

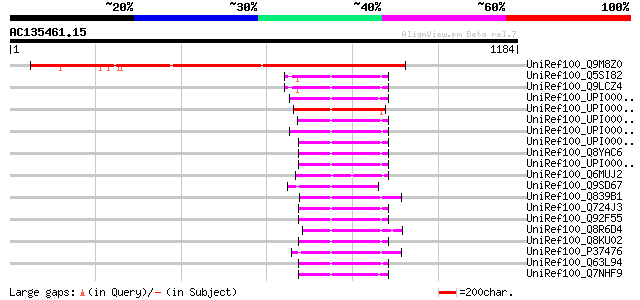
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.15 + phase: 0 /pseudo
(1184 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M8Z0 T6K12.4 protein [Arabidopsis thaliana] 839 0.0
UniRef100_Q5SI82 Cell division protein FtsH [Thermus thermophilu... 162 7e-38
UniRef100_Q9LCZ4 FtsH [Thermus thermophilus] 162 7e-38
UniRef100_UPI000027C449 UPI000027C449 UniRef100 entry 161 1e-37
UniRef100_UPI0000430168 UPI0000430168 UniRef100 entry 157 2e-36
UniRef100_UPI00002ECB14 UPI00002ECB14 UniRef100 entry 156 3e-36
UniRef100_UPI00003383E0 UPI00003383E0 UniRef100 entry 156 4e-36
UniRef100_UPI00003CA6AA UPI00003CA6AA UniRef100 entry 155 5e-36
UniRef100_Q8YAC6 FtsH protein [Listeria monocytogenes] 155 5e-36
UniRef100_UPI00002C3DD1 UPI00002C3DD1 UniRef100 entry 155 7e-36
UniRef100_Q6MUJ2 ATP-dependent zinc metallopeptidase FtsH [Mycop... 155 9e-36
UniRef100_Q9SD67 FtsH metalloprotease-like protein [Arabidopsis ... 155 9e-36
UniRef100_Q839B1 Cell division protein FtsH [Enterococcus faecalis] 154 1e-35
UniRef100_Q724J3 ATP-dependent metalloprotease FtsH [Listeria mo... 154 1e-35
UniRef100_Q92F55 FtsH protein [Listeria innocua] 154 1e-35
UniRef100_Q8R6D4 Cell division protein ftsH [Fusobacterium nucle... 154 1e-35
UniRef100_Q8KU02 FtsH [Listeria monocytogenes] 154 1e-35
UniRef100_P37476 Cell division protein ftsH homolog [Bacillus su... 154 1e-35
UniRef100_Q63L94 FtsH-2 protease [Burkholderia pseudomallei] 154 2e-35
UniRef100_Q7NHF9 Cell division protein [Gloeobacter violaceus] 154 2e-35
>UniRef100_Q9M8Z0 T6K12.4 protein [Arabidopsis thaliana]
Length = 1293
Score = 839 bits (2168), Expect = 0.0
Identities = 469/952 (49%), Positives = 638/952 (66%), Gaps = 87/952 (9%)
Query: 48 QNNHTVLSQPTNSTLVLSQCC--LTKQLILRALFCFAVGVSTFGTFQIAPAFALPTIPWV 105
+ N VLS NS +TK L+ ALFC A+G+S +FQ APA A+P + V
Sbjct: 87 EGNELVLSSEYNSAKTRESVIQFVTKPLVY-ALFCIAIGLSPIRSFQ-APALAVPFVSDV 144
Query: 106 QFLSKNKENKNQ--------HEYSDCTQKVLDTVPSLLRTIEEVRKGNGDFEDVKRALEF 157
+ K + + + HE+SD T+++L+TV LL+TIE VRK NG+ +V AL+
Sbjct: 145 IWKKKKERVREKEVVLKAVDHEFSDYTRRLLETVSVLLKTIEIVRKENGEVAEVGAALDA 204
Query: 158 VKLKKYEMEKEILERMHPVLMDLKEELRLLQIKEGEISWQMAEVNREHRKLMG------- 210
VK++K +++KEI+ ++ + L++E LL + +I + + ++ KL+
Sbjct: 205 VKVEKEKLQKEIMSGLYRDMRRLRKERDLLMKRADKIVDEALSLKKQSEKLLRKGAREKM 264
Query: 211 -----------------WE-MDMKDNVVNEVEKKVL----------DKRMVELEKKWNEI 242
WE +D D+++ + E L ++ VEL K +N
Sbjct: 265 EKLEESVDIMESEYNKIWERIDEIDDIILKKETTTLSFGVRELIFIERECVELVKSFNRE 324
Query: 243 LVKIDEMEDV-------ISRKETVA-------------LSYGVLEI----CFIQRECENL 278
L + E V +SR E + VLE+ F R+ +
Sbjct: 325 LNQ-KSFESVPESSITKLSRSEIKQELVNAQRKHLEQMILPNVLELEEVDPFFDRDSVDF 383
Query: 279 VERFKQEIKQ-KKIGSSFASSVNKLSKSVIQEDLETVQRKQIEQTILPSIVDVDDLGPFF 337
R K+ +++ KK+ + + K K +E L +K E + + + F
Sbjct: 384 SLRIKKRLEESKKLQRDLQNRIRKRMKKFGEEKLFV--QKTPEGEAVKGFPEAEVKWMFG 441
Query: 338 HQDSV---DFAQHLERSLKDSREQQK-NLEAQIRKDMQYDKEKRSVVYSPEEEERILLDR 393
++ V HL K +E+ K +L+ ++ +D+ + K+ Y + +E++LLDR
Sbjct: 442 EKEVVVPKAIQLHLRHGWKKWQEEAKADLKQKLLEDVDFGKQ-----YIAQRQEQVLLDR 496
Query: 394 DRVVSKTWYNEEKNRWEMDPVAVPHAVSKKLIEHVRIRYDGRAMYIALKGEDKEFYVDIK 453
DRVVSKTWYNE+K+RWEMDP+AVP+AVS+KLI+ RIR+D MY+ALKG+DKEFYVDIK
Sbjct: 497 DRVVSKTWYNEDKSRWEMDPMAVPYAVSRKLIDSARIRHDYAVMYVALKGDDKEFYVDIK 556
Query: 454 EFERLFEYIGGFDVLYRKMLACGIPTAVHLMWIPLSELSVHQRISVILRFPLRFLSGRWN 513
E+E LFE GGFD LY KMLACGIPT+VHLMWIP+SELS+ Q+ ++ R R +
Sbjct: 557 EYEMLFEKFGGFDALYLKMLACGIPTSVHLMWIPMSELSLQQQFLLVTRVVSRVFNALRK 616
Query: 514 SETVLTTTNLIFDNIKEMTDDIMTVIGFPIVEYILPNPVRVKLGMAWPEE--ETMNTPWY 571
++ V + + + I+ + DDIM + FP++E+I+P +R++LGMAWPEE +T+ + WY
Sbjct: 617 TQVVSNAKDTVLEKIRNINDDIMMAVVFPVIEFIIPYQLRLRLGMAWPEEIEQTVGSTWY 676
Query: 572 LNWQLNAEARVQSRRADGDFRWIMLFIARAAISGFVLINVFQFMRRKIPRLLGYGPIQKN 631
L WQ AE +SR + DF+W + F+ R++I GFVL +VF+F++RK+PRLLGYGP +++
Sbjct: 677 LQWQSEAEMNFKSRNTE-DFQWFLWFLIRSSIYGFVLYHVFRFLKRKVPRLLGYGPFRRD 735
Query: 632 PNRRKLEQMAYYFDERKGRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEE 691
PN RK ++ YF RK R++ +R+ G+DPIKTAF+ MKRVK PPIPL NF+SI+SM+EE
Sbjct: 736 PNVRKFWRVKSYFTYRKRRIKQKRKAGIDPIKTAFDRMKRVKNPPIPLKNFASIESMREE 795
Query: 692 ISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEA 751
I+EVVAFLQNP+AFQEMGARAPRGVLIVGERGTGKTSLA+AIAAEA+VPVV ++AQ+LEA
Sbjct: 796 INEVVAFLQNPKAFQEMGARAPRGVLIVGERGTGKTSLALAIAAEARVPVVNVEAQELEA 855
Query: 752 GMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVEL 811
G+WVGQSA+NVRELFQTARDLAPVI+FVEDFDLFAGVRGKF+HT+ QDHE+FINQLLVEL
Sbjct: 856 GLWVGQSAANVRELFQTARDLAPVIIFVEDFDLFAGVRGKFVHTKQQDHESFINQLLVEL 915
Query: 812 DGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQ 871
DGFEKQDGVVLMATTRN KQIDEAL+RPGRMDR+FHLQ PT+ ERE IL++AA+ETMD +
Sbjct: 916 DGFEKQDGVVLMATTRNHKQIDEALRRPGRMDRVFHLQSPTEMERERILHNAAEETMDRE 975
Query: 872 LVEYVDWKKVAEKTALLRPIELKLVPIALEGSAFRSKVLDTDEIMSYCSFFA 923
LV+ VDW+KV+EKT LLRPIELKLVP+ALE SAFRSK LDTDE++SY S+FA
Sbjct: 976 LVDLVDWRKVSEKTTLLRPIELKLVPMALESSAFRSKFLDTDELLSYVSWFA 1027
>UniRef100_Q5SI82 Cell division protein FtsH [Thermus thermophilus HB8]
Length = 624
Score = 162 bits (409), Expect = 7e-38
Identities = 98/250 (39%), Positives = 134/250 (53%), Gaps = 17/250 (6%)
Query: 641 AYYFDERKGRMRDRRREGVDPIKTAFEHMKR-----VKKPPIPLNNFSSIDSMKEEISEV 695
A Y+ R GR P +AF K + P + + + + KEE+ E+
Sbjct: 120 ALYYFSRNGR--------AGPSDSAFSFTKSRARVLTEAPKVTFKDVAGAEEAKEELKEI 171
Query: 696 VAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWV 755
V FL+NP F EMGAR P+GVL+VG G GKT LA A+A EA+VP + M+V
Sbjct: 172 VEFLKNPSRFHEMGARIPKGVLLVGPPGVGKTHLARAVAGEARVPFITASGSDF-VEMFV 230
Query: 756 GQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFE 815
G A+ VR+LF+TA+ AP I+F+++ D RG + N + E +NQLLVE+DGFE
Sbjct: 231 GVGAARVRDLFETAKRHAPCIVFIDEIDAVGRKRGSGVGGGNDEREQTLNQLLVEMDGFE 290
Query: 816 KQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEY 875
K +V+MA T +D AL RPGR DR + P RE IL A+ L E
Sbjct: 291 KDTAIVVMAATNRPDILDPALLRPGRFDRQIAIDAPDVKGREQILRIHAR---GKPLAED 347
Query: 876 VDWKKVAEKT 885
VD +A++T
Sbjct: 348 VDLALLAKRT 357
>UniRef100_Q9LCZ4 FtsH [Thermus thermophilus]
Length = 624
Score = 162 bits (409), Expect = 7e-38
Identities = 98/250 (39%), Positives = 134/250 (53%), Gaps = 17/250 (6%)
Query: 641 AYYFDERKGRMRDRRREGVDPIKTAFEHMKR-----VKKPPIPLNNFSSIDSMKEEISEV 695
A Y+ R GR P +AF K + P + + + + KEE+ E+
Sbjct: 120 ALYYFSRNGR--------AGPSDSAFSFTKSRARVLTEAPKVTFKDVAGAEEAKEELKEI 171
Query: 696 VAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWV 755
V FL+NP F EMGAR P+GVL+VG G GKT LA A+A EA+VP + M+V
Sbjct: 172 VEFLKNPSRFHEMGARIPKGVLLVGPPGVGKTHLARAVAGEARVPFITASGSDF-VEMFV 230
Query: 756 GQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFE 815
G A+ VR+LF+TA+ AP I+F+++ D RG + N + E +NQLLVE+DGFE
Sbjct: 231 GVGAARVRDLFETAKRHAPCIVFIDEIDAVGRKRGSGVGGGNDEREQTLNQLLVEMDGFE 290
Query: 816 KQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEY 875
K +V+MA T +D AL RPGR DR + P RE IL A+ L E
Sbjct: 291 KDTAIVVMAATNRPDILDPALLRPGRFDRQIAIDAPDVKGREQILRIHAR---GKPLAED 347
Query: 876 VDWKKVAEKT 885
VD +A++T
Sbjct: 348 VDLALLAKRT 357
>UniRef100_UPI000027C449 UPI000027C449 UniRef100 entry
Length = 637
Score = 161 bits (407), Expect = 1e-37
Identities = 93/235 (39%), Positives = 136/235 (57%), Gaps = 7/235 (2%)
Query: 654 RRREGVDPIKTAFEHMKRV---KKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGA 710
RR GV + + RV ++ I ++ + ID K E+ ++VAFL+NP +Q +G
Sbjct: 140 RRPGGVRDLSGMGKSQARVYVQQETGITFDDIAGIDEAKAELQQIVAFLRNPERYQRLGG 199
Query: 711 RAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTAR 770
+ P+GVLIVG GTGKT LA A+A EA VP I M+VG A+ VR+LF+ A+
Sbjct: 200 KIPKGVLIVGAPGTGKTLLARAVAGEAAVPFFTISGSAF-VEMFVGVGAARVRDLFEQAK 258
Query: 771 DLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLK 830
AP I+F+++ D RG + + N + E +NQLLVE+DGF+ GV++MA T +
Sbjct: 259 QKAPCIVFIDELDALGKARGVGLMSGNDEREQTLNQLLVEMDGFQANSGVIIMAATNRPE 318
Query: 831 QIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
+D AL RPGR DR + RP R+ IL A +T +L VD ++A++T
Sbjct: 319 ILDPALLRPGRFDRHIAIDRPDLTGRKQIL---AVQTKRVKLAPEVDLAELAQRT 370
>UniRef100_UPI0000430168 UPI0000430168 UniRef100 entry
Length = 719
Score = 157 bits (397), Expect = 2e-36
Identities = 87/222 (39%), Positives = 134/222 (60%), Gaps = 11/222 (4%)
Query: 664 TAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERG 723
++F +K + K I ++ + + K+EI E+V FL++P+ FQ++GA+ P+G L+VG G
Sbjct: 244 SSFSQVKNMNKN-IKFSDIAGMKEAKQEIYELVEFLKDPKRFQDLGAKIPKGALLVGPPG 302
Query: 724 TGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFD 783
TGKT LA A+A EA VP I ++VG AS VRELF AR L+P I+F+++ D
Sbjct: 303 TGKTLLAKAVAGEANVPFFYISGSDF-IEIFVGMGASRVRELFSQARKLSPSIVFIDEID 361
Query: 784 LFAGVRGK---FIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPG 840
R K F + N + E+ +NQ+LVE+DGF + +GV+++A T +D AL RPG
Sbjct: 362 AVGRKRAKGGGFAASSNDERESTLNQILVEMDGFTENNGVIVLAGTNRSDVLDPALTRPG 421
Query: 841 RMDRIFHLQRPTQAERENILYSAAK------ETMDDQLVEYV 876
R DRI +++RP ER+ I K + D+L++Y+
Sbjct: 422 RFDRIINIERPNLEERKEIFKIHLKPLKLNEKLNKDELIKYL 463
>UniRef100_UPI00002ECB14 UPI00002ECB14 UniRef100 entry
Length = 317
Score = 156 bits (395), Expect = 3e-36
Identities = 85/213 (39%), Positives = 128/213 (59%), Gaps = 4/213 (1%)
Query: 673 KKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMA 732
+ P + + + ++ KEE+ E+ FL++P F ++GA+ P+GVL+VG GTGKT LA A
Sbjct: 43 ESPKVTFKDVAGVEEAKEELEEIKEFLKSPEKFNKLGAKIPKGVLLVGPPGTGKTLLAKA 102
Query: 733 IAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKF 792
+A EA VP I M+VG AS VR+LF+ A++ +P I+F+++ D +RG
Sbjct: 103 VAGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFKKAKEASPAIIFIDEIDAVGRMRGAG 161
Query: 793 IHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPT 852
+ + + E +NQLLVE+DGFE GV+LMA T +D AL RPGR DR + RP
Sbjct: 162 LGGGHDEREQTLNQLLVEMDGFEANQGVILMAATNRPDVLDPALLRPGRFDRQVIVGRPD 221
Query: 853 QAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
+ R IL AK D L + V+++ +A++T
Sbjct: 222 LSGRTQILKVHAK---DKPLAKNVNFETLAKQT 251
>UniRef100_UPI00003383E0 UPI00003383E0 UniRef100 entry
Length = 658
Score = 156 bits (394), Expect = 4e-36
Identities = 93/235 (39%), Positives = 132/235 (55%), Gaps = 7/235 (2%)
Query: 654 RRREGVDPIKTAFEHMKRV---KKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGA 710
RRR G+ + RV ++ I ++ + ID K E+ ++VAFL+NP +Q +G
Sbjct: 140 RRRGGLQDFTGMGKSRARVYVQQETGITFDDIAGIDEAKAELQQLVAFLRNPERYQRLGG 199
Query: 711 RAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTAR 770
+ P+GVL+VG GTGKT LA A+A EA VP I M+VG A+ VR+LF+ A+
Sbjct: 200 KIPKGVLVVGAPGTGKTLLARAVAGEAAVPFFSISGSAF-VEMFVGVGAARVRDLFEQAQ 258
Query: 771 DLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLK 830
AP I+FV++ D VRG + N + E +NQLLVE+DGF+ GV++MA T +
Sbjct: 259 QKAPCIVFVDELDALGKVRGVGPMSGNDEREQTLNQLLVEMDGFQAGSGVIIMAATNRPE 318
Query: 831 QIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
+D AL RPGR DR + RP R IL K +L VD ++A +T
Sbjct: 319 ILDPALMRPGRFDRHIAIDRPDVNGRRQILGVHVKRV---KLAADVDLGELASRT 370
>UniRef100_UPI00003CA6AA UPI00003CA6AA UniRef100 entry
Length = 691
Score = 155 bits (393), Expect = 5e-36
Identities = 87/212 (41%), Positives = 125/212 (58%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + + + D K+E+ EVV FL++PR F E+GAR P+GVL+VG GTGKT LA A+
Sbjct: 174 KKKVRFTDVAGADEEKQELVEVVEFLKDPRKFAELGARIPKGVLLVGPPGTGKTLLARAV 233
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I M+VG AS VR+LF+ A+ AP I+F+++ D RG +
Sbjct: 234 AGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCIIFIDEIDAVGRQRGAGM 292
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ + E +NQLLVE+DGF +G++++A T +D AL RPGR DR + RP
Sbjct: 293 GGGHDEREQTLNQLLVEMDGFGGNEGIIIIAATNRADVLDPALLRPGRFDRQIMVDRPDV 352
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RE +L A+ + L + VD K +A++T
Sbjct: 353 KGREAVLRVHAR---NKPLAKSVDLKAIAQRT 381
>UniRef100_Q8YAC6 FtsH protein [Listeria monocytogenes]
Length = 691
Score = 155 bits (393), Expect = 5e-36
Identities = 87/212 (41%), Positives = 125/212 (58%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + + + D K+E+ EVV FL++PR F E+GAR P+GVL+VG GTGKT LA A+
Sbjct: 174 KKKVRFTDVAGADEEKQELVEVVEFLKDPRKFAELGARIPKGVLLVGPPGTGKTLLARAV 233
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I M+VG AS VR+LF+ A+ AP I+F+++ D RG +
Sbjct: 234 AGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCIIFIDEIDAVGRQRGAGM 292
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ + E +NQLLVE+DGF +G++++A T +D AL RPGR DR + RP
Sbjct: 293 GGGHDEREQTLNQLLVEMDGFGGNEGIIIIAATNRADVLDPALLRPGRFDRQIMVDRPDV 352
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RE +L A+ + L + VD K +A++T
Sbjct: 353 KGREAVLRVHAR---NKPLAKSVDLKAIAQRT 381
>UniRef100_UPI00002C3DD1 UPI00002C3DD1 UniRef100 entry
Length = 604
Score = 155 bits (392), Expect = 7e-36
Identities = 86/211 (40%), Positives = 124/211 (58%), Gaps = 4/211 (1%)
Query: 675 PPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIA 734
P + + + + KEE+ E+ FL++P F +GA+ P+GVL+VG GTGKT LA A+A
Sbjct: 142 PKVTFKDVAGAEEAKEELEEIKEFLKSPEKFNNLGAKIPKGVLLVGPPGTGKTLLARAVA 201
Query: 735 AEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIH 794
E++VP I M+VG AS VR+LF+ A++ AP I+F+++ D +RG +
Sbjct: 202 GESEVPFYSISGSDF-VEMFVGVGASRVRDLFKKAKESAPSIIFIDEIDAVGRMRGAGLG 260
Query: 795 TENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQA 854
+ + E +NQLLVE+DGFE GV+LMA T +D AL RPGR DR + RP
Sbjct: 261 GGHDEREQTLNQLLVEMDGFESNQGVILMAATNRPDVLDPALLRPGRFDRQVIVDRPDLN 320
Query: 855 ERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
R IL AK D L + ++ K VA++T
Sbjct: 321 GRTEILKVHAK---DKPLAKNINLKTVAKQT 348
>UniRef100_Q6MUJ2 ATP-dependent zinc metallopeptidase FtsH [Mycoplasma mycoides]
Length = 648
Score = 155 bits (391), Expect = 9e-36
Identities = 88/219 (40%), Positives = 126/219 (57%), Gaps = 6/219 (2%)
Query: 667 EHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGK 726
++ R +K + ++ + I+ K E+ E+V +L+ P + GARAP+GVL+ G GTGK
Sbjct: 157 QNKARREKSNVKFSDVAGIEEEKLELVELVDYLKQPAKYASAGARAPKGVLMEGPPGTGK 216
Query: 727 TSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFA 786
T LA A+A EA V I + E M+VG AS VRE+F A+ AP I+F+++ D
Sbjct: 217 TLLAKAVAGEANVSFFSIAGSEFEE-MFVGVGASRVREMFNEAKKAAPAIIFIDEIDAVG 275
Query: 787 GVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIF 846
R I T +E +NQLLVELDGFE G+++MA T + +D AL RPGR DR+
Sbjct: 276 RKRNSAIGTGT--NEQTLNQLLVELDGFETNSGIIVMAATNRVDVLDPALLRPGRFDRVI 333
Query: 847 HLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
+ P ERE IL A+ D +DW ++AE+T
Sbjct: 334 QVSLPDIKEREQILKLHARNKKID---PSIDWHRIAERT 369
>UniRef100_Q9SD67 FtsH metalloprotease-like protein [Arabidopsis thaliana]
Length = 802
Score = 155 bits (391), Expect = 9e-36
Identities = 86/213 (40%), Positives = 125/213 (58%), Gaps = 8/213 (3%)
Query: 649 GRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEM 708
G++R R+ G D K + I + + +D KEE+ E+V FL+NP + +
Sbjct: 300 GQLRTRKAGGPDGGKVSGGG------ETITFADVAGVDEAKEELEEIVEFLRNPEKYVRL 353
Query: 709 GARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQT 768
GAR PRGVL+VG GTGKT LA A+A EA+VP + A + ++VG AS VR+LF
Sbjct: 354 GARPPRGVLLVGLPGTGKTLLAKAVAGEAEVPFISCSASEF-VELYVGMGASRVRDLFAR 412
Query: 769 ARDLAPVILFVEDFDLFAGVR-GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTR 827
A+ AP I+F+++ D A R GKF N + E +NQLL E+DGF+ V+++ T
Sbjct: 413 AKKEAPSIIFIDEIDAVAKSRDGKFRMGSNDEREQTLNQLLTEMDGFDSNSAVIVLGATN 472
Query: 828 NLKQIDEALQRPGRMDRIFHLQRPTQAERENIL 860
+D AL+RPGR DR+ ++ P + RE+IL
Sbjct: 473 RADVLDPALRRPGRFDRVVTVETPDKIGRESIL 505
>UniRef100_Q839B1 Cell division protein FtsH [Enterococcus faecalis]
Length = 718
Score = 154 bits (390), Expect = 1e-35
Identities = 95/241 (39%), Positives = 137/241 (56%), Gaps = 6/241 (2%)
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + + K+E+ EVV FL++PR F E+GAR P GVL+ G GTGKT LA A+A E
Sbjct: 184 VRFSDVAGAEEEKQELVEVVEFLKDPRRFAELGARIPAGVLLEGPPGTGKTLLAKAVAGE 243
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP I M+VG AS VR+LF+TA+ AP I+F+++ D RG +
Sbjct: 244 AGVPFYSISGSDF-VEMFVGVGASRVRDLFETAKKNAPAIIFIDEIDAVGRQRGAGMGGG 302
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
+ + E +NQLLVE+DGF+ +GV+++A T +D AL RPGR DR + RP R
Sbjct: 303 HDEREQTLNQLLVEMDGFDGNEGVIVIAATNRSDVLDPALLRPGRFDRQILVGRPDVKGR 362
Query: 857 ENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELK--LVPIALEGSAFRSKVLDTDE 914
E IL AK + L + VD K VA++T +L+ L AL + K +D +
Sbjct: 363 EAILRVHAK---NKPLADDVDLKVVAQQTPGFAGADLENVLNEAALVAARRNKKKIDASD 419
Query: 915 I 915
+
Sbjct: 420 V 420
>UniRef100_Q724J3 ATP-dependent metalloprotease FtsH [Listeria monocytogenes]
Length = 691
Score = 154 bits (390), Expect = 1e-35
Identities = 86/212 (40%), Positives = 125/212 (58%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + + + D K+E+ EVV FL++PR F ++GAR P+GVL+VG GTGKT LA A+
Sbjct: 174 KKKVRFTDVAGADEEKQELVEVVEFLKDPRKFADLGARIPKGVLLVGPPGTGKTLLARAV 233
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I M+VG AS VR+LF+ A+ AP I+F+++ D RG +
Sbjct: 234 AGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCIIFIDEIDAVGRQRGAGM 292
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ + E +NQLLVE+DGF +G++++A T +D AL RPGR DR + RP
Sbjct: 293 GGGHDEREQTLNQLLVEMDGFGGNEGIIIIAATNRADVLDPALLRPGRFDRQIMVDRPDV 352
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RE +L A+ + L + VD K +A++T
Sbjct: 353 KGREAVLRVHAR---NKPLAKSVDLKAIAQRT 381
>UniRef100_Q92F55 FtsH protein [Listeria innocua]
Length = 690
Score = 154 bits (390), Expect = 1e-35
Identities = 86/212 (40%), Positives = 125/212 (58%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + + + D K+E+ EVV FL++PR F ++GAR P+GVL+VG GTGKT LA A+
Sbjct: 174 KKKVRFTDVAGADEEKQELVEVVEFLKDPRKFADLGARIPKGVLLVGPPGTGKTLLARAV 233
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I M+VG AS VR+LF+ A+ AP I+F+++ D RG +
Sbjct: 234 AGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCIIFIDEIDAVGRQRGAGM 292
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ + E +NQLLVE+DGF +G++++A T +D AL RPGR DR + RP
Sbjct: 293 GGGHDEREQTLNQLLVEMDGFGGNEGIIIIAATNRADVLDPALLRPGRFDRQIMVDRPDV 352
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RE +L A+ + L + VD K +A++T
Sbjct: 353 KGREAVLRVHAR---NKPLAKSVDLKAIAQRT 381
>UniRef100_Q8R6D4 Cell division protein ftsH [Fusobacterium nucleatum]
Length = 714
Score = 154 bits (390), Expect = 1e-35
Identities = 93/235 (39%), Positives = 136/235 (57%), Gaps = 6/235 (2%)
Query: 683 SSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVV 742
+ ID K+E+ EVV FL+ P F+++GA+ P+GVL++GE GTGKT LA A+A EAKVP
Sbjct: 276 AGIDEAKQELKEVVDFLKEPEKFRKIGAKIPKGVLLLGEPGTGKTLLAKAVAGEAKVPFF 335
Query: 743 EIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEA 802
+ + M+VG AS VR+LF AR AP I+F+++ D RG N + E
Sbjct: 336 SMSGSEF-VEMFVGVGASRVRDLFNKARKNAPCIVFIDEIDAVGRKRGTGQGGGNDEREQ 394
Query: 803 FINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYS 862
+NQLLVE+DGF + ++++A T +D+AL+RPGR DR + P RE IL
Sbjct: 395 TLNQLLVEMDGFGTDETIIVLAATNRADVLDKALRRPGRFDRQVVVDMPDVKGREEILKV 454
Query: 863 AAKETMDDQLVEYVDWKKVAEKTALLRPIELKLVPIALEGSAFRSKVLDTDEIMS 917
AK + VD+K +A+KTA + + L I EG+ ++ T+ M+
Sbjct: 455 HAK---GKKFAPDVDFKIIAKKTAGMAGAD--LANILNEGAILAARAGRTEITMA 504
>UniRef100_Q8KU02 FtsH [Listeria monocytogenes]
Length = 687
Score = 154 bits (390), Expect = 1e-35
Identities = 86/212 (40%), Positives = 125/212 (58%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + + + D K+E+ EVV FL++PR F ++GAR P+GVL+VG GTGKT LA A+
Sbjct: 174 KKKVRFTDVAGADEEKQELVEVVEFLKDPRKFADLGARIPKGVLLVGPPGTGKTLLARAV 233
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I M+VG AS VR+LF+ A+ AP I+F+++ D RG +
Sbjct: 234 AGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCIIFIDEIDAVGRQRGAGM 292
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ + E +NQLLVE+DGF +G++++A T +D AL RPGR DR + RP
Sbjct: 293 GGGHDEREQTLNQLLVEMDGFGGNEGIIIIAATNRADVLDPALLRPGRFDRQIMVDRPDV 352
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RE +L A+ + L + VD K +A++T
Sbjct: 353 KGREAVLRVHAR---NKPLAKSVDLKAIAQRT 381
>UniRef100_P37476 Cell division protein ftsH homolog [Bacillus subtilis]
Length = 637
Score = 154 bits (390), Expect = 1e-35
Identities = 99/260 (38%), Positives = 140/260 (53%), Gaps = 11/260 (4%)
Query: 658 GVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVL 717
G K E KRVK + + D K+E+ EVV FL++PR F E+GAR P+GVL
Sbjct: 144 GKSKAKLYTEEKKRVK-----FKDVAGADEEKQELVEVVEFLKDPRKFAELGARIPKGVL 198
Query: 718 IVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVIL 777
+VG GTGKT LA A A EA VP I M+VG AS VR+LF+ A+ AP ++
Sbjct: 199 LVGPPGTGKTLLAKACAGEAGVPFFSISGSDF-VEMFVGVGASRVRDLFENAKKNAPCLI 257
Query: 778 FVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQ 837
F+++ D RG + + + E +NQLLVE+DGF +G++++A T +D AL
Sbjct: 258 FIDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGFSANEGIIIIAATNRADILDPALL 317
Query: 838 RPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELK--L 895
RPGR DR + RP RE +L A+ + L E V+ K +A +T +L+ L
Sbjct: 318 RPGRFDRQITVDRPDVIGREAVLKVHAR---NKPLDETVNLKSIAMRTPGFSGADLENLL 374
Query: 896 VPIALEGSAFRSKVLDTDEI 915
AL + K +D +I
Sbjct: 375 NEAALVAARQNKKKIDARDI 394
>UniRef100_Q63L94 FtsH-2 protease [Burkholderia pseudomallei]
Length = 666
Score = 154 bits (389), Expect = 2e-35
Identities = 87/212 (41%), Positives = 122/212 (57%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K I ++ + ID K E+ ++VAFL+ P +Q +G + P+GVLIVG GTGKT LA A+
Sbjct: 164 KTGIDFDDIAGIDEAKAELQQIVAFLRAPARYQRLGGKIPKGVLIVGAPGTGKTLLAKAV 223
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP M+VG A+ VR+LF+ A+ AP I+F+++ D VRG +
Sbjct: 224 AGEAGVPFFSTSGSSF-VEMFVGVGAARVRDLFEQAQQKAPCIIFIDELDALGKVRGAGL 282
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ N + E +NQLLVE+DGF+ GV+LMA T + +D AL RPGR DR + RP
Sbjct: 283 ASGNDEREQTLNQLLVEMDGFQANSGVILMAATNRPEILDPALLRPGRFDRHIAIDRPDL 342
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
R IL K +L VD ++A +T
Sbjct: 343 TGRRQILSVHVKHV---KLGPDVDLGELASRT 371
>UniRef100_Q7NHF9 Cell division protein [Gloeobacter violaceus]
Length = 630
Score = 154 bits (388), Expect = 2e-35
Identities = 87/212 (41%), Positives = 123/212 (57%), Gaps = 4/212 (1%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K + ++ + ID KEE+ EVV FL+ P F +GA+ P+GVL+VG GTGKT LA AI
Sbjct: 168 KTGVKFDDVAGIDEAKEELQEVVQFLKRPERFTAVGAKIPKGVLLVGPPGTGKTLLAKAI 227
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A EA VP I + M+VG AS VR+LF+ A++ AP I+F+++ D RG I
Sbjct: 228 AGEAGVPFFSISGSEF-VEMFVGVGASRVRDLFKKAKENAPCIVFIDEIDAVGRQRGAGI 286
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
N + E +NQLLVE+DGFE G++++A T +D A+ RPGR DR + RP
Sbjct: 287 GGGNDEREQTLNQLLVEMDGFEGNTGIIIIAATNRPDVLDAAILRPGRFDRQITVDRPDM 346
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
A R IL + + +L +D +A +T
Sbjct: 347 AGRLEIL---KVHSRNKKLAPDIDLDVIARRT 375
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.332 0.143 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,852,789,058
Number of Sequences: 2790947
Number of extensions: 77475721
Number of successful extensions: 337793
Number of sequences better than 10.0: 4963
Number of HSP's better than 10.0 without gapping: 2244
Number of HSP's successfully gapped in prelim test: 2806
Number of HSP's that attempted gapping in prelim test: 325357
Number of HSP's gapped (non-prelim): 10453
length of query: 1184
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1045
effective length of database: 460,108,200
effective search space: 480813069000
effective search space used: 480813069000
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 81 (35.8 bits)
Medicago: description of AC135461.15


