] 71 2e-11
UniRef100_UPI000042D37F UPI000042D37F UniRef100 entry 70 4e-11
UniRef100_UPI000004DF56 UPI000004DF56 UniRef100 entry 69 2e-10
UniRef100_UPI00002968D1 UPI00002968D1 UniRef100 entry 68 3e-10
UniRef100_Q9M3G8 Putative proline-rich protein [Arabidopsis thal... 67 3e-10
UniRef100_UPI0000456BB2 UPI0000456BB2 UniRef100 entry 66 7e-10
UniRef100_Q7X8E2 OJ990528_30.2 protein [Oryza sativa] 65 2e-09
>UniRef100_Q41122 Proline-rich protein precursor [Phaseolus vulgaris]
Length = 297
Score = 226 bits (577), Expect = 3e-58
Identities = 138/307 (44%), Positives = 165/307 (52%), Gaps = 64/307 (20%)
Query: 1 MAKIILALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHA-PHHH 59
MAK +AL+ L+L +T VF+EEL TL P+ PLHPPT P A P H PHHH
Sbjct: 1 MAKA-MALMLLMLLVT---VFSEELHTL---PSAPLHPPTYPP-----ANAPHHPHPHHH 48
Query: 60 HHHSPSHAPLP------------PPHPAKPLTHHHHQHQH-------------------- 87
HHH P+ AP P PP PA P THHHH H
Sbjct: 49 HHHPPAPAPAPLHPPSPPSHPHYPPAPAHPPTHHHHHHPSAPVHPPLNPPLVPVHPPLKP 108
Query: 88 ----HSPA--PSPYHVPT----PLQRPAKPPTLHH---------HQHPPAHAPTHMPRVS 128
H P P P H P P+ P KPP H H P H P +
Sbjct: 109 PVPIHPPLNPPVPVHPPVKPPVPVHPPVKPPVPVHPPVKPPVPVHPPVPVHPPVPVHPFR 168
Query: 129 RSSIAVEGVVYVKSCHHAGFDTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIG 188
RS +AV+GVVYVKSC +AG DTL GAT L GAVVK QC+N KYK V +K++K GY Y
Sbjct: 169 RSFVAVQGVVYVKSCKYAGVDTLLGATPLLGAVVKVQCNNTKYKLVETSKSDKNGYFYFE 228
Query: 189 SSKNISSYASGHCNVVLESAPNGLKPSNLHGGLTGAHPKSVKRIVSKGVSLIRYTVDPLA 248
+ K+I++Y + CNVVL AP GLKPSNLH G+TGA + K +SK + + YTV PLA
Sbjct: 229 APKSITTYGAHKCNVVLVGAPYGLKPSNLHSGVTGAVLRPEKPFLSKKLPFVLYTVGPLA 288
Query: 249 FEPKCNH 255
FEPKC +
Sbjct: 289 FEPKCKN 295
>UniRef100_Q9FSW6 Arabinogalactan protein precursor [Daucus carota]
Length = 242
Score = 152 bits (385), Expect = 6e-36
Identities = 97/231 (41%), Positives = 125/231 (53%), Gaps = 30/231 (12%)
Query: 40 TKSPVHKPLAKPPTHAPHHH----HHHSPSHAPLPPPHP-AKPLTHHHHQHQHHSPAPSP 94
T + P A P APHHH HHH HAP P P P KP TH +PAP+P
Sbjct: 27 TATIAQPPTALAPAPAPHHHKPGGHHHHHKHAPTPAPAPLTKPPTH--------APAPAP 78
Query: 95 YHVPT-PLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTLKG 153
P P +P KPP P+HAPT +P +R +AV+GVVY K C++ G +TL G
Sbjct: 79 VKPPVKPPVQPVKPPV-----KAPSHAPTPLP--ARKLVAVQGVVYCKPCNYTGVETLLG 131
Query: 154 ATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESAPNGL- 212
AT L GAVVK QC+N KY V++ KT+K GY + + K I++Y C V + S+P
Sbjct: 132 ATPLLGAVVKLQCNNTKYPLVVQGKTDKNGYFSLNAPKTITTYGVHKCRVFVVSSPEKKC 191
Query: 213 -KPSNLHGGLTGA-HPKSVKRIVS--KGVSLIRYTVDPLAFEPK----CNH 255
KP+NL G+ GA KS K VS + ++V P AFEP C+H
Sbjct: 192 DKPTNLRYGVKGAILEKSTKPPVSTKTPATFEMFSVGPFAFEPSTKKPCSH 242
>UniRef100_Q9XES6 Putative hybrid proline-rich protein PRP1 [Trifolium subterraneum]
Length = 97
Score = 124 bits (311), Expect = 2e-27
Identities = 62/95 (65%), Positives = 71/95 (74%)
Query: 159 GAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESAPNGLKPSNLH 218
GAVVK QC+N KYK V +T+K GY +I K+I+SYA+ CNVVL SAPNGLKPSNLH
Sbjct: 1 GAVVKLQCNNTKYKLVQTHETDKNGYFFIEGPKSITSYAAHKCNVVLVSAPNGLKPSNLH 60
Query: 219 GGLTGAHPKSVKRIVSKGVSLIRYTVDPLAFEPKC 253
GGLTGA + K VSKG+ I YTV PLAFEPKC
Sbjct: 61 GGLTGAGLRPGKPYVSKGLPFIVYTVGPLAFEPKC 95
>UniRef100_Q9FZA2 F3H9.6 protein [Arabidopsis thaliana]
Length = 359
Score = 117 bits (294), Expect = 2e-25
Identities = 80/233 (34%), Positives = 112/233 (47%), Gaps = 32/233 (13%)
Query: 35 PLHPPTKSPVH---KPLAKPPTHAPHHHHHHSPSHAPLPPP--HPAKPLTHHHHQHQHHS 89
P++PPTK+PV KP KPP + P P+ P+ PP PAKP
Sbjct: 143 PVYPPTKAPVKPPTKPPVKPPVYPPTKAPVKPPTKPPVKPPVSPPAKP------------ 190
Query: 90 PAPSPYHVPT--PLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAG 147
P P + PT P++ P PPT PP P + P+ +RS +AV G VY KSC +A
Sbjct: 191 PVKPPVYPPTKAPVKPPVSPPT-----KPPVTPPVYPPKFNRSLVAVRGTVYCKSCKYAA 245
Query: 148 FDTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLES 207
F+TL GA + GA VK C +K + T+K GY + + K ++++ C V L
Sbjct: 246 FNTLLGAKPIEGATVKLVC-KSKKNITAETTTDKNGYFLLLAPKTVTNFGFRGCRVYLVK 304
Query: 208 APN--GLKPSNLHGGLTGAHPKSVKRIVSKGVSLIR-----YTVDPLAFEPKC 253
+ + K S L GG GA K K++ V + + + V P AF P C
Sbjct: 305 SKDYKCSKVSKLFGGDVGAELKPEKKLGKSTVVVNKLVYGLFNVGPFAFNPSC 357
Score = 59.7 bits (143), Expect = 7e-08
Identities = 46/130 (35%), Positives = 54/130 (41%), Gaps = 39/130 (30%)
Query: 1 MAKIILALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHH 60
+ K +L + L TS V F EE+ +HK TP P +P H H H
Sbjct: 4 IGKSVLVSLVALWCFTSSV-FTEEV---NHKTQTPSLAPAPAPYH------------HGH 47
Query: 61 HHSPSHAPLPPPHPAKPLTHHHHQHQH---HSPAPSPYHVP--TPLQRPAKPPTLHHHQH 115
HH PHP HHHH H H H PA SP P P+ PAKPP
Sbjct: 48 HH---------PHPP----HHHHPHPHPHPHPPAKSPVKPPVKAPVSPPAKPPV-----K 89
Query: 116 PPAHAPTHMP 125
PP + PT P
Sbjct: 90 PPVYPPTKAP 99
>UniRef100_Q07894 Proline-rich protein [Nicotiana alata]
Length = 255
Score = 114 bits (285), Expect = 2e-24
Identities = 80/222 (36%), Positives = 105/222 (47%), Gaps = 38/222 (17%)
Query: 58 HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPT------PLQRPAKPPT-- 109
HH H SP+ AP P H H H HSPAPSP PT P++ P KPPT
Sbjct: 36 HHDHLSPAQAPKP---------HKGHHHPKHSPAPSPTKPPTYSPSKPPVKPPVKPPTKP 86
Query: 110 -LHHHQHPPAHAPTHMPR---------VSRSSIAVEGVVYVKSCHHAGFDTLKGATLLFG 159
+ PPA AP P ++R +AV G+VY K C G TL A+ L G
Sbjct: 87 PTYSPSKPPAKAPVKPPTPSPYPAPAPITRKPVAVRGLVYCKPCKFRGVKTLNQASPLLG 146
Query: 160 AVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESAPNGL--KPSNL 217
AVV+ C+N K V + KT+K G+ +I K +SS A C V L S+ N P++
Sbjct: 147 AVVELVCNNTKKTLVEQGKTDKNGFFWI-MPKFLSSAAYHKCKVFLVSSNNTYCDVPTDY 205
Query: 218 HGGLTGA--------HPKSVKRIVSKGVSLIRYTVDPLAFEP 251
+GG +GA P + + K + +TV P FEP
Sbjct: 206 NGGKSGALLKYTPLPKPPAASSLPVKLPTFDVFTVGPFGFEP 247
>UniRef100_Q6QNA3 Proline-rich protein 1 [Capsicum annuum]
Length = 260
Score = 107 bits (266), Expect = 4e-22
Identities = 84/231 (36%), Positives = 106/231 (45%), Gaps = 35/231 (15%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH P P + P SP+ P PP +P SP A P P PAKP
Sbjct: 49 HHPP--PKNSPAPSPIDTP-TPPPAKSP------SPPPAKPPTPPPAKP----------- 88
Query: 89 SPAPSPYHVPT--PLQRPAKPPTLHHHQHPPAHAPTHMPRV-SRSSIAVEGVVYVKSCHH 145
P+P P PT P + P+ PP PP PT P SR + V G+VY K C +
Sbjct: 89 -PSPPPSKPPTKPPAKSPSPPPA-----KPPTKPPTPSPYYPSRKPVIVRGLVYCKPCKY 142
Query: 146 AGFDTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVL 205
G +TL A L GAVVK C N+K V + KT+K GY +I K ++S A C V L
Sbjct: 143 RGIETLYRAKPLEGAVVKLVCKNSKKTLVEQGKTDKNGYFWI-MPKLLTSGAYHKCKVFL 201
Query: 206 ESAPNGL--KPSNLHGGLTGA---HPKSVKRIVSKGVSLIRYTVDPLAFEP 251
S+ N P+N +GG +GA + K S +TV P FEP
Sbjct: 202 VSSNNSYCNVPTNFNGGKSGALLKYTPPSKPTPSAITKFDVFTVGPFGFEP 252
>UniRef100_P93013 Putative proline-rich protein [Arabidopsis thaliana]
Length = 239
Score = 101 bits (251), Expect = 2e-20
Identities = 68/203 (33%), Positives = 102/203 (49%), Gaps = 12/203 (5%)
Query: 61 HHSPSHAPLPPPH-PAKPLTHHHHQHQHHSPAPSPYHVPT--PLQRPAKPPTLHHHQHPP 117
H P H PLPP P P + + PA +P +PT P + P K PTL + PP
Sbjct: 37 HSLPQHLPLPPIKLPTLPPAKAPIKLPAYPPAKAPIKLPTLPPAKAPIKLPTLPPIK-PP 95
Query: 118 AHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTLKGATLLFGAVVKFQCHNAKYKFVLKA 177
P + P+ +++ +AV GVVY K+C +AG + ++GA + AVV+ C N K + +
Sbjct: 96 VLPPVYPPKYNKTLVAVRGVVYCKACKYAGVNNVQGAKPVKDAVVRLVCKNKK-NSISET 154
Query: 178 KTNKEGYIYIGSSKNISSYASGHCNVVLESAPNG--LKPSNLHGGLTGAHPKSVKR---- 231
KT+K GY + + K +++Y C L +P+ K S+LH G G+ K V +
Sbjct: 155 KTDKNGYFMLLAPKTVTNYDIKGCRAFLVKSPDTKCSKVSSLHDGGKGSVLKPVLKPGFS 214
Query: 232 -IVSKGVSLIRYTVDPLAFEPKC 253
+ + Y V P AFEP C
Sbjct: 215 STIMRWFKYSVYNVGPFAFEPTC 237
>UniRef100_O64586 Putative proline-rich glycoprotein [Arabidopsis thaliana]
Length = 175
Score = 91.3 bits (225), Expect = 2e-17
Identities = 52/143 (36%), Positives = 76/143 (52%), Gaps = 6/143 (4%)
Query: 117 PAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTLKGATLLFGAVVKFQCHNAKYKFVLK 176
P P+ ++SR +AVEG+VY KSC ++G DTL A+ L GA VK C+N K ++
Sbjct: 31 PMTPPSSPAKMSRRLVAVEGMVYCKSCKYSGVDTLLEASPLQGATVKLACNNTKRGVTME 90
Query: 177 AKTNKEGYIYIGSSKNISSYASGHCNV------VLESAPNGLKPSNLHGGLTGAHPKSVK 230
KT+K GY ++ + K +++YA C + PS L+ G+TGA K K
Sbjct: 91 TKTDKNGYFFMLAPKKLTTYAFHTCRAWPTNPGPTTATMTCTVPSKLNNGITGAMLKPSK 150
Query: 231 RIVSKGVSLIRYTVDPLAFEPKC 253
I + ++V P AFEP C
Sbjct: 151 TINIGEHDYVLFSVGPFAFEPAC 173
>UniRef100_Q03211 Pistil-specific extensin-like protein precursor [Nicotiana tabacum]
Length = 426
Score = 86.7 bits (213), Expect = 5e-16
Identities = 76/231 (32%), Positives = 105/231 (44%), Gaps = 20/231 (8%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
K P PP K+P P +PPT P S PL PP P P+ + SP
Sbjct: 195 KQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKS--PLLPPPP--PVAYPPVMTPSPSP 250
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDT 150
A P + PA PP + PP P +P + + I V G+VY KSC+ G T
Sbjct: 251 AAEPPIIAPFPSPPANPPLIPRRPAPPVVKP--LPPLGKPPI-VSGLVYCKSCNSYGVPT 307
Query: 151 LKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESAPN 210
L A+LL GAVVK C+ K V A T+ +G I K++++ G C V L +PN
Sbjct: 308 LLNASLLQGAVVKLICYGKK-TMVQWATTDNKGEFRI-MPKSLTTADVGKCKVYLVKSPN 365
Query: 211 G--LKPSNLHGGLTGAHPKSV---KRIVSKGVSLIR------YTVDPLAFE 250
P+N +GG +G K + K+ ++ V ++ Y V P FE
Sbjct: 366 PNCNVPTNFNGGKSGGLLKPLLPPKQPITPAVVPVQPPMSDLYGVGPFIFE 416
Score = 45.1 bits (105), Expect = 0.002
Identities = 30/101 (29%), Positives = 39/101 (37%), Gaps = 2/101 (1%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P P + SP+ KP PP+ S P PPP AK + +P+
Sbjct: 133 PVNQPKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPP--AKQPSPPPPPPPVKAPS 190
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSI 132
PSP P P P K P+ PP P PR +S +
Sbjct: 191 PSPAKQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSPL 231
Score = 37.0 bits (84), Expect = 0.48
Identities = 21/71 (29%), Positives = 25/71 (34%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
PP + P P PPP P KP Q P P+ P P P K P+
Sbjct: 132 PPVNQPKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPPAKQPSPPPPPPPVKAPSP 191
Query: 111 HHHQHPPAHAP 121
+ PP P
Sbjct: 192 SPAKQPPPPPP 202
>UniRef100_Q40385 Pistil extensin-like protein [Nicotiana alata]
Length = 430
Score = 85.1 bits (209), Expect = 2e-15
Identities = 76/234 (32%), Positives = 105/234 (44%), Gaps = 28/234 (11%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P PP ++P P +PPT P S P PPP A P +P+
Sbjct: 200 PAKQSPPPPRAPSPSPATQPPTKQPPPPPRAKKSPPPPPPPPVAYPPVM--------APS 251
Query: 92 PSPYHVP---TPLQRP-AKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAG 147
PSP P P P A PP + PP P +P + + I V G+VY KSC+ G
Sbjct: 252 PSPAAEPPIVAPFPSPTANPPHVPRRPAPPVVKP--LPPLGKPPI-VNGLVYCKSCNSYG 308
Query: 148 FDTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLES 207
TL A+LL GAVVK C+ K V A T+ +G I K++++ G C V L
Sbjct: 309 VPTLLNASLLQGAVVKLICYGKK-AMVQWATTDNKGEFRI-MPKSLTTTDVGKCKVYLVK 366
Query: 208 APNG--LKPSNLHGGLTGAHPKSV---KRIVSKGVSLIR------YTVDPLAFE 250
+PN P+N +GG +G K + K+ ++ V ++ Y V P FE
Sbjct: 367 SPNPNCNVPTNFNGGKSGGLLKPLLPPKQPITPAVVPVQPPMSDLYGVGPFIFE 420
Score = 37.0 bits (84), Expect = 0.48
Identities = 24/75 (32%), Positives = 27/75 (36%), Gaps = 1/75 (1%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
PP + P P P PPP P KP Q PA P P P A P+
Sbjct: 142 PPVNRPKPSAPSPPVKPPPPPPSPCKPSPPDQSAKQ-PPPAKQPPPPPPPPPVKAPSPSP 200
Query: 111 HHHQHPPAHAPTHMP 125
PP AP+ P
Sbjct: 201 AKQSPPPPRAPSPSP 215
>UniRef100_Q40552 Pistil extensin like protein [Nicotiana tabacum]
Length = 393
Score = 83.6 bits (205), Expect = 5e-15
Identities = 77/232 (33%), Positives = 101/232 (43%), Gaps = 15/232 (6%)
Query: 31 KPTTPLHPPTKSPVHKPLAK--PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
K P PP KSP P + PP AP P PPP K
Sbjct: 155 KQPPPPPPPVKSPSPSPAKQSPPPPRAPSPSPATQPPIKQPPPPSAKKSPPPPVAYPPVM 214
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPT---HMPRVSRSSIAVEGVVYVKSCHH 145
+P+PSP P P+ P PT + P AP +P + + I V G+VY KSC+
Sbjct: 215 APSPSPAAEP-PIIAPFPSPTANLPLIPRRPAPPVVKPLPPLGKPPI-VNGLVYCKSCNS 272
Query: 146 AGFDTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVL 205
GF TL +LL GAVVK C+N K V A T+ +G I K+++ G C + L
Sbjct: 273 YGFPTLLNTSLLPGAVVKLVCYNGKKTMVQSATTDNKGEFRI-IPKSLTRADVGKCKLYL 331
Query: 206 ESAPNG--LKPSNLHGGLTGAHPKSV----KRIVSKGVSLI-RYTVDPLAFE 250
+PN P+N +GG +G K + + I V L Y V P FE
Sbjct: 332 VKSPNPNCNVPTNFNGGKSGGLLKPLLPPKQPITPAAVPLSDLYGVGPFIFE 383
Score = 38.1 bits (87), Expect = 0.22
Identities = 22/81 (27%), Positives = 28/81 (34%)
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
PP + P P P PPP P KP Q P PS P P + + P
Sbjct: 95 PPVNRPKPSSPSPPVKPPPPPPSPCKPSPPDQSTKQPPPPPPSKQPPPPPPVKASSPSPA 154
Query: 111 HHHQHPPAHAPTHMPRVSRSS 131
PP + P ++ S
Sbjct: 155 KQPPPPPPPVKSPSPSPAKQS 175
>UniRef100_Q40549 Pistil extensin like protein, partial CDS [Nicotiana tabacum]
Length = 264
Score = 82.8 bits (203), Expect = 8e-15
Identities = 75/229 (32%), Positives = 101/229 (43%), Gaps = 26/229 (11%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P PP ++P P +PP P PS PPP A P +P+
Sbjct: 42 PAKQSPPPPRAPSPSPATQPPIKQPP-----PPSAKKSPPPPVAYPPVM--------APS 88
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPT---HMPRVSRSSIAVEGVVYVKSCHHAGF 148
PSP P P+ P PT + P AP +P + + I V G+VY KSC+ GF
Sbjct: 89 PSPAAEP-PIIAPFPSPTANLPLIPRRPAPPVVKPLPPLGKPPI-VNGLVYCKSCNSYGF 146
Query: 149 DTLKGATLLFGAVVKFQCHNAKYKFVLKAKTNKEGYIYIGSSKNISSYASGHCNVVLESA 208
TL +LL GAVVK C+N K V A T+ +G I K+++ G C + L +
Sbjct: 147 PTLLNTSLLPGAVVKLVCYNGKKTMVQSATTDNKGEFRI-IPKSLTRADVGKCKLYLVKS 205
Query: 209 PNG--LKPSNLHGGLTGAHPKSV----KRIVSKGVSLI-RYTVDPLAFE 250
PN P+N +GG +G K + + I V L Y V P FE
Sbjct: 206 PNPNCNVPTNFNGGKSGGLLKPLLPPKQPITPAAVPLSDLYGVGPFIFE 254
>UniRef100_O49986 120 kDa style glycoprotein [Nicotiana alata]
Length = 461
Score = 71.6 bits (174), Expect = 2e-11
Identities = 70/231 (30%), Positives = 92/231 (39%), Gaps = 33/231 (14%)
Query: 31 KPTTPLHPPTKSP----VHKPLAKPPTHAPHHHHHHSPSHAPL---PPPHPA-------- 75
KP P P + P P+ KPP A +P P+ PPP A
Sbjct: 193 KPPPPAQLPIRQPPPPATQLPIRKPPPPAYTQPPFKTPPPPPIRSSPPPQDAYDEPPVVE 252
Query: 76 -KPLTHHHHQHQHHSPAPSPYHVPT-----PLQRPA----KPPTLHHHQHPPAHAPTHMP 125
P H P PS Y P P +P PPTL PP P +P
Sbjct: 253 SPPAPPSDHDPPVVEPPPSDYEPPIIEPTPPTDQPPIIIPSPPTLAPPFIPPITPPVKLP 312
Query: 126 RVS----RSSIAVEGVVYVKSCHHAGFDTLKGATLLFGAVVKFQCH-NAKYKFVLKAKTN 180
S + + G V+ KSC+ G TL A+ L GAVVK CH NA+ V A T+
Sbjct: 313 PPSIHPAGKPLIIVGHVHCKSCNSRGLPTLYKASPLQGAVVKLVCHNNARKANVQTAMTD 372
Query: 181 KEGYIYIGSSKNISSYASGHCNVVLESAPNGL--KPSNLHGGLTGAHPKSV 229
K G I +++ C V L +P + P+N +GG +GA K +
Sbjct: 373 KNGEFVI-MPMSLTRADVHKCKVYLGKSPKPICNVPTNFNGGKSGALLKPI 422
Score = 56.2 bits (134), Expect = 8e-07
Identities = 42/141 (29%), Positives = 60/141 (41%), Gaps = 24/141 (17%)
Query: 4 IILALIFLLLQITSFVVFAEELETL------HHKPTTPLH----------PPTKSPVHKP 47
I++ L+ ++L S + F E +E HH + +H P + +PV+KP
Sbjct: 10 ILIQLLVVILSSFSELSFGERIERSLIDKGQHHPIFSTVHLFFGKSPKKSPSSPTPVNKP 69
Query: 48 LAKPPTHAPHHHHHHSP---SHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRP 104
PP +P SP + +P PPP + PL Q SP PSP P P
Sbjct: 70 SPSPPAKSPPPQVKSSPPPPAKSPPPPPAKSPPLPPPPVQPPKQSPPPSPAKQPPPPPPS 129
Query: 105 AKPPTLHHHQHPPAHAPTHMP 125
AKPP PP+ +P P
Sbjct: 130 AKPPV-----KPPSPSPAAQP 145
Score = 37.4 bits (85), Expect = 0.37
Identities = 29/94 (30%), Positives = 37/94 (38%), Gaps = 15/94 (15%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P P+ PP +SP P +PP P + P+ PP P+ Q
Sbjct: 104 PPPPVQPPKQSPPPSPAKQPPPPPPS-------AKPPVKPPSPSPAAQPPATQR------ 150
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
+P P P+QR A PP L PPA P P
Sbjct: 151 ATPPSQPPPMQR-APPPKL-PLPPPPAQLPIRQP 182
Score = 35.0 bits (79), Expect = 1.8
Identities = 26/102 (25%), Positives = 33/102 (31%), Gaps = 27/102 (26%)
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHH----------------HHHSPSHAPLPPPHPA 75
P+ PP P KP KPP+ +P P PLPPP
Sbjct: 117 PSPAKQPPPPPPSAKPPVKPPSPSPAAQPPATQRATPPSQPPPMQRAPPPKLPLPPPPAQ 176
Query: 76 KPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPP 117
P+ P P P+++P P L Q PP
Sbjct: 177 LPIRQ-----------PPPPATQLPIRKPPPPAQLPIRQPPP 207
>UniRef100_Q8MP30 Hypothetical protein [Dictyostelium discoideum]
Length = 233
Score = 71.2 bits (173), Expect = 2e-11
Identities = 32/66 (48%), Positives = 33/66 (49%), Gaps = 4/66 (6%)
Query: 54 HAPHHHHHHSPSHAPLPPPHP-AKPLTH---HHHQHQHHSPAPSPYHVPTPLQRPAKPPT 109
H HHHHHH P H P P PHP P H H H H H P P P+H P P P P
Sbjct: 145 HHHHHHHHHHPHHHPHPHPHPHPHPHLHPNPHPHPHPHPHPHPHPHHHPNPNPHPHPHPH 204
Query: 110 LHHHQH 115
HHH H
Sbjct: 205 PHHHHH 210
Score = 67.0 bits (162), Expect = 4e-10
Identities = 38/101 (37%), Positives = 39/101 (37%), Gaps = 5/101 (4%)
Query: 29 HHKPTTPLHPPTKSPV----HKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQ 84
HH P P H P H H HHHHHH H H HHHH
Sbjct: 94 HHHPHHPHHHPHHHHHPHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH 153
Query: 85 HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H HH P P P+ P P P P H H HP H P H P
Sbjct: 154 HPHHHPHPHPHPHPHPHLHPNPHPHPHPHPHPHPH-PHHHP 193
Score = 53.1 bits (126), Expect = 7e-06
Identities = 32/86 (37%), Positives = 35/86 (40%), Gaps = 14/86 (16%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH H P H P P H PH H H +P P P PHP H H HH
Sbjct: 141 HHHHHHHHHHHHHHPHHHPHPHPHPH-PHPHLHPNPHPHPHPHPHP--------HPHPHH 191
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQ 114
P P+P+ P P P HHHQ
Sbjct: 192 HPNPNPHPHPHP-----HPHHHHHHQ 212
Score = 52.8 bits (125), Expect = 9e-06
Identities = 30/95 (31%), Positives = 33/95 (34%), Gaps = 17/95 (17%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
H+ P HP +H H HHHHHH H P HP HHHH H HH
Sbjct: 58 HNPNNNPHHP---HHLHHHHHHHHHHHHHHHHHHHHHHHHHHPHHPHHHPHHHHHPHHHH 114
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+H HHH H H H
Sbjct: 115 HHHHHHHH--------------HHHHHHHHHHHHH 135
Score = 52.4 bits (124), Expect = 1e-05
Identities = 28/62 (45%), Positives = 29/62 (46%), Gaps = 3/62 (4%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHP-AKPLTHHHHQHQH 87
HH P HP +H P P H PH H H P H P P PHP P HHHH HQ
Sbjct: 156 HHHPHPHPHPHPHPHLH-PNPHPHPH-PHPHPHPHPHHHPNPNPHPHPHPHPHHHHHHQE 213
Query: 88 HS 89
S
Sbjct: 214 AS 215
Score = 50.4 bits (119), Expect = 4e-05
Identities = 31/116 (26%), Positives = 49/116 (41%), Gaps = 5/116 (4%)
Query: 10 FLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHH--HSPSHA 67
+ L+ + F++ + + K T + K +++ P H P+++ H H+P++
Sbjct: 5 YFLVLLVGFLLVLPSIVNPYRKGVTITNQQPKINIYQLDVNNP-HNPNNNPHNPHNPNNN 63
Query: 68 PLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
P P H HHHH H HH +H P P HHH HP H H
Sbjct: 64 PHHPHHLHHHHHHHHHHHHHHHHHHHHHHHHHHPHHPHHHP--HHHHHPHHHHHHH 117
>UniRef100_UPI000042D37F UPI000042D37F UniRef100 entry
Length = 983
Score = 70.5 bits (171), Expect = 4e-11
Identities = 41/100 (41%), Positives = 46/100 (46%), Gaps = 13/100 (13%)
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH--PAKPLTHHHHQHQH- 87
KP P HPP H+P +PP PH H P H P PPH P KP H+ H
Sbjct: 372 KPKKPEHPP-----HEPPHEPPHEPPHEPPHEPPHHPPHEPPHEPPHKPPHEPPHEPPHK 426
Query: 88 --HSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H P P H P P + P +PP HH H P H P H P
Sbjct: 427 PPHEPPHKPPHEP-PHKPPHEPP--HHPPHHPPHHPPHDP 463
Score = 60.8 bits (146), Expect = 3e-08
Identities = 35/100 (35%), Positives = 38/100 (38%), Gaps = 15/100 (15%)
Query: 26 ETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH--PAKPLTHHHH 83
E H P P H P P H P +PP PH H P P PPH P KP H
Sbjct: 382 EPPHEPPHEPPHEPPHEPPHHPPHEPPHEPPHKPPHEPPHEPPHKPPHEPPHKPPHEPPH 441
Query: 84 QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+ H P P+H P HH H P H P H
Sbjct: 442 KPPHEPPHHPPHHPP------------HHPPHDP-HDPNH 468
Score = 58.5 bits (140), Expect = 2e-07
Identities = 33/109 (30%), Positives = 47/109 (42%), Gaps = 9/109 (8%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH---HQH 85
H P P H P P HKP +PP PH HH P H P PPH HHH H+
Sbjct: 417 HEPPHEPPHKPPHEPPHKPPHEPPHKPPHEPPHHPPHHPPHHPPHDPHDPNHHHGKYHEE 476
Query: 86 QHHSPAPSPYHVPTPL---QRPAKPPTLHHHQHPPAHAPTHMPRVSRSS 131
+ P PL + P + P ++ + ++P+VS+++
Sbjct: 477 RRIFTEAKPADEIYPLTFNKNPNQAPVVYLASN---KLSVNLPKVSKNT 522
Score = 57.8 bits (138), Expect = 3e-07
Identities = 33/88 (37%), Positives = 36/88 (40%), Gaps = 11/88 (12%)
Query: 26 ETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
E H P P H P P HKP +PP PH H P P PPH KP H+
Sbjct: 394 EPPHEPPHHPPHEPPHEPPHKPPHEPPHEPPHKPPHEPPHKPPHEPPH--KP----PHEP 447
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHH 113
HH P P+H P P HHH
Sbjct: 448 PHHPPHHPPHHPPHDPHDPN-----HHH 470
Score = 57.0 bits (136), Expect = 5e-07
Identities = 33/88 (37%), Positives = 36/88 (40%), Gaps = 6/88 (6%)
Query: 26 ETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH--PAKPLTH--H 81
E HH P P H P P H+P +PP PH H P P PPH P P H H
Sbjct: 398 EPPHHPPHEPPHEPPHKPPHEPPHEPPHKPPHEPPHKPPHEPPHKPPHEPPHHPPHHPPH 457
Query: 82 HHQHQHHSP--APSPYHVPTPLQRPAKP 107
H H H P YH + AKP
Sbjct: 458 HPPHDPHDPNHHHGKYHEERRIFTEAKP 485
Score = 55.1 bits (131), Expect = 2e-06
Identities = 38/105 (36%), Positives = 41/105 (38%), Gaps = 11/105 (10%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH--PAKPLTHHHHQHQ 86
HHK H K K PP PH H P P PPH P +P H+
Sbjct: 359 HHKDDK--HHKDKDGKPKKPEHPPHEPPHEPPHEPPHEPPHEPPHHPPHEPPHEPPHKPP 416
Query: 87 H---HSPAPSPYHVP---TPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H H P P H P P + P KPP H HPP H P H P
Sbjct: 417 HEPPHEPPHKPPHEPPHKPPHEPPHKPPHEPPH-HPPHHPPHHPP 460
Score = 46.6 bits (109), Expect = 6e-04
Identities = 29/88 (32%), Positives = 30/88 (33%), Gaps = 9/88 (10%)
Query: 45 HKPLAKPPTHAPHHH----HHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVP-- 98
H K H HH HH P P HP H H P P H P
Sbjct: 346 HDDHHKDKHHKGKHHKDDKHHKDKDGKPKKPEHPPHEPPHEPPHEPPHEPPHEPPHHPPH 405
Query: 99 -TPLQRPAKPPTLHHHQHPPAHAPTHMP 125
P + P KPP H H P H P H P
Sbjct: 406 EPPHEPPHKPP--HEPPHEPPHKPPHEP 431
>UniRef100_UPI000004DF56 UPI000004DF56 UniRef100 entry
Length = 1130
Score = 68.6 bits (166), Expect = 2e-10
Identities = 35/89 (39%), Positives = 38/89 (42%), Gaps = 4/89 (4%)
Query: 37 HPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA----P 92
H P P P PP P H HH + P PPP P PL HHHH H P P
Sbjct: 278 HQPGIFPPPPPPPPPPPPHPDHLHHQYYGYPPPPPPPPPPPLHHHHHYPPSHHPGFGFPP 337
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
P P P P PP H+HQ P +P
Sbjct: 338 PPPGPPGPPGPPPVPPPPHYHQSAPPESP 366
Score = 58.9 bits (141), Expect = 1e-07
Identities = 32/89 (35%), Positives = 35/89 (38%), Gaps = 13/89 (14%)
Query: 40 TKSPVHKPLAKPPTHAP-------HHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAP 92
T S V P PP P H H P P PPP P P H H H +
Sbjct: 249 TTSSVQSPPPPPPPPQPPRGMGIQFHEASHQPGIFPPPPPPPPPPPPHPDHLHHQY---- 304
Query: 93 SPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
Y P P P PP HHH +PP+H P
Sbjct: 305 --YGYPPPPPPPPPPPLHHHHHYPPSHHP 331
Score = 47.0 bits (110), Expect = 5e-04
Identities = 31/108 (28%), Positives = 34/108 (30%), Gaps = 37/108 (34%)
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH------- 82
H P PP P P P PH+H PP P K H H
Sbjct: 329 HHPGFGFPPPPPGPPGPPGPPPVPPPPHYHQ-------SAPPESPPKEFKHRHPKYFHKQ 381
Query: 83 ----HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
H+H+HH P LHHH PP H H PR
Sbjct: 382 NNKGHRHRHH-------------------PHLHHHHPPPPHHHHHHPR 410
Score = 34.7 bits (78), Expect = 2.4
Identities = 18/58 (31%), Positives = 22/58 (37%), Gaps = 7/58 (12%)
Query: 29 HHKPTTPLHPPTK-SPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+H+ P PP + H + H H HH H PPP HHHH H
Sbjct: 357 YHQSAPPESPPKEFKHRHPKYFHKQNNKGHRHRHHPHLHHHHPPP------PHHHHHH 408
Score = 33.5 bits (75), Expect = 5.3
Identities = 20/61 (32%), Positives = 24/61 (38%), Gaps = 4/61 (6%)
Query: 68 PLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH---PPAHAPTHM 124
P PPP P + + H+ H P P P P P P LHH + PP P
Sbjct: 259 PPPPPQPPRGMGIQFHEASHQ-PGIFPPPPPPPPPPPPHPDHLHHQYYGYPPPPPPPPPP 317
Query: 125 P 125
P
Sbjct: 318 P 318
>UniRef100_UPI00002968D1 UPI00002968D1 UniRef100 entry
Length = 220
Score = 67.8 bits (164), Expect = 3e-10
Identities = 40/117 (34%), Positives = 46/117 (39%), Gaps = 20/117 (17%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
HH P +TP T H P A P P H H SH P P + P H H
Sbjct: 57 HHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTH 116
Query: 84 QHQHHSPAPSP-------------YHVPTPLQR--PAKPPTLHHHQHPPAHAPTHMP 125
HH PAP+P +H P P R P++ T H HPPA AP P
Sbjct: 117 DASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHHASHHPPAPAPRSTP 173
Score = 64.3 bits (155), Expect = 3e-09
Identities = 39/117 (33%), Positives = 45/117 (38%), Gaps = 20/117 (17%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
HH P +TP T H P A P P H H SH P P + P H H
Sbjct: 36 HHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTH 95
Query: 84 QHQHHSPAPSP-------------YHVPTPLQR--PAKPPTLHHHQHPPAHAPTHMP 125
HH PAP+P +H P P R P++ T HPPA AP P
Sbjct: 96 DASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTP 152
Score = 64.3 bits (155), Expect = 3e-09
Identities = 39/117 (33%), Positives = 45/117 (38%), Gaps = 20/117 (17%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
HH P +TP T H P A P P H H SH P P + P H H
Sbjct: 99 HHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTH 158
Query: 84 QHQHHSPAPSP-------------YH--VPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
HH PAP+P +H VP P P++ T HPPA AP P
Sbjct: 159 HASHHPPAPAPRSTPSQQHTHDASHHPPVPAPRSTPSQQHTHDASHHPPAPAPRSTP 215
Score = 62.0 bits (149), Expect = 1e-08
Identities = 37/109 (33%), Positives = 42/109 (37%), Gaps = 17/109 (15%)
Query: 33 TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAP 92
+TP T H P A P P H H SH P P + P H H HH PAP
Sbjct: 3 STPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAP 62
Query: 93 SP-------------YHVPTPLQRPAKPPTLHHHQ---HPPAHAPTHMP 125
+P +H P P R + P H H HPPA AP P
Sbjct: 63 APRSTPSQQHTHDASHHPPAPAPR-STPSQQHTHDASHHPPAPAPRSTP 110
Score = 55.1 bits (131), Expect = 2e-06
Identities = 34/99 (34%), Positives = 39/99 (39%), Gaps = 9/99 (9%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
HH P +TP T H P A P P H H SH P P + P H H
Sbjct: 15 HHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTH 74
Query: 84 QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPT 122
HH PAP+P TP Q+ + HH PA T
Sbjct: 75 DASHHPPAPAPRS--TPSQQHTHDAS--HHPPAPAPRST 109
Score = 53.5 bits (127), Expect = 5e-06
Identities = 27/76 (35%), Positives = 31/76 (40%), Gaps = 5/76 (6%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
HH P +TP T H P A P P H H SH P P + P H H
Sbjct: 141 HHPPAPAPRSTPSQQHTHHASHHPPAPAPRSTPSQQHTHDASHHPPVPAPRSTPSQQHTH 200
Query: 84 QHQHHSPAPSPYHVPT 99
HH PAP+P P+
Sbjct: 201 DASHHPPAPAPRSTPS 216
Score = 53.5 bits (127), Expect = 5e-06
Identities = 30/89 (33%), Positives = 35/89 (38%), Gaps = 15/89 (16%)
Query: 52 PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP-------------YHVP 98
P P H H SH P P + P H H HH PAP+P +H P
Sbjct: 1 PRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQHTHDASHHPP 60
Query: 99 TPLQR--PAKPPTLHHHQHPPAHAPTHMP 125
P R P++ T HPPA AP P
Sbjct: 61 APAPRSTPSQQHTHDASHHPPAPAPRSTP 89
Score = 33.5 bits (75), Expect = 5.3
Identities = 18/58 (31%), Positives = 20/58 (34%), Gaps = 5/58 (8%)
Query: 29 HHKP-----TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHH 81
HH P +TP T H P P P H H SH P P + P H
Sbjct: 162 HHPPAPAPRSTPSQQHTHDASHHPPVPAPRSTPSQQHTHDASHHPPAPAPRSTPSQQH 219
>UniRef100_Q9M3G8 Putative proline-rich protein [Arabidopsis thaliana]
Length = 219
Score = 67.4 bits (163), Expect = 3e-10
Identities = 39/100 (39%), Positives = 44/100 (44%), Gaps = 12/100 (12%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHP---AKPLTHHH 82
HH+ + P PP P P PP T A HHHH SP P PPP P P+T
Sbjct: 115 HHRRSPP--PPPPPPPPPPTITPPVTTTTAGHHHHRRSPPPPPPPPPPPPTITPPVTTTT 172
Query: 83 HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPT 122
H HH P P P TP+ + HH HPP A T
Sbjct: 173 TGHHHHRPPPPPPATTTPITNTSD----HHQLHPPPPATT 208
Score = 65.1 bits (157), Expect = 2e-09
Identities = 40/122 (32%), Positives = 47/122 (37%), Gaps = 17/122 (13%)
Query: 27 TLHHKPTTPLHPPTKSPVHKPLAKPPTHAP---------HHHHHHSPSHAPLPPPHP--- 74
T H P +P T + H L PP P HHHH SP P PPP P
Sbjct: 74 TGHRHCRPPSNPATTNSGHHQLRPPPPPPPPLSAITTTGHHHHRRSPPPPPPPPPPPPTI 133
Query: 75 AKPLT-----HHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSR 129
P+T HHHH+ P P P PT T HHH PP P ++
Sbjct: 134 TPPVTTTTAGHHHHRRSPPPPPPPPPPPPTITPPVTTTTTGHHHHRPPPPPPATTTPITN 193
Query: 130 SS 131
+S
Sbjct: 194 TS 195
Score = 51.6 bits (122), Expect = 2e-05
Identities = 35/116 (30%), Positives = 45/116 (38%), Gaps = 15/116 (12%)
Query: 29 HHK----PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQ 84
HH+ P+ P+ T +P + P T HH P P PPP A T HHH
Sbjct: 59 HHRQLRPPSIPVTTNTGHRHCRPPSNPATTNSGHHQLRPPP--PPPPPLSAITTTGHHHH 116
Query: 85 HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH---------PPAHAPTHMPRVSRSS 131
+ P P P P + P T HH H PP PT P V+ ++
Sbjct: 117 RRSPPPPPPPPPPPPTITPPVTTTTAGHHHHRRSPPPPPPPPPPPPTITPPVTTTT 172
Score = 34.7 bits (78), Expect = 2.4
Identities = 20/66 (30%), Positives = 23/66 (34%), Gaps = 15/66 (22%)
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSP---------------SHAPLPPPH 73
HH +P PP P + P T HHHH P H L PP
Sbjct: 145 HHHRRSPPPPPPPPPPPPTITPPVTTTTTGHHHHRPPPPPPATTTPITNTSDHHQLHPPP 204
Query: 74 PAKPLT 79
PA +T
Sbjct: 205 PATTIT 210
>UniRef100_UPI0000456BB2 UPI0000456BB2 UniRef100 entry
Length = 700
Score = 66.2 bits (160), Expect = 7e-10
Identities = 43/107 (40%), Positives = 49/107 (45%), Gaps = 15/107 (14%)
Query: 30 HKPTTPL-HPPTKSPVHKPLAKPPTHAPHHHHHH-------SPSHAPL--PPPHPA-KPL 78
H PT PL +PPT VH P PPTH P H H PS PL PP HP+ P
Sbjct: 95 HPPTHPLIYPPTHPSVH-PSTHPPTHPPIHSSTHPSIHPSIHPSTHPLIHPPTHPSIHPP 153
Query: 79 THHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H + H P H PT P+ P++H HPP H P H P
Sbjct: 154 IHPAIRPPVHPPIHPSTHPPT---HPSTHPSIHPFTHPPTHPPVHPP 197
Score = 65.9 bits (159), Expect = 1e-09
Identities = 34/102 (33%), Positives = 43/102 (41%), Gaps = 4/102 (3%)
Query: 28 LHHKPTTPLHPPTKSPVH---KPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQ 84
+H P HP T +H P PP H P H H +H P+ PP P H +
Sbjct: 166 IHPSTHPPTHPSTHPSIHPFTHPPTHPPVHPPTHPSSHPSTHPPIHPPPSIHPPIHPSIR 225
Query: 85 HQHHSPAPSPYHVPT-PLQRPAKPPTLHHHQHPPAHAPTHMP 125
H P P H P P P P++ HPP H+ TH+P
Sbjct: 226 PSIHPPIHPPIHPPIHPSIHPPAHPSICPSIHPPIHSSTHLP 267
Score = 65.9 bits (159), Expect = 1e-09
Identities = 42/110 (38%), Positives = 50/110 (45%), Gaps = 19/110 (17%)
Query: 30 HKPTTPL-HPPTKSPVHKPL---AKPPTHAPHHHHHHSPSHAPLPPPHPA-KPLTHHH-- 82
H T PL HPPT +H P+ +PP H P H H P+H P HP+ P TH
Sbjct: 135 HPSTHPLIHPPTHPSIHPPIHPAIRPPVHPPIHPSTHPPTH---PSTHPSIHPFTHPPTH 191
Query: 83 ---HQHQHHSPAPS---PYHVPTPLQRPAKP---PTLHHHQHPPAHAPTH 123
H H S PS P H P + P P P++H HPP H P H
Sbjct: 192 PPVHPPTHPSSHPSTHPPIHPPPSIHPPIHPSIRPSIHPPIHPPIHPPIH 241
Score = 65.5 bits (158), Expect = 1e-09
Identities = 39/107 (36%), Positives = 47/107 (43%), Gaps = 9/107 (8%)
Query: 28 LHHKPT-----TPLHPPTKSPVHKPL---AKPPTHAPHHHHHHSPSHAPL-PPPHPAKPL 78
L H PT P+HP + PVH P+ PPTH H H +H P PP HP
Sbjct: 141 LIHPPTHPSIHPPIHPAIRPPVHPPIHPSTHPPTHPSTHPSIHPFTHPPTHPPVHPPTHP 200
Query: 79 THHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
+ H H P PS + P RP+ P +H HPP H H P
Sbjct: 201 SSHPSTHPPIHPPPSIHPPIHPSIRPSIHPPIHPPIHPPIHPSIHPP 247
Score = 65.5 bits (158), Expect = 1e-09
Identities = 38/101 (37%), Positives = 47/101 (45%), Gaps = 11/101 (10%)
Query: 30 HKPT-TPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPL-----PPPHP-AKPLTHHH 82
H PT + +HPPT + +H P+ PP H H H P H P+ PP HP P TH
Sbjct: 51 HPPTHSSIHPPTHASIHPPI-HPPVHPSIHPPIHPPIHPPVHPSIHPPTHPLIYPPTHPS 109
Query: 83 HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
H P P H T P+ P++H HP H PTH
Sbjct: 110 VHPSTHPPTHPPIHSST---HPSIHPSIHPSTHPLIHPPTH 147
Score = 62.4 bits (150), Expect = 1e-08
Identities = 36/98 (36%), Positives = 43/98 (43%), Gaps = 8/98 (8%)
Query: 30 HKPTTPLHPPTKSPVHK---PLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQ 86
H P+ +HPP S H P PPTH+ H H+ H P+ P P P H
Sbjct: 30 HPPS--IHPPIHSSTHPSIHPFIHPPTHSSIHPPTHASIHPPIHP--PVHPSIHPPIHPP 85
Query: 87 HHSPAPSPYHVPT-PLQRPAKPPTLHHHQHPPAHAPTH 123
H P H PT PL P P++H HPP H P H
Sbjct: 86 IHPPVHPSIHPPTHPLIYPPTHPSVHPSTHPPTHPPIH 123
Score = 62.4 bits (150), Expect = 1e-08
Identities = 42/108 (38%), Positives = 48/108 (43%), Gaps = 13/108 (12%)
Query: 30 HKPT-TPLHPPTKSPVH---KPLAKPPTHAPHHHHHHSPSHAPL--PPPHPA-KPLTHHH 82
H PT +HPP PVH P PP H P H H P+H PL PP HP+ P TH
Sbjct: 59 HPPTHASIHPPIHPPVHPSIHPPIHPPIHPPVHPSIHPPTH-PLIYPPTHPSVHPSTHPP 117
Query: 83 -----HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H H S PS + PL P P++H HP P H P
Sbjct: 118 THPPIHSSTHPSIHPSIHPSTHPLIHPPTHPSIHPPIHPAIRPPVHPP 165
Score = 58.5 bits (140), Expect = 2e-07
Identities = 36/107 (33%), Positives = 42/107 (38%), Gaps = 10/107 (9%)
Query: 27 TLHHKPTTPLHPPTKSPVH-------KPLAKPPTHAPHHHHHHSPSHAPL-PPPHPAKPL 78
++H P HPP S H P P H P H H P H + PP HP
Sbjct: 109 SVHPSTHPPTHPPIHSSTHPSIHPSIHPSTHPLIHPPTHPSIHPPIHPAIRPPVHPPIHP 168
Query: 79 THHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
+ H H P+ P+ P P P PPT H HP H P H P
Sbjct: 169 STHPPTHPSTHPSIHPFTHP-PTHPPVHPPT-HPSSHPSTHPPIHPP 213
Score = 51.2 bits (121), Expect = 2e-05
Identities = 31/91 (34%), Positives = 38/91 (41%), Gaps = 3/91 (3%)
Query: 36 LHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP-HP-AKPLTHHHHQHQHHSPAPS 93
+HPPT P+ P+ P + H H P H+ P HP P TH H+
Sbjct: 9 IHPPTHLPICPPIQPPICPSIHPPSIHPPIHSSTHPSIHPFIHPPTHSSIHPPTHASIHP 68
Query: 94 PYHVPT-PLQRPAKPPTLHHHQHPPAHAPTH 123
P H P P P P +H HP H PTH
Sbjct: 69 PIHPPVHPSIHPPIHPPIHPPVHPSIHPPTH 99
Score = 49.7 bits (117), Expect = 7e-05
Identities = 33/93 (35%), Positives = 37/93 (39%), Gaps = 9/93 (9%)
Query: 35 PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
P+HP P H P+ PP P H PS P P TH H P S
Sbjct: 4 PIHPSIHPPTHLPIC-PPIQPPICPSIHPPSIHP-----PIHSSTHPSIHPFIHPPTHSS 57
Query: 95 YHVPT--PLQRPAKPPTLHHHQHPPAHAPTHMP 125
H PT + P PP +H HPP H P H P
Sbjct: 58 IHPPTHASIHPPIHPP-VHPSIHPPIHPPIHPP 89
Score = 49.3 bits (116), Expect = 9e-05
Identities = 35/109 (32%), Positives = 43/109 (39%), Gaps = 26/109 (23%)
Query: 30 HKPT-----TPLHPPTKSPVHKPLAKPP----THAPHHHHHHSPSHAPLPPPHPAKPLTH 80
H PT P+ PP +H P PP TH H H P+H+ + PP A
Sbjct: 10 HPPTHLPICPPIQPPICPSIHPPSIHPPIHSSTHPSIHPFIHPPTHSSIHPPTHASI--- 66
Query: 81 HHHQHQHHSPAPSPYH--VPTPLQRPAKPPTLHHHQHPPAH----APTH 123
H P P H + P+ P PP +H HPP H PTH
Sbjct: 67 -------HPPIHPPVHPSIHPPIHPPIHPP-VHPSIHPPTHPLIYPPTH 107
Score = 48.5 bits (114), Expect = 2e-04
Identities = 38/113 (33%), Positives = 47/113 (40%), Gaps = 18/113 (15%)
Query: 30 HKPTTP-LHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPL-PPPHPAKPLTHHHHQHQH 87
H P P + P P+H PP H P H H P+H + P HP P+ H H
Sbjct: 217 HPPIHPSIRPSIHPPIH-----PPIHPPIHPSIHPPAHPSICPSIHP--PIHSSTHLPTH 269
Query: 88 HSPAPSPYHVPT------PLQRPAKPPTLHH--HQHPPAHAPTHMPRVSRSSI 132
S PS H T P P+ P++HH HP H TH P + SI
Sbjct: 270 PSIHPSTLHPSTHPSIHCPSACPSIYPSIHHPLTHHPSVHPLTH-PSIHLPSI 321
Score = 46.2 bits (108), Expect = 8e-04
Identities = 30/100 (30%), Positives = 41/100 (41%), Gaps = 10/100 (10%)
Query: 27 TLHHKPTTPLHPPTKSPVHKPLAK---PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
++H P+HPP +H P P H P H H P+H P HP+ T H
Sbjct: 227 SIHPPIHPPIHPPIHPSIHPPAHPSICPSIHPPIHSSTHLPTH---PSIHPS---TLHPS 280
Query: 84 QHQH-HSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPT 122
H H P+ P P+ P++H HP H P+
Sbjct: 281 THPSIHCPSACPSIYPSIHHPLTHHPSVHPLTHPSIHLPS 320
Score = 45.1 bits (105), Expect = 0.002
Identities = 35/117 (29%), Positives = 45/117 (37%), Gaps = 5/117 (4%)
Query: 35 PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
P+H T P H + H H H PS P P PLTHH H P+
Sbjct: 259 PIHSSTHLPTHPSIHPSTLHPSTHPSIHCPSACPSIYPSIHHPLTHHPSVHPLTHPS--- 315
Query: 95 YHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHAGFDTL 151
H+P+ L PP + HPP P + RS +G S H +G D +
Sbjct: 316 IHLPSILSF-IHPPAIFLSVHPPIMHPFNRRAGPRSPHTPQGRTPFPS-HPSGQDPI 370
Score = 40.0 bits (92), Expect = 0.057
Identities = 27/97 (27%), Positives = 35/97 (35%), Gaps = 12/97 (12%)
Query: 36 LHPPTKSPVHKPLAKPPTHAPHHHH---HHSPSHAPLPPPHPA-----KPLTHHHHQHQH 87
+HPP P ++ H P H P+PPPHP+ P
Sbjct: 334 VHPPIMHPFNRRAGPRSPHTPQGRTPFPSHPSGQDPIPPPHPSGQDPVPPTPLRAGPRSP 393
Query: 88 HSP---APSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
H+P PSP H P + P+ P T P H P
Sbjct: 394 HTPQGRTPSPPHTPRD-RTPSPPHTPQGRTPSPPHTP 429
Score = 37.7 bits (86), Expect = 0.28
Identities = 33/116 (28%), Positives = 44/116 (37%), Gaps = 31/116 (26%)
Query: 34 TPLHPP---TKSPVHKPLAKPPTHAPHHHHHHSP------------------SHAPLPPP 72
+PLH P T SP+H P + P P H P P+PPP
Sbjct: 435 SPLHTPQGRTPSPLHTPQGRTPF--PPHPSGQDPVPPSTPLGAGPVPPSDPSGQDPIPPP 492
Query: 73 HPAKPLTHHHHQHQHHSPAPSPYH------VPTPLQRPAKPPTLHHHQHP-PAHAP 121
HP+ Q +P+P P+ PTPL+ +PP + P P H P
Sbjct: 493 HPSGQDPSPPQTPQGRTPSP-PHPSGQDPVPPTPLRAGPRPPHTPQGRTPCPPHTP 547
>UniRef100_Q7X8E2 OJ990528_30.2 protein [Oryza sativa]
Length = 235
Score = 65.1 bits (157), Expect = 2e-09
Identities = 42/162 (25%), Positives = 62/162 (37%), Gaps = 29/162 (17%)
Query: 6 LALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKP--PTHAPHHHH--- 60
+AL+ +L +S + A HH P +P +P P P H H HH
Sbjct: 1 MALVMSMLLASSLLAAASAARADHHSPAYAPYPHHHAPWPARAQSPSAPDHGAHGHHAPA 60
Query: 61 ----------------HHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRP 104
HH+P+ AP+ P+ HHHH H HH A + P
Sbjct: 61 PAPVHADQPAQAPEWHHHAPAPAPVRDDKPSPSHHHHHHGHHHHRHATAT--------AP 112
Query: 105 AKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGVVYVKSCHHA 146
A P+ HH +H PA + P + + V++ + H A
Sbjct: 113 AHAPSSHHDRHAPAPVHSSWPWPAHAPAPAPAVIHGTNSHLA 154
Score = 46.2 bits (108), Expect = 8e-04
Identities = 41/144 (28%), Positives = 55/144 (37%), Gaps = 44/144 (30%)
Query: 37 HPPTKSPVHKPLAKPPTHAPHHHHHH-------SPSHAPLP--------PPHPAKPLTHH 81
H P +PV P H HH HHH +P+HAP P H + P H
Sbjct: 78 HAPAPAPVRDDKPSPSHHHHHHGHHHHRHATATAPAHAPSSHHDRHAPAPVHSSWPWPAH 137
Query: 82 ---------HHQHQHHSPAPSP-YHV-----PTP----LQRPAKPPT----------LHH 112
H + H +PAP+P HV PTP Q P PP+ +
Sbjct: 138 APAPAPAVIHGTNSHLAPAPAPSSHVQYSPAPTPGDGRHQSPPPPPSPPSADEGAQAPSY 197
Query: 113 HQHPPAHAPTHMPRVSRSSIAVEG 136
+ H P+ AP S +++A G
Sbjct: 198 YGHYPSPAPAPAQESSSAAVAFAG 221
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,305,962
Number of Sequences: 2790947
Number of extensions: 24202828
Number of successful extensions: 279920
Number of sequences better than 10.0: 7365
Number of HSP's better than 10.0 without gapping: 1608
Number of HSP's successfully gapped in prelim test: 6159
Number of HSP's that attempted gapping in prelim test: 178908
Number of HSP's gapped (non-prelim): 39052
length of query: 255
length of database: 848,049,833
effective HSP length: 125
effective length of query: 130
effective length of database: 499,181,458
effective search space: 64893589540
effective search space used: 64893589540
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC135464.6


