

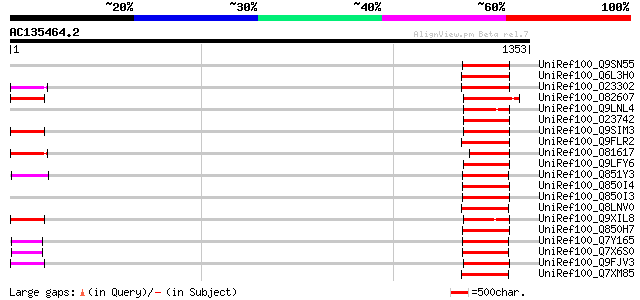
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.2 - phase: 0 /pseudo
(1353 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SN55 Putative retrotransposon polyprotein [Arabidops... 158 1e-36
UniRef100_Q6L3H0 Putative receptor kinase [Solanum demissum] 149 4e-34
UniRef100_O23302 Retrovirus-related like polyprotein [Arabidopsi... 148 1e-33
UniRef100_O82607 T2L5.9 protein [Arabidopsis thaliana] 148 1e-33
UniRef100_Q9LNL4 F12K21.14 [Arabidopsis thaliana] 147 2e-33
UniRef100_O23742 SLG-Sc and SLA-Sc genes and Melmoth retrotransp... 147 2e-33
UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidop... 144 2e-32
UniRef100_Q9FLR2 Polyprotein-like [Arabidopsis thaliana] 142 7e-32
UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana] 141 1e-31
UniRef100_Q9LFY6 T7N9.5 [Arabidopsis thaliana] 140 3e-31
UniRef100_Q851Y3 Putative polyprotein [Oryza sativa] 138 1e-30
UniRef100_Q850I4 Gag-pol polyprotein [Vitis vinifera] 137 2e-30
UniRef100_Q850I3 Gag-pol polyprotein [Vitis vinifera] 137 2e-30
UniRef100_Q8LNV0 Putative copia-like polyprotein [Oryza sativa] 136 4e-30
UniRef100_Q9XIL8 Putative retroelement pol polyprotein [Arabidop... 136 5e-30
UniRef100_Q850H7 Gag-pol polyprotein [Vitis vinifera] 135 7e-30
UniRef100_Q7Y165 Putative retrotransposon protein [Oryza sativa] 135 7e-30
UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa] 135 7e-30
UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis ... 135 9e-30
UniRef100_Q7XM85 OSJNBb0060E08.14 protein [Oryza sativa] 135 1e-29
>UniRef100_Q9SN55 Putative retrotransposon polyprotein [Arabidopsis thaliana]
Length = 1203
Score = 158 bits (399), Expect = 1e-36
Identities = 71/122 (58%), Positives = 96/122 (78%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+PG GL + S L L G+SDADWG C D+RRS+TG+C ++G SL++WKSKKQ +VSRSS
Sbjct: 866 NPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSS 925
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+E+EYR+L +TCE+ L LL DL+VT + L CD++SALH+A+NP+FHERTKH++I
Sbjct: 926 TESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEI 985
Query: 1300 DC 1301
DC
Sbjct: 986 DC 987
>UniRef100_Q6L3H0 Putative receptor kinase [Solanum demissum]
Length = 1358
Score = 149 bits (377), Expect = 4e-34
Identities = 68/124 (54%), Positives = 91/124 (72%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+++PG GL F H++GY+DADW G P RRS +GYC +G +LVSWKSKKQ +V+R
Sbjct: 1183 KSAPGKGLLFEDQGHEHIIGYTDADWAGSPSDRRSTSGYCVLVGGNLVSWKSKKQNVVAR 1242
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
SS+E+EYRA+ +TCEL + LL +L L+CD+Q+ALHIASNP+FHERTKH+
Sbjct: 1243 SSAESEYRAMATATCELVWIKQLLGELKFGKVDKMELVCDNQAALHIASNPVFHERTKHI 1302
Query: 1298 DIDC 1301
+IDC
Sbjct: 1303 EIDC 1306
>UniRef100_O23302 Retrovirus-related like polyprotein [Arabidopsis thaliana]
Length = 1489
Score = 148 bits (374), Expect = 1e-33
Identities = 66/124 (53%), Positives = 92/124 (73%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ +PG GL + D + L G+SDADW C DTRRSI+G+C ++G SL+SWKSKKQ + SR
Sbjct: 1309 KANPGQGLMYSADYEICLNGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASR 1368
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
SS+E+EYR++ +TCE+ L LL DL++ + L CD++SALH + NP+FHERTKH+
Sbjct: 1369 SSTESEYRSMAQATCEIIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHI 1428
Query: 1298 DIDC 1301
+IDC
Sbjct: 1429 EIDC 1432
Score = 87.0 bits (214), Expect = 3e-15
Identities = 41/97 (42%), Positives = 58/97 (59%), Gaps = 1/97 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V +WLMNSV I QSL+Y + WN+L +RF + D RI +++ L Q
Sbjct: 96 RCNDIVSTWLMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSKIEQG 155
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEA 98
S+ I Y+T L + WEE + +P CTC RC C+A
Sbjct: 156 SMDISTYYTALLTLWEEHRNYVELPVCTCG-RCECDA 191
>UniRef100_O82607 T2L5.9 protein [Arabidopsis thaliana]
Length = 1244
Score = 148 bits (373), Expect = 1e-33
Identities = 73/148 (49%), Positives = 98/148 (65%), Gaps = 3/148 (2%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ D L L G++D+DWG CPDTRRS TG F+G SL++W+SKKQ VSRSS+E
Sbjct: 1073 GQGLFYSSDPDLTLKGFADSDWGTCPDTRRSTTGLTMFLGSSLITWRSKKQPTVSRSSAE 1132
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL ++CE+ L LL DL + VP + DS +A++IA+NP+FHERTKH++IDC
Sbjct: 1133 AEYRALALASCEMVWLASLLLDLKIITGSVPIVFSDSTAAIYIATNPVFHERTKHIEIDC 1192
Query: 1302 QCSEGETSKWTHGIVSYL*SRADCRYLD 1329
K G++ L R + + D
Sbjct: 1193 HLVRERLDK---GLIRMLHVRTEDQVAD 1217
Score = 79.7 bits (195), Expect = 6e-13
Identities = 37/90 (41%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SWL+NSV+P I +S++ + A D W D+ RF + R L +++ FRQ
Sbjct: 108 RCNSMVKSWLLNSVSPQIYRSILRMNDATDIWRDIYGRFHMTNLPRTYNLTQEIQDFRQG 167
Query: 62 SLSIIEYFTELKSYWEELE-VFCPIPNCTC 90
SLS+ EY+T+L+ W+ L+ P CTC
Sbjct: 168 SLSLSEYYTQLRILWDLLDSTEEPDDPCTC 197
>UniRef100_Q9LNL4 F12K21.14 [Arabidopsis thaliana]
Length = 427
Score = 147 bits (372), Expect = 2e-33
Identities = 73/120 (60%), Positives = 90/120 (74%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ DS L L GY+DADWG CPD+RRS TG+ F+G SL+SW+SKKQ VSRSS+E
Sbjct: 257 GQGLFYSADSDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAE 316
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL ++CE+ L LL L V S VP L DS +A++IA NP+FHERTKH++IDC
Sbjct: 317 AEYRALALASCEMAWLFTLLIALRVATS-VPILYSDSTAAVYIAINPVFHERTKHIEIDC 375
>UniRef100_O23742 SLG-Sc and SLA-Sc genes and Melmoth retrotransposon sequence
[Brassica oleracea]
Length = 253
Score = 147 bits (371), Expect = 2e-33
Identities = 68/120 (56%), Positives = 90/120 (74%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L G++DADW C D+RRS TG+ F+G SL+SW+SKKQ VSRSS+E
Sbjct: 82 GQGLFYSAHSDLTLKGFADADWAACVDSRRSTTGFTMFVGDSLISWRSKKQPTVSRSSAE 141
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL ++CE+ L LL +L+V S VP L DS +A++IA+NP+FHERTKH+++DC
Sbjct: 142 AEYRALALASCEMMWLCILLRELHVASSSVPVLFSDSTAAIYIATNPVFHERTKHIELDC 201
>UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1461
Score = 144 bits (363), Expect = 2e-32
Identities = 71/120 (59%), Positives = 90/120 (74%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ + L L GY+DADWG CPD+RRS TG+ F+G SL+SW+SKKQ VSRSS+E
Sbjct: 1290 GQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAE 1349
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL ++CE+ L LL L V S VP L DS +A++IA+NP+FHERTKH++IDC
Sbjct: 1350 AEYRALALASCEMAWLSTLLLALRVH-SGVPILYSDSTAAVYIATNPVFHERTKHIEIDC 1408
Score = 77.0 bits (188), Expect = 4e-12
Identities = 35/90 (38%), Positives = 58/90 (63%), Gaps = 1/90 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SWL+NSV+P I +S++ + A D W DL RF+ + R L +++ RQ
Sbjct: 127 RCNSMVKSWLLNSVSPQIYRSILRLNDATDIWRDLFDRFNLTNLPRTYNLTQEIQDLRQG 186
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPN-CTC 90
++S+ EY+T LK+ W++L+ + + CTC
Sbjct: 187 TMSLSEYYTLLKTLWDQLDSTEALDDPCTC 216
>UniRef100_Q9FLR2 Polyprotein-like [Arabidopsis thaliana]
Length = 509
Score = 142 bits (358), Expect = 7e-32
Identities = 65/124 (52%), Positives = 92/124 (73%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ +PG GLF + L+L G++DADWG C D+RRS++G C F+G SL++WKSKKQ++ S
Sbjct: 325 KNNPGQGLFSSAGTELYLNGFADADWGTCLDSRRSVSGVCVFLGTSLITWKSKKQEVASG 384
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
SS+EAEYR++ +T EL L +L DL+V L CD++SA+HIA+N +FHERTKH+
Sbjct: 385 SSTEAEYRSMAVATKELLWLAQMLKDLHVEMEFQVKLFCDNKSAMHIANNSVFHERTKHV 444
Query: 1298 DIDC 1301
+IDC
Sbjct: 445 EIDC 448
>UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana]
Length = 1633
Score = 141 bits (356), Expect = 1e-31
Identities = 62/102 (60%), Positives = 84/102 (81%)
Query: 1200 DADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSEAEYRALVASTCELQ*LVY 1259
DADWG C D+RRS+TG+C ++G SL++WKSKKQ +VSRSS+E+EYR+L +TCE+ L
Sbjct: 1284 DADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQ 1343
Query: 1260 LLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
LL DL+VT + L CD++SALH+A+NP+FHERTKH++IDC
Sbjct: 1344 LLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDC 1385
Score = 87.0 bits (214), Expect = 3e-15
Identities = 39/97 (40%), Positives = 61/97 (62%), Gaps = 1/97 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN+ V +WLMNSV+ I QSL++ +A W ++ +RF + D R+ +++ L Q
Sbjct: 97 RCNDTVSTWLMNSVSKKIGQSLLFIPTAEGIWKNMLSRFKQDDAPRVYDIEQRLSKIEQG 156
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEA 98
S+ I Y+TEL++ WEE + + +P CTC RC C+A
Sbjct: 157 SMDISAYYTELQTLWEEHKNYVDLPVCTCG-RCECDA 192
>UniRef100_Q9LFY6 T7N9.5 [Arabidopsis thaliana]
Length = 1436
Score = 140 bits (353), Expect = 3e-31
Identities = 65/120 (54%), Positives = 88/120 (73%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ D++ L G+SD+DW CPDTRR +TG+ F+G SLVSW+SKKQ +VS SS+E
Sbjct: 1264 GQGLFYGADTNFDLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAE 1323
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRA+ +T EL L Y+LT + + L CD+++ALHIA+N +FHERTKH++ DC
Sbjct: 1324 AEYRAMSVATKELIWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDC 1383
>UniRef100_Q851Y3 Putative polyprotein [Oryza sativa]
Length = 1299
Score = 138 bits (347), Expect = 1e-30
Identities = 65/121 (53%), Positives = 90/121 (73%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SPG GL+F ++ L + GY DADW C D RRS +GYC ++G +LVSW+SKKQ +VSRS+
Sbjct: 1126 SPGKGLWFKKNGHLKIEGYCDADWASCLDDRRSTSGYCVYVGGNLVSWRSKKQSVVSRST 1185
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ AS EL L LL +L + + LLCD++SA++IA+NP+ H+RTKH++I
Sbjct: 1186 AEAEYRAMAASLSELLWLRNLLVELKILGNTPMKLLCDNKSAINIANNPVQHDRTKHVEI 1245
Query: 1300 D 1300
D
Sbjct: 1246 D 1246
Score = 52.4 bits (124), Expect = 1e-04
Identities = 31/102 (30%), Positives = 49/102 (47%), Gaps = 5/102 (4%)
Query: 4 NNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRIST-LQRDLHSFRQNS 62
N+LV +WL+ S+ P+I ++ SA + W L +S V + Q + RQ
Sbjct: 99 NSLVVAWLLTSLIPAIATTVETISSASEMWKTLTNLYSGEGNVMLMVEAQEKISVLRQGE 158
Query: 63 LSIIEYFTELKSYWEELEVFCPI----PNCTCNVRCSCEAMR 100
S+ EY ELK W +L+ + P+ P+C +R E R
Sbjct: 159 RSVAEYVAELKHLWSDLDHYDPLGLEHPDCIAKMRKWIERRR 200
>UniRef100_Q850I4 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 137 bits (346), Expect = 2e-30
Identities = 61/122 (50%), Positives = 87/122 (71%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+PG G+ + + +SDADW G RRS TGYC F G +LV+WKSKKQ +VSRSS
Sbjct: 153 APGLGILYSSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVAWKSKKQSVVSRSS 212
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+E+EYRA+ +TCE+ + LL ++ + C+ L CD+Q+ALHIA+NP++HERTKH+++
Sbjct: 213 AESEYRAMSRATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAANPVYHERTKHIEV 272
Query: 1300 DC 1301
DC
Sbjct: 273 DC 274
>UniRef100_Q850I3 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 137 bits (346), Expect = 2e-30
Identities = 61/122 (50%), Positives = 87/122 (71%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+PG G+ + + +SDADW G RRS TGYC F G +LV+WKSKKQ +VSRSS
Sbjct: 153 APGLGILYSSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVAWKSKKQSVVSRSS 212
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+E+EYRA+ +TCE+ + LL ++ + C+ L CD+Q+ALHIA+NP++HERTKH+++
Sbjct: 213 AESEYRAMSQATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAANPVYHERTKHIEV 272
Query: 1300 DC 1301
DC
Sbjct: 273 DC 274
>UniRef100_Q8LNV0 Putative copia-like polyprotein [Oryza sativa]
Length = 894
Score = 136 bits (343), Expect = 4e-30
Identities = 64/123 (52%), Positives = 91/123 (73%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ SPG G++F ++ L + GY DADWG C D RRS +GYC FIG +LVSW+SKK+ +VSR
Sbjct: 719 KASPGKGIWFKKNGHLDMEGYCDADWGSCLDDRRSTSGYCVFIGGNLVSWRSKKESVVSR 778
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
S++EAEYR++ S EL L LL +L ++ S L CD++SA++IA+NP+ H+RTKH+
Sbjct: 779 STAEAEYRSMSMSLSELLWLKNLLAELKLSTSTSMKLWCDNKSAINIANNPVQHDRTKHV 838
Query: 1298 DID 1300
+ID
Sbjct: 839 EID 841
>UniRef100_Q9XIL8 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1264
Score = 136 bits (342), Expect = 5e-30
Identities = 66/120 (55%), Positives = 86/120 (71%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L G++D+DW CPD+R S TG+ F+G SL+S +SKKQ +VSRSS+E
Sbjct: 1093 GQGLFYSASSDLTLKGFADSDWASCPDSRHSTTGFTMFVGDSLISLRSKKQHVVSRSSAE 1152
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL +TCEL L LL L + +P L DS +A++IA NP+FHERTKH++IDC
Sbjct: 1153 AEYRALALATCELVWLHTLLASL-TAATTIPILFSDSTAAIYIAINPVFHERTKHIEIDC 1211
Score = 76.6 bits (187), Expect = 5e-12
Identities = 34/91 (37%), Positives = 57/91 (62%), Gaps = 1/91 (1%)
Query: 1 IRCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQ 60
I CN +V SWL+NS++P I +S++ + D W DL +RF+ + R L +++ RQ
Sbjct: 96 ISCNTMVKSWLLNSMSPQIYRSILRMNDVSDIWRDLNSRFNMTNLPRTYNLTQEIQDLRQ 155
Query: 61 NSLSIIEYFTELKSYWEELEVFCPIPN-CTC 90
++LS+ EY+T LK+ W++L + + CTC
Sbjct: 156 STLSLSEYYTRLKTLWDQLNSTEELDDPCTC 186
>UniRef100_Q850H7 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 135 bits (341), Expect = 7e-30
Identities = 60/122 (49%), Positives = 86/122 (70%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+P G+ + + +SDADW G RRS TGYC F G +LV+WKSKKQ +VSRSS
Sbjct: 153 APSLGILYRSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVAWKSKKQSVVSRSS 212
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+E+EYRA+ +TCE+ + LL ++ + C+ L CD+Q+ALHIA+NP++HERTKH+++
Sbjct: 213 AESEYRAMAQATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAANPVYHERTKHIEV 272
Query: 1300 DC 1301
DC
Sbjct: 273 DC 274
>UniRef100_Q7Y165 Putative retrotransposon protein [Oryza sativa]
Length = 1176
Score = 135 bits (341), Expect = 7e-30
Identities = 64/121 (52%), Positives = 88/121 (71%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SPG GL+F ++ L + GY DADW CPD RRS +GYC F+G +LVSW+SKKQ +VSRS+
Sbjct: 1003 SPGKGLWFKKNGHLEVEGYCDADWASCPDDRRSTSGYCVFVGGNLVSWRSKKQPVVSRST 1062
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ S EL L LL++L + L CD++SA+ IA+NP+ H+RTKH+++
Sbjct: 1063 AEAEYRAMSVSLSELLWLRNLLSELMLPVDTPMKLWCDNKSAISIANNPVQHDRTKHVEL 1122
Query: 1300 D 1300
D
Sbjct: 1123 D 1123
Score = 52.8 bits (125), Expect = 7e-05
Identities = 27/83 (32%), Positives = 44/83 (52%), Gaps = 1/83 (1%)
Query: 4 NNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRIST-LQRDLHSFRQNS 62
N+LV +WL+ S+ P+I ++ SA + W L +S V + Q + + RQ
Sbjct: 99 NSLVVAWLLTSLIPAIATTVETISSASEMWKTLTKLYSGEGNVMLMVEAQEKISALRQGE 158
Query: 63 LSIIEYFTELKSYWEELEVFCPI 85
S+ EY ELKS W +L+ + P+
Sbjct: 159 RSVAEYVAELKSLWSDLDHYDPL 181
>UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa]
Length = 1262
Score = 135 bits (341), Expect = 7e-30
Identities = 64/121 (52%), Positives = 88/121 (71%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SPG GL+F ++ L + GY DADW CPD RRS +GYC F+G +LVSW+SKKQ +VSRS+
Sbjct: 1089 SPGKGLWFKKNGHLEVEGYCDADWASCPDDRRSTSGYCVFVGGNLVSWRSKKQPVVSRST 1148
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ S EL L LL++L + L CD++SA+ IA+NP+ H+RTKH+++
Sbjct: 1149 AEAEYRAMSVSLSELLWLRNLLSELMLPVDTPMKLWCDNKSAISIANNPVQHDRTKHVEL 1208
Query: 1300 D 1300
D
Sbjct: 1209 D 1209
Score = 52.8 bits (125), Expect = 7e-05
Identities = 27/83 (32%), Positives = 44/83 (52%), Gaps = 1/83 (1%)
Query: 4 NNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRIST-LQRDLHSFRQNS 62
N+LV +WL+ S+ P+I ++ SA + W L +S V + Q + + RQ
Sbjct: 98 NSLVVAWLLTSLIPAIATTVETISSASEMWKTLTKLYSGEGNVMLMVEAQEKISALRQGE 157
Query: 63 LSIIEYFTELKSYWEELEVFCPI 85
S+ EY ELKS W +L+ + P+
Sbjct: 158 RSVAEYVAELKSLWSDLDHYDPL 180
>UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1475
Score = 135 bits (340), Expect = 9e-30
Identities = 64/120 (53%), Positives = 84/120 (69%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L Y+DADWG C D+RRS +G C F+G SL+SWKSKKQ + S SS+E
Sbjct: 1304 GLGLFYSSKSDLCLKAYTDADWGSCVDSRRSTSGICMFLGDSLISWKSKKQNMASSSSAE 1363
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
+EYRA+ + E+ LV LL + V +K L CDS +A+HIA+N +FHERTKH++ DC
Sbjct: 1364 SEYRAMAMGSREIAWLVKLLAEFQVKQTKPVPLFCDSTAAIHIANNAVFHERTKHIENDC 1423
Score = 72.8 bits (177), Expect = 7e-11
Identities = 33/92 (35%), Positives = 55/92 (58%), Gaps = 3/92 (3%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SW++NSVT I +S++ + A + W DL RF + R L + + S +Q
Sbjct: 125 RCNSMVKSWILNSVTKQIYKSILRFNDAAEIWKDLDTRFHITNLPRSYQLTQQIWSLQQG 184
Query: 62 SLSIIEYFTELKSYWEELE---VFCPIPNCTC 90
++S+ +Y+T LK+ W++L+ NCTC
Sbjct: 185 TMSLSDYYTALKTLWDDLDGASCVSTCKNCTC 216
>UniRef100_Q7XM85 OSJNBb0060E08.14 protein [Oryza sativa]
Length = 917
Score = 135 bits (339), Expect = 1e-29
Identities = 64/123 (52%), Positives = 90/123 (73%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ SPG G++F ++ L + GY DADWG C D RS +GYC FIG +LVSW+SKKQ +VSR
Sbjct: 408 KASPGKGIWFKKNGHLDVEGYCDADWGSCLDDMRSTSGYCVFIGGNLVSWRSKKQSVVSR 467
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
S++EAEYR++ S EL L LL +L ++ S L CD++SA++IA+NP+ H+RTKH+
Sbjct: 468 STAEAEYRSMSMSLSELLWLKNLLAELKLSTSTSMKLWCDNKSAINIANNPVQHDRTKHV 527
Query: 1298 DID 1300
+ID
Sbjct: 528 EID 530
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.360 0.159 0.618
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,896,863,694
Number of Sequences: 2790947
Number of extensions: 68275668
Number of successful extensions: 282808
Number of sequences better than 10.0: 663
Number of HSP's better than 10.0 without gapping: 590
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 281519
Number of HSP's gapped (non-prelim): 1101
length of query: 1353
length of database: 848,049,833
effective HSP length: 140
effective length of query: 1213
effective length of database: 457,317,253
effective search space: 554725827889
effective search space used: 554725827889
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 81 (35.8 bits)
Medicago: description of AC135464.2


