

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.13 + phase: 0
(560 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
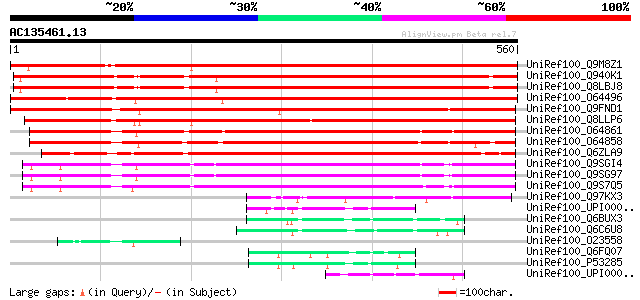
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M8Z1 T6K12.3 protein [Arabidopsis thaliana] 793 0.0
UniRef100_Q940K1 Hypothetical protein [Arabidopsis thaliana] 757 0.0
UniRef100_Q8LBJ8 Hypothetical protein [Arabidopsis thaliana] 756 0.0
UniRef100_O64496 F20D22.14 protein [Arabidopsis thaliana] 657 0.0
UniRef100_Q9FND1 Arabidopsis thaliana genomic DNA, chromosome 5,... 640 0.0
UniRef100_Q8LLP6 OSJNBa0031O09.02 [Oryza sativa] 580 e-164
UniRef100_O64861 Expressed protein [Arabidopsis thaliana] 498 e-139
UniRef100_O64858 Expressed protein [Arabidopsis thaliana] 493 e-138
UniRef100_Q6ZLA9 Hypothetical protein OJ1065_B06.1 [Oryza sativa] 464 e-129
UniRef100_Q9SGI4 F28J7.20 protein [Arabidopsis thaliana] 421 e-116
UniRef100_Q9SG97 F1C9.35 protein [Arabidopsis thaliana] 421 e-116
UniRef100_Q9S7Q5 F1C9.34 protein [Arabidopsis thaliana] 419 e-116
UniRef100_Q97KX3 Uncharacterized conserved protein of probably e... 105 3e-21
UniRef100_UPI000042E5AF UPI000042E5AF UniRef100 entry 64 1e-08
UniRef100_Q6BUX3 Similar to CA0982|IPF7397 Candida albicans [Deb... 59 5e-07
UniRef100_Q6C6U8 Similar to sp|P53285 Saccharomyces cerevisiae Y... 55 7e-06
UniRef100_O23558 Hypothetical protein AT4g17130 [Arabidopsis tha... 52 5e-05
UniRef100_Q6FQ07 Similar to sp|P53285 Saccharomyces cerevisiae Y... 52 5e-05
UniRef100_P53285 Hypothetical 54.5 kDa protein in CBF2-SKN1 inte... 52 5e-05
UniRef100_UPI0000235C44 UPI0000235C44 UniRef100 entry 52 6e-05
>UniRef100_Q9M8Z1 T6K12.3 protein [Arabidopsis thaliana]
Length = 567
Score = 793 bits (2049), Expect = 0.0
Identities = 373/572 (65%), Positives = 450/572 (78%), Gaps = 17/572 (2%)
Query: 1 MFGFECFCWDSFPEFFDSD---PLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFE 57
MFG +CF W DS+ P PFSLP+PLP WPQG GFA GRISLG+IEV K+ KF
Sbjct: 1 MFGCDCFYWSRGISELDSESSEPKPFSLPAPLPSWPQGKGFATGRISLGEIEVVKITKFH 60
Query: 58 KVWRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCS 117
+VW +S+ K+ TFYR +IP+GF CLG+YC DQPLRG+VL AR TSK+ +
Sbjct: 61 RVWSSDSSHDKSKRATFYRADDIPEGFHCLGHYCQPTDQPLRGYVLAAR--TSKA---VN 115
Query: 118 ESESPALKKPLNYSLIWCMDSHDECV-YFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCV 176
+ P LKKP++YSL+W DS YFWLPNPP GY+A+G++VT P EP+ EEVRCV
Sbjct: 116 ADDFPPLKKPVSYSLVWSADSEKNGGGYFWLPNPPVGYRAMGVIVTHEPGEPETEEVRCV 175
Query: 177 RTDLTEVCETSDLLLTIKSKKNS------FQVWNTQPCDRGMLARGVSVGTFFCGTY-FD 229
R DLTE CETS+++L + S K S F VW+T+PC+RGML++GV+VG+FFC TY
Sbjct: 176 REDLTESCETSEMILEVGSSKKSNGSSSPFSVWSTRPCERGMLSQGVAVGSFFCCTYDLS 235
Query: 230 SEQVV-DVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAI 288
SE+ V D+ CLKNLD LHAMPNL+Q+HA+IEH+GPTVYFHP+E YMPSSV WFFKNGA+
Sbjct: 236 SERTVPDIGCLKNLDPTLHAMPNLDQVHAVIEHFGPTVYFHPEEAYMPSSVQWFFKNGAL 295
Query: 289 LYTAGNAKGKAIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPA 348
LY +G ++G+ I+ G+NLP GG ND FWIDLP DE+A+SNLKKGN+ES+ELYVHVKPA
Sbjct: 296 LYRSGKSEGQPINSTGSNLPAGGCNDMDFWIDLPEDEEAKSNLKKGNLESSELYVHVKPA 355
Query: 349 LGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFF 408
LGG FTDI MW+FCPFNGPATLK+ L + M +IGEHVGDWEHFT R+ NF+GELW +FF
Sbjct: 356 LGGTFTDIVMWIFCPFNGPATLKIGLFTLPMTRIGEHVGDWEHFTFRICNFSGELWQMFF 415
Query: 409 SEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNF 468
S+HSGG WV+A D+EF+K+NKP VYSS+HGHAS+PH G YLQGSSKLGIGVRND AKS +
Sbjct: 416 SQHSGGGWVDASDIEFVKDNKPAVYSSKHGHASFPHPGMYLQGSSKLGIGVRNDVAKSKY 475
Query: 469 ILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSV 528
I+DSS RY IVAAEYLG G + EPCWLQYMREWGPTI YDS SEI KI+++LP+ VRFS+
Sbjct: 476 IVDSSQRYVIVAAEYLGKGAVIEPCWLQYMREWGPTIAYDSGSEINKIMNLLPLVVRFSI 535
Query: 529 ENLFELFPTELSGEEGPTGPKEKDNWLGDEYC 560
EN+ +LFP L GEEGPTGPKEKDNW GDE C
Sbjct: 536 ENIVDLFPIALYGEEGPTGPKEKDNWEGDEMC 567
>UniRef100_Q940K1 Hypothetical protein [Arabidopsis thaliana]
Length = 553
Score = 757 bits (1954), Expect = 0.0
Identities = 357/563 (63%), Positives = 436/563 (77%), Gaps = 20/563 (3%)
Query: 5 ECFCWD---SFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWR 61
+CF W+ S E S+ PFSLPSPLPQWPQG GFA GRISLG+I+V KV +F++VW+
Sbjct: 4 DCFYWNKGFSELESESSESKPFSLPSPLPQWPQGRGFATGRISLGEIQVVKVTEFDRVWK 63
Query: 62 CTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESES 121
C S GK +FY+P+ IP+GF CLG+YC N+QPLRG VL AR AD
Sbjct: 64 CGTSRGKLRCASFYKPVGIPEGFHCLGHYCQPNNQPLRGFVLAARANKPGHLAD---DHR 120
Query: 122 PALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLT 181
P LKKPLNYSL+W DS +C YFWLPNPP GY+AVG++VT +EP+ +EVRCVR DLT
Sbjct: 121 PPLKKPLNYSLVWSSDS--DC-YFWLPNPPVGYRAVGVIVTDGSEEPEVDEVRCVREDLT 177
Query: 182 EVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTY---FDSEQVVDVVC 238
E CET + +L + SF VW+T+PC+RG+ +RGV VG+F C T D++ +++ C
Sbjct: 178 ESCETGEKVLGV----GSFNVWSTKPCERGIWSRGVEVGSFVCSTNDLSSDNKAAMNIAC 233
Query: 239 LKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGK 298
LKNLD L MPNL+Q+HALI HYGP VYFHP+E YMPSSV WFFKNGA+L+ G ++G+
Sbjct: 234 LKNLDPSLQGMPNLDQVHALIHHYGPMVYFHPEETYMPSSVPWFFKNGALLHRFGKSQGE 293
Query: 299 AIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAM 358
I+ G+NLP GG NDG+FWIDLP DE+ RSNLKKGNIES+ELYVHVKPALGG FTD+ M
Sbjct: 294 PINSAGSNLPAGGENDGSFWIDLPEDEEVRSNLKKGNIESSELYVHVKPALGGIFTDVVM 353
Query: 359 WVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVN 418
W+FCPFNGPATLK+ L+ + MN++GEHVGDWEHFT R+SNF G+L +FFS+HSGG WV+
Sbjct: 354 WIFCPFNGPATLKIGLLTVPMNRLGEHVGDWEHFTFRISNFNGDLTQMFFSQHSGGGWVD 413
Query: 419 AFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYK 477
DLEF+K NKP+VYSS+HGHAS+PH G YLQG SKLGIGVRND AKS +++DSS RY+
Sbjct: 414 VSDLEFVKGSNKPVVYSSKHGHASFPHPGMYLQGPSKLGIGVRNDVAKSKYMVDSSQRYR 473
Query: 478 IVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPT 537
IVAAEYLG+G ++EP WLQ+MREWGPTIVYDS +EI KIID+LP+ +R S E+ LFP
Sbjct: 474 IVAAEYLGEGAVSEPYWLQFMREWGPTIVYDSAAEINKIIDLLPLILRNSFES---LFPI 530
Query: 538 ELSGEEGPTGPKEKDNWLGDEYC 560
EL GEEGPTGPKEKDNW GDE C
Sbjct: 531 ELYGEEGPTGPKEKDNWEGDEIC 553
>UniRef100_Q8LBJ8 Hypothetical protein [Arabidopsis thaliana]
Length = 553
Score = 756 bits (1951), Expect = 0.0
Identities = 356/563 (63%), Positives = 436/563 (77%), Gaps = 20/563 (3%)
Query: 5 ECFCWD---SFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWR 61
+CF W+ S E S+ PFSLPSPLPQWPQG GFA GRI+LG+I+V V +F++VW+
Sbjct: 4 DCFYWNKGFSELESESSESKPFSLPSPLPQWPQGRGFATGRINLGEIQVVNVTEFDRVWK 63
Query: 62 CTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESES 121
C S GK +FY+P+ IP+GF CLG+YC N+QPLRG VL AR AD
Sbjct: 64 CGTSRGKLRCASFYKPVGIPEGFHCLGHYCQPNNQPLRGFVLAARANEPGHLAD---DHL 120
Query: 122 PALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLT 181
P LKKPLNYSL+W DS +C YFWLPNPP GY+AVG++VT +EP+ +EVRCVR DLT
Sbjct: 121 PPLKKPLNYSLVWSSDS--DC-YFWLPNPPVGYRAVGVIVTDGSEEPEVDEVRCVREDLT 177
Query: 182 EVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTY---FDSEQVVDVVC 238
E CET + +L + SF VW+T+PC+RG+ +RGV VG+FFC T D++ +++ C
Sbjct: 178 ESCETGEKVLGV----GSFNVWSTKPCERGIWSRGVEVGSFFCSTNDLSSDNKAAMNIAC 233
Query: 239 LKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGK 298
LKNLD L MPNL+Q+HALI HYGP VYFHP+E YMPSSV WFFKNGA+L+ ++G+
Sbjct: 234 LKNLDPSLQGMPNLDQVHALIHHYGPMVYFHPEETYMPSSVPWFFKNGALLHRFEKSQGE 293
Query: 299 AIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAM 358
I+ G+NLP GG NDG+FWIDLP D++ RSNLKKGNIES+ELYVHVKPALGG FTDI M
Sbjct: 294 PINSTGSNLPAGGENDGSFWIDLPEDDEVRSNLKKGNIESSELYVHVKPALGGIFTDIVM 353
Query: 359 WVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVN 418
W+FCPFNGPATLK+ L+ + MN++GEHVGDWEHFT R+SNF G+L +FFS+HSGG WV+
Sbjct: 354 WIFCPFNGPATLKIGLLTVPMNRLGEHVGDWEHFTFRISNFNGDLTQMFFSQHSGGGWVD 413
Query: 419 AFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYK 477
DLEF+K NKP+VYSS+HGHAS+PH G YLQG SKLGIGVRND AKS +++DSS RYK
Sbjct: 414 VSDLEFVKGSNKPVVYSSKHGHASFPHPGMYLQGPSKLGIGVRNDVAKSKYMVDSSQRYK 473
Query: 478 IVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPT 537
IVAAEYLG+G ++EP WLQ+MREWGPTIVYDS +EI KIID+LP+ +R+S E+ LFP
Sbjct: 474 IVAAEYLGEGAVSEPYWLQFMREWGPTIVYDSAAEINKIIDLLPLILRYSFES---LFPI 530
Query: 538 ELSGEEGPTGPKEKDNWLGDEYC 560
EL GEEGPTGPKEKDNW GDE C
Sbjct: 531 ELYGEEGPTGPKEKDNWEGDEIC 553
>UniRef100_O64496 F20D22.14 protein [Arabidopsis thaliana]
Length = 1345
Score = 657 bits (1696), Expect = 0.0
Identities = 310/577 (53%), Positives = 410/577 (70%), Gaps = 22/577 (3%)
Query: 1 MFGFECFCWDSFPEFFD-SDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKV 59
M G++C W++ + DP FSLPS +P WP G GF G I+LGK++V K+ FE +
Sbjct: 774 MLGYKCLHWNNLIDLPPLKDPETFSLPSSIPHWPPGQGFGSGTINLGKLQVIKITDFEFI 833
Query: 60 WRCTNSNGKALGFTFYRPLEI-PDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSE 118
WR S K +FY+P + P F CLG+YC S+ PLRG+VL AR+ + +
Sbjct: 834 WRY-RSTEKKKNISFYKPKGLLPKDFHCLGHYCQSDSHPLRGYVLAARDLVDSLE----Q 888
Query: 119 SESPALKKPLNYSLIWCMD--------SHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKA 170
E PAL +P++++L+W + S EC YFWLP PP+GY+++G VVT +P+
Sbjct: 889 VEKPALVEPVDFTLVWSSNDSAENECSSKSECGYFWLPQPPEGYRSIGFVVTKTSVKPEL 948
Query: 171 EEVRCVRTDLTEVCETSDLLLTIKSKKNSFQ--VWNTQPCDRGMLARGVSVGTFFCGTYF 228
EVRCVR DLT++CE ++++T S+ +W T+P DRGM +GVS GTFFC T
Sbjct: 949 NEVRCVRADLTDICEPHNVIVTAVSESLGVPLFIWRTRPSDRGMWGKGVSAGTFFCRTRL 1008
Query: 229 DSEQV---VDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKN 285
+ + + + CLKNLD LHAMPN++QI ALI+HYGPT+ FHP E Y+PSSVSWFFKN
Sbjct: 1009 VAAREDLGIGIACLKNLDLSLHAMPNVDQIQALIQHYGPTLVFHPGETYLPSSVSWFFKN 1068
Query: 286 GAILYTAGNAKGKAIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHV 345
GA+L GN + ID +G+NLP GG ND FWIDLP D+ R +K+GN+ES++LY+H+
Sbjct: 1069 GAVLCEKGNPIEEPIDENGSNLPQGGSNDKQFWIDLPCDDQQRDFVKRGNLESSKLYIHI 1128
Query: 346 KPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWS 405
KPALGG FTD+ W+FCPFNGPATLK+ L++I + IG+HV DWEHFTLR+SNF+GEL+S
Sbjct: 1129 KPALGGTFTDLVFWIFCPFNGPATLKLGLVDISLISIGQHVCDWEHFTLRISNFSGELYS 1188
Query: 406 VFFSEHSGGKWVNAFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAA 464
++ S+HSGG+W+ A+DLE I NK +VYSS+HGHAS+P AGTYLQGS+ LGIG+RND A
Sbjct: 1189 IYLSQHSGGEWIEAYDLEIIPGSNKAVVYSSKHGHASFPRAGTYLQGSTMLGIGIRNDTA 1248
Query: 465 KSNFILDSSFRYKIVAAEYL-GDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIF 523
+S ++DSS RY+I+AAEYL G+ V+ EP WLQYMREWGP +VYDSR EIE++++ P
Sbjct: 1249 RSELLVDSSSRYEIIAAEYLSGNSVLAEPPWLQYMREWGPKVVYDSREEIERLVNRFPRT 1308
Query: 524 VRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDEYC 560
VR S+ + P ELSGEEGPTGPKEK+NW GDE C
Sbjct: 1309 VRVSLATVLRKLPVELSGEEGPTGPKEKNNWYGDERC 1345
>UniRef100_Q9FND1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRH10
[Arabidopsis thaliana]
Length = 566
Score = 640 bits (1652), Expect = 0.0
Identities = 308/573 (53%), Positives = 409/573 (70%), Gaps = 24/573 (4%)
Query: 1 MFGFECFCWDSFPEFFD-SDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKV 59
MFG +C W++ E+ +P FSLP+ LPQWP G GF GRI+LG++EV ++ FE V
Sbjct: 1 MFGCKCLYWNNLKEYPPLKEPETFSLPASLPQWPSGQGFGLGRINLGELEVAEITSFEFV 60
Query: 60 WRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSES 119
WR + +FY+P ++P+ F CLG+YC S+ LRG +LVAR+ S+
Sbjct: 61 WRYCSRRDNKKSVSFYKPDKLPEDFHCLGHYCQSDSHLLRGFLLVARQVNKSSE------ 114
Query: 120 ESPALKKPLNYSLIWCMDSHDE------CVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEV 173
PAL +PL+Y+L+W + E YFWLP PP+GYK +G +VTT+P +P+ ++V
Sbjct: 115 --PALVQPLDYTLVWSSNDLSEERQSESYGYFWLPQPPQGYKPIGYLVTTSPAKPELDQV 172
Query: 174 RCVRTDLTEVCETSDLLLTIKSKKNSFQ--VWNTQPCDRGMLARGVSVGTFFCGTYFDSE 231
RCVR DLT+ CE +++T S S +W T+P DRGM +GVS GTFFC T E
Sbjct: 173 RCVRADLTDKCEAHKVIITAISDSLSIPMFIWKTRPSDRGMRGKGVSTGTFFCTTQSPEE 232
Query: 232 QVVDVV-CLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILY 290
+ + CLKNLDS LHAMPN+ QIHA+I+HYGP VYFHP+E Y+PSSVSWFFKNGA+L
Sbjct: 233 DHLSTIACLKNLDSSLHAMPNIEQIHAMIQHYGPRVYFHPNEVYLPSSVSWFFKNGALLC 292
Query: 291 TAGNAK---GKAIDYHGTNLPGGGYNDGAFWIDLP-TDEDARSNLKKGNIESAELYVHVK 346
+ N+ + ID G+NLP GG ND +WIDLP D+ R +K+G++ES++LYVHVK
Sbjct: 293 SNSNSSVINNEPIDETGSNLPHGGTNDKRYWIDLPINDQQRREFIKRGDLESSKLYVHVK 352
Query: 347 PALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSV 406
PA GG FTD+A W+FCPFNGPATLK+ LM++ + K G+HV DWEHFT+R+SNF+GEL+S+
Sbjct: 353 PAFGGTFTDLAFWIFCPFNGPATLKLGLMDLSLAKTGQHVCDWEHFTVRISNFSGELYSI 412
Query: 407 FFSEHSGGKWVNAFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAK 465
+FS+HSGG+W+ +LEF++ NK +VYSS++GHAS+ +G YLQGS+ LGIG+RND+AK
Sbjct: 413 YFSQHSGGEWIKPENLEFVEGSNKAVVYSSKNGHASFSKSGMYLQGSALLGIGIRNDSAK 472
Query: 466 SNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVR 525
S+ +DSS +Y+IVAAEYL G + EP WL YMREWGP IVY+SRSEIEK+ + LP +R
Sbjct: 473 SDLFVDSSLKYEIVAAEYL-RGAVVEPPWLGYMREWGPKIVYNSRSEIEKLNERLPWRLR 531
Query: 526 FSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
V+ + P ELSGEEGPTGPKEK+NW GDE
Sbjct: 532 SWVDAVLRKIPVELSGEEGPTGPKEKNNWFGDE 564
>UniRef100_Q8LLP6 OSJNBa0031O09.02 [Oryza sativa]
Length = 576
Score = 580 bits (1494), Expect = e-164
Identities = 289/567 (50%), Positives = 365/567 (63%), Gaps = 31/567 (5%)
Query: 17 DSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFYR 76
+++P F+ LP WPQGGGFA GRI +G++E+ V FEK+ + + G G TFYR
Sbjct: 14 ETEPARFAFADALPAWPQGGGFATGRICVGELELAAVTAFEKICALSATKGGGGGVTFYR 73
Query: 77 PLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESES-PALKKPLNYSLIWC 135
P +P GFS LG+YC SN +PL GH+LVA+ K ESES P L+ P +Y L+
Sbjct: 74 PAGVPGGFSVLGHYCQSNTRPLHGHLLVAKAVAGKP-----ESESLPPLRPPHDYELVCA 128
Query: 136 M------DSHDEC-------VYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTE 182
+ C YFWLP P GY+A+G++VT PD+P EV C R DLT+
Sbjct: 129 FRADGVGEDRKSCRGYGRTGAYFWLPVPTDGYRALGLLVTAEPDKPPLREVACARADLTD 188
Query: 183 VCETSDLLLTIKSKKNS-----------FQVWNTQPCDRGMLARGVSVGTFFCGTYFDSE 231
CE LL ++ S F + +P RGM RG+ GTF CG S
Sbjct: 189 ECEPHGSLLQLQLVGQSACWSSSTVPAAFALRGIRPTHRGMWGRGIGAGTFCCGAVGLSP 248
Query: 232 QVVDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYT 291
+ + CLKN+D L AMP L Q HA+I HYGPT+YFHP E Y+PSSVSWFFKNGA L
Sbjct: 249 REQGMACLKNVDLDLSAMPTLEQAHAVIRHYGPTLYFHPKEVYLPSSVSWFFKNGAALCK 308
Query: 292 AGNAKGKAIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGG 351
G +D G++LP G NDG +WI LP + S + G+I+SAELY HVKPA+GG
Sbjct: 309 KGEDAAVELDVEGSHLPCGECNDGEYWIGLPDGKRGESIIY-GDIDSAELYAHVKPAMGG 367
Query: 352 AFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEH 411
TD+AMWVFCPFNGPA K+ + I + K G+H+GDWEHFTLRVSNFTGEL +V+FS+H
Sbjct: 368 TCTDVAMWVFCPFNGPARFKLGPITIPLGKTGQHIGDWEHFTLRVSNFTGELMAVYFSQH 427
Query: 412 SGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILD 471
SGG+WV+A LE+ NKP VYSSR+GHASYP G YLQGS+ LGIG+RNDAA+S +D
Sbjct: 428 SGGRWVDASALEYTAGNKPAVYSSRNGHASYPFPGVYLQGSAALGIGIRNDAARSELAVD 487
Query: 472 SSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENL 531
SS +Y+IVAAEYLG+G + EP WL +MR WGPT+VY SR +E++ + +R E +
Sbjct: 488 SSAKYRIVAAEYLGEGAVEEPRWLNFMRVWGPTVVYKSRQRMERMTSAMHRRLRSPAERM 547
Query: 532 FELFPTELSGEEGPTGPKEKDNWLGDE 558
P ELS EEGPTGPKEK+NW GDE
Sbjct: 548 LNKLPNELSREEGPTGPKEKNNWEGDE 574
>UniRef100_O64861 Expressed protein [Arabidopsis thaliana]
Length = 553
Score = 498 bits (1283), Expect = e-139
Identities = 250/544 (45%), Positives = 343/544 (62%), Gaps = 28/544 (5%)
Query: 23 FSLPSPLPQWPQGGGFAGGRISLGK-IEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIP 81
F PSPLP + +G GFA G I LG +EV++V+ F KVW LG TF+ P IP
Sbjct: 29 FKFPSPLPTFTRGDGFAKGTIDLGGGLEVSQVSTFNKVWSTYEGGPDNLGATFFEPSSIP 88
Query: 82 DGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWCMDS--- 138
GFS LGYY N++ L G VL AR+ +S + LK P++Y+L+ +S
Sbjct: 89 SGFSILGYYAQPNNRNLFGWVLTARDLSSNT-----------LKPPVDYTLVGNTESLKI 137
Query: 139 -HDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKK 197
D YFW P PP GY+AVG++VT +P +++RC+R+DLTE CE +
Sbjct: 138 KQDGTGYFWQPVPPDGYQAVGLIVTNYSQKPPLDKLRCIRSDLTEQCEADTWIWGT---- 193
Query: 198 NSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHA 257
N + N +P RG A GV VGTF T S + CLKN MPN +QI
Sbjct: 194 NGVNISNLKPTTRGTQATGVYVGTFTWQTQNSSPPSLS--CLKNTKLDFSTMPNGSQIEE 251
Query: 258 LIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGN-AKGKAIDYHGTNLPGGGYNDGA 316
L + + P +YFHPDE+Y+PSSV+W+F NGA+LY G +K I+ +G+NLP GG NDG+
Sbjct: 252 LFQTFSPCIYFHPDEEYLPSSVTWYFNNGALLYKKGEESKPIPIESNGSNLPQGGSNDGS 311
Query: 317 FWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMN 376
+W+DLP D++ + +KKG+++S ++Y+H+KP LG FTDI++W+F PFNGPA KV +N
Sbjct: 312 YWLDLPIDKNGKERVKKGDLQSTKVYLHIKPMLGATFTDISIWIFYPFNGPAKAKVKFVN 371
Query: 377 IEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK--ENKPIVYS 434
+ + +IGEH+GDWEH TLR+SNFTGELW VF S+HSGG W++A DLEF NK + Y+
Sbjct: 372 LPLGRIGEHIGDWEHTTLRISNFTGELWRVFLSQHSGGIWIDACDLEFQDGGNNKFVAYA 431
Query: 435 SRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCW 494
S HGHA YP G LQG G+G+RND K +LD+ Y+++AAEY G GV+ EP W
Sbjct: 432 SLHGHAMYPKPGLVLQGDD--GVGIRNDTGKGKKVLDTGLGYEVIAAEYDGGGVV-EPPW 488
Query: 495 LQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNW 554
++Y R+WGP I Y+ E++ + +LP ++ + + P E+ GE+GPTGPK K NW
Sbjct: 489 VKYFRKWGPKIDYNVDDEVKSVERILPGLLKKAFVKFVKKIPDEVYGEDGPTGPKLKSNW 548
Query: 555 LGDE 558
GDE
Sbjct: 549 AGDE 552
>UniRef100_O64858 Expressed protein [Arabidopsis thaliana]
Length = 542
Score = 493 bits (1270), Expect = e-138
Identities = 252/549 (45%), Positives = 349/549 (62%), Gaps = 40/549 (7%)
Query: 23 FSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIPD 82
F+LPSPLP WP G GFA GRI LG +EV++V+ F KVW LG TF+ P +P+
Sbjct: 20 FNLPSPLPSWPSGEGFAKGRIDLGGLEVSQVDTFNKVWTVYEGGQDNLGATFFEPSSVPE 79
Query: 83 GFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWCMDSHD-- 140
GFS LG+Y N++ L G LV ++ + S L+ P++Y L+W S
Sbjct: 80 GFSILGFYAQPNNRKLFGWTLVGKDLSGDS-----------LRPPVDYLLLWSGKSTKVE 128
Query: 141 ----ECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSK 196
E YFW P PP GY AVG++VTT+ ++P +++RCVR+DLT+ E L+ +
Sbjct: 129 NNKVETGYFWQPVPPDGYNAVGLIVTTSDEKPPLDKIRCVRSDLTDQSEPDALIW----E 184
Query: 197 KNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIH 256
N F V +++P +RG A GVSVGTFF + + + CLKN + MP+ QI
Sbjct: 185 TNGFSVSSSKPVNRGTQASGVSVGTFFSNSPNPA-----LPCLKNNNFDFSCMPSKPQID 239
Query: 257 ALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTNLPGGGYNDG 315
AL + Y P +YFH DEKY+PSSV+WFF NGA+LY G+ ++ +G NLP G +NDG
Sbjct: 240 ALFQTYAPWIYFHKDEKYLPSSVNWFFSNGALLYKKGDESNPVPVEPNGLNLPQGEFNDG 299
Query: 316 AFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLM 375
+W+DLP DAR ++ G+++S E+Y+H+KP GG FTDIA+W+F PFNGP+ K+
Sbjct: 300 LYWLDLPVASDARKRVQCGDLQSMEVYLHIKPVFGGTFTDIAVWMFYPFNGPSRAKLKAA 359
Query: 376 NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFI-KENKPIVYS 434
+I + +IGEH+GDWEHFTLR+SNF+G+L ++ S+HSGG W +A ++EF NKP+ Y+
Sbjct: 360 SIPLGRIGEHIGDWEHFTLRISNFSGKLHRMYLSQHSGGSWADASEIEFQGGGNKPVAYA 419
Query: 435 SRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCW 494
S +GHA Y G LQG K +G+RND KS ++D++ R+++VAAEY+ G + EP W
Sbjct: 420 SLNGHAMYSKPGLVLQG--KDNVGIRNDTGKSEKVIDTAVRFRVVAAEYM-RGELEEPAW 476
Query: 495 LQYMREWGPTIVYDSRSEI---EKII--DMLPIFVRFSVENLFELFPTELSGEEGPTGPK 549
L YMR WGP I Y +EI EKI+ + L R +++ L P E+ GEEGPTGPK
Sbjct: 477 LNYMRHWGPKIDYGHENEIRGVEKIMVGESLKTTFRSAIKGL----PNEVFGEEGPTGPK 532
Query: 550 EKDNWLGDE 558
K NWLGDE
Sbjct: 533 LKRNWLGDE 541
>UniRef100_Q6ZLA9 Hypothetical protein OJ1065_B06.1 [Oryza sativa]
Length = 522
Score = 464 bits (1193), Expect = e-129
Identities = 244/529 (46%), Positives = 324/529 (61%), Gaps = 29/529 (5%)
Query: 36 GGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSND 95
GGFA G I LG +EV +V F KVW G G TF+RP ++P GFS LG+Y ND
Sbjct: 13 GGFAKGSIDLGGLEVRQVTTFAKVWSTGQDGG---GATFFRPEQVPAGFSALGHYAQRND 69
Query: 96 QPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYK 155
+PL GHVLVAR+ S L PL+Y+ +W S D+ +FWLP PP GY+
Sbjct: 70 RPLFGHVLVARDV----------SGGGLLAPPLDYAPVW--SSQDDAAHFWLPTPPDGYR 117
Query: 156 AVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLAR 215
A+G+ VT +PD+P +EV CVR D T+ CE + K+ F +P RG+ AR
Sbjct: 118 AIGVAVTASPDKPPRDEVACVRADFTDACEAEATVWD----KDGFSAVALRPAVRGVDAR 173
Query: 216 GVSVGTFFCG-TYFDSEQVVDVVCLKNLDSLLHA-MPNLNQIHALIEHYGPTVYFHPDEK 273
GV GTF + + + CLKN + + MP+L Q++AL+ Y P ++ HPDE
Sbjct: 174 GVHAGTFVLARSDATAASASALACLKNNGAAYTSCMPDLAQVNALLAAYAPQLFLHPDEP 233
Query: 274 YMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKK 333
Y+PSSV+WFF+NGA+LY G+ + G+NLP GG NDG +W+DLP D R +KK
Sbjct: 234 YLPSSVTWFFQNGALLYQKGSQTPTPVAADGSNLPQGGGNDGGYWLDLPVDNFQRERVKK 293
Query: 334 GNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFT 393
G++ A++YV KP LG TD+A+W F PFNGPA KV + I + KIGEHVGDWEH T
Sbjct: 294 GDLAGAKVYVQAKPMLGATATDLAVWFFYPFNGPARAKVGPLTIPLGKIGEHVGDWEHVT 353
Query: 394 LRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGS 452
LRVSNF+GEL ++FS+HS G WV+A LE++ N+P YSS HGHA YP AG LQG
Sbjct: 354 LRVSNFSGELLRMYFSQHSAGAWVDAPQLEYLDGGNRPSAYSSLHGHALYPRAGLVLQGD 413
Query: 453 SKLGIGVRNDAAK-SNFILDSSFRYKIVAAEYL--GDGVITEPCWLQYMREWGPTIVYDS 509
++LG+G+RND + S + R ++V+AEYL G G + EP WL + REWGP YD
Sbjct: 414 ARLGVGIRNDCDRGSRLDTGGAGRCEVVSAEYLGGGGGGVAEPTWLLFDREWGPREEYDI 473
Query: 510 RSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
EI ++ +LP R + E L +L + G EGPTGP+ K +W DE
Sbjct: 474 GREINRVAKLLP---RSTRERLRKLVESVFVG-EGPTGPRMKGSWRNDE 518
>UniRef100_Q9SGI4 F28J7.20 protein [Arabidopsis thaliana]
Length = 583
Score = 421 bits (1083), Expect = e-116
Identities = 234/561 (41%), Positives = 319/561 (56%), Gaps = 40/561 (7%)
Query: 15 FFDSDPLP----FSLPSPLPQWPQGGG-FAGGRISLGKIEVTKVN----KFEKVWRCTNS 65
F S LP F+ PS LP P GGG F GRI LG +EV +V+ ++VWR
Sbjct: 46 FLSSTSLPVETAFTFPSALPVIPSGGGNFGKGRIDLGGLEVIQVSISTSTSQRVWRTYEG 105
Query: 66 NGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALK 125
+G + ++P+ +P FS LG+Y N++ L G VL AR+ + S L+
Sbjct: 106 GPDNMGLSIFQPINLPPSFSTLGFYGQPNNRLLFGWVLAARDVSGNS-----------LR 154
Query: 126 KPLNYSLIWCMDS----HDECVYFWLPNPPKGYKAVGIVVTTNPDEPKA--EEVRCVRTD 179
P++Y + S + +FW P P GY+AVG+ VTT+P +P E + CVR+D
Sbjct: 155 PPVDYIQVINTTSMNINQEGAAFFWQPLCPNGYQAVGLYVTTSPIKPSLSQESISCVRSD 214
Query: 180 LTEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCL 239
LTE ET + + S + +P +RG A GV GTF C + + CL
Sbjct: 215 LTEQSETDTWVWGTEEMTLS----SLRPANRGTEATGVHTGTFSCQP-LNIPPPPPLFCL 269
Query: 240 KNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA 299
KN L +MP+ NQ L + Y P +Y HPDE ++ SSV WFF NGA+L+ GN
Sbjct: 270 KNTKFDLSSMPSHNQTTVLFQSYSPWIYLHPDEDFISSSVDWFFSNGALLFQKGNESNPV 329
Query: 300 -IDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAM 358
+ G+NLP GG +DG FW+D P D++A+ +K+G++ ++Y+H+KP GG FTDI +
Sbjct: 330 PVQPDGSNLPQGGSDDGLFWLDYPADKNAKEWVKRGDLGHTKVYLHIKPMFGGTFTDIVV 389
Query: 359 WVFCPFNGPATLK-VSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWV 417
W+F PFNG A LK + ++ + IGEH+GDWEH TLR+SNF GELW +FSEHSGG V
Sbjct: 390 WIFYPFNGNARLKFLFFKSLSLGDIGEHIGDWEHVTLRISNFNGELWRAYFSEHSGGTLV 449
Query: 418 NAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYK 477
A DLEF NK + YSS HGHA + G LQG G G+RND A+SN D+ Y+
Sbjct: 450 EACDLEFQGGNKLVSYSSLHGHAMFSKPGLVLQGDD--GNGIRNDMARSNKFFDAGVAYE 507
Query: 478 IVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPT 537
+VA G G I EP WL Y R+WGP + +D + +E I LP +R NL P
Sbjct: 508 LVA----GPG-IQEPPWLNYFRKWGPLVPHDIQKNLEGIAKSLPGLLRKKFRNLINKIPR 562
Query: 538 ELSGEEGPTGPKEKDNWLGDE 558
E+ E+GPTGPK K +W GD+
Sbjct: 563 EVLEEDGPTGPKVKRSWTGDD 583
>UniRef100_Q9SG97 F1C9.35 protein [Arabidopsis thaliana]
Length = 549
Score = 421 bits (1083), Expect = e-116
Identities = 234/561 (41%), Positives = 319/561 (56%), Gaps = 40/561 (7%)
Query: 15 FFDSDPLP----FSLPSPLPQWPQGGG-FAGGRISLGKIEVTKVN----KFEKVWRCTNS 65
F S LP F+ PS LP P GGG F GRI LG +EV +V+ ++VWR
Sbjct: 12 FLSSTSLPVETAFTFPSALPVIPSGGGNFGKGRIDLGGLEVIQVSISTSTSQRVWRTYEG 71
Query: 66 NGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALK 125
+G + ++P+ +P FS LG+Y N++ L G VL AR+ + S L+
Sbjct: 72 GPDNMGLSIFQPINLPPSFSTLGFYGQPNNRLLFGWVLAARDVSGNS-----------LR 120
Query: 126 KPLNYSLIWCMDS----HDECVYFWLPNPPKGYKAVGIVVTTNPDEPKA--EEVRCVRTD 179
P++Y + S + +FW P P GY+AVG+ VTT+P +P E + CVR+D
Sbjct: 121 PPVDYIQVINTTSMNINQEGAAFFWQPLCPNGYQAVGLYVTTSPIKPSLSQESISCVRSD 180
Query: 180 LTEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCL 239
LTE ET + + S + +P +RG A GV GTF C + + CL
Sbjct: 181 LTEQSETDTWVWGTEEMTLS----SLRPANRGTEATGVHTGTFSCQP-LNIPPPPPLFCL 235
Query: 240 KNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA 299
KN L +MP+ NQ L + Y P +Y HPDE ++ SSV WFF NGA+L+ GN
Sbjct: 236 KNTKFDLSSMPSHNQTTVLFQSYSPWIYLHPDEDFISSSVDWFFSNGALLFQKGNESNPV 295
Query: 300 -IDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAM 358
+ G+NLP GG +DG FW+D P D++A+ +K+G++ ++Y+H+KP GG FTDI +
Sbjct: 296 PVQPDGSNLPQGGSDDGLFWLDYPADKNAKEWVKRGDLGHTKVYLHIKPMFGGTFTDIVV 355
Query: 359 WVFCPFNGPATLK-VSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWV 417
W+F PFNG A LK + ++ + IGEH+GDWEH TLR+SNF GELW +FSEHSGG V
Sbjct: 356 WIFYPFNGNARLKFLFFKSLSLGDIGEHIGDWEHVTLRISNFNGELWRAYFSEHSGGTLV 415
Query: 418 NAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYK 477
A DLEF NK + YSS HGHA + G LQG G G+RND A+SN D+ Y+
Sbjct: 416 EACDLEFQGGNKLVSYSSLHGHAMFSKPGLVLQGDD--GNGIRNDMARSNKFFDAGVAYE 473
Query: 478 IVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPT 537
+VA G G I EP WL Y R+WGP + +D + +E I LP +R NL P
Sbjct: 474 LVA----GPG-IQEPPWLNYFRKWGPLVPHDIQKNLEGIAKSLPGLLRKKFRNLINKIPR 528
Query: 538 ELSGEEGPTGPKEKDNWLGDE 558
E+ E+GPTGPK K +W GD+
Sbjct: 529 EVLEEDGPTGPKVKRSWTGDD 549
>UniRef100_Q9S7Q5 F1C9.34 protein [Arabidopsis thaliana]
Length = 592
Score = 419 bits (1078), Expect = e-116
Identities = 232/562 (41%), Positives = 322/562 (57%), Gaps = 40/562 (7%)
Query: 15 FFDSDPLP----FSLPSPLPQWPQGGGFAGGR-ISLGKIEVTKVN----KFEKVWRCTNS 65
+ S+ LP F PSPLP P GG G R I +G +EVT+++ +VWR
Sbjct: 50 YLSSNGLPVETSFKFPSPLPSMPSDGGNFGKRSIDMGGLEVTQISISNSTSHRVWRTYEG 109
Query: 66 NGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALK 125
+G + + P IP F LG+Y N++ L G +LVA++ + + L+
Sbjct: 110 GPDNMGVSIFEPTTIPRNFFKLGFYAQPNNRQLFGWILVAKDVSGSN-----------LR 158
Query: 126 KPLNYSLIW----CMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEE--VRCVRTD 179
P++Y+ + + + YFW P P GY AVG+ VTT+P +P + + CVR+D
Sbjct: 159 PPVDYTEVGNTTTLLIKQEGPAYFWQPLCPNGYHAVGLYVTTSPMKPSLGQNSISCVRSD 218
Query: 180 LTEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCL 239
LTE E + IK S + +P RG+ A GV GTF C + CL
Sbjct: 219 LTEQSEADTWVWRIKDMTIS----SLRPATRGVEATGVFTGTFSCKQLNFLPHPPPLFCL 274
Query: 240 KNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA 299
KN L +MP+ NQ L + Y P +Y HP E ++PSSV+W F NGA+L+ GN
Sbjct: 275 KNTKFDLSSMPSENQTRVLFKTYSPWIYLHPKEDFLPSSVNWVFANGALLHKKGNESIPV 334
Query: 300 -IDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAM 358
I +G+NLP GG ND FW+D D+ AR +K+G++ES ++Y+H+KP G FTDI +
Sbjct: 335 PIHPNGSNLPQGGCNDDLFWLDYLVDKKAREKVKRGDLESTKVYLHIKPMFGATFTDIVV 394
Query: 359 WVFCPFNGPATLK-VSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWV 417
W+F P+NG A LK + + ++ + IGEHVGDWEH TLR+SNF GELW V+FSEHSGG V
Sbjct: 395 WLFFPYNGNAHLKFLFIKSLSLGNIGEHVGDWEHVTLRISNFNGELWRVYFSEHSGGTLV 454
Query: 418 NAFDLEFIK-ENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRY 476
+A DLEF++ NKP+VYSS HGHA + G LQG K GI RND A+S+ D+ Y
Sbjct: 455 DACDLEFMQGGNKPVVYSSLHGHAMFSKPGVVLQGGGKSGI--RNDMARSDKCFDAGIGY 512
Query: 477 KIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFP 536
+++A G GV+ EP WL Y R+WGP + Y + + +LPIF+R + L P
Sbjct: 513 EVIA----GPGVV-EPPWLNYFRKWGPRVHYRIDIFLNSVAKILPIFLRKGLRKLINKIP 567
Query: 537 TELSGEEGPTGPKEKDNWLGDE 558
E+ G++GPTGPK K W GDE
Sbjct: 568 LEMRGQDGPTGPKVKVTWTGDE 589
>UniRef100_Q97KX3 Uncharacterized conserved protein of probably eukaryotic origin
[Clostridium acetobutylicum]
Length = 348
Score = 105 bits (262), Expect = 3e-21
Identities = 85/312 (27%), Positives = 138/312 (43%), Gaps = 31/312 (9%)
Query: 262 YGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDGA----F 317
Y P VYFHPDEK P +V F + ++ N K + G + G + D F
Sbjct: 45 YAPIVYFHPDEKCFPITVEEFLECTDVM----NEKNEKF-CDGKEILSGSFKDALGNQKF 99
Query: 318 WIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPAT---LKVSL 374
++ + TD +++++GN+ SA+ YV+V+ G + D+ + F +NGP L +
Sbjct: 100 YLKI-TD----TSVREGNLNSAKCYVYVRKRNKGRYLDLQYFFFYGYNGPTINIHLNKNK 154
Query: 375 MNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYS 434
N + IGE+ G+WEH T+ + + G + F +H + + LE+ ++ +VYS
Sbjct: 155 KNYSTSNIGENYGNWEHVTVVIDSTDGSILGAIFDQHGKSCFYDIDQLEY-EKGHIVVYS 213
Query: 435 SRHGHASYPHAGTYLQGSSKLGIGV-----------RNDAAKSNFILDSSFRYKIVAAEY 483
+ HASYP AG + +GV +DA D+S ++V+
Sbjct: 214 ALSSHASYPSAGKSVTEYDSQELGVPPVSVGIVKFELHDATDKGRRWDTSLVCQLVSYNI 273
Query: 484 LGDGVITEPCWLQYMREWG-PTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGE 542
G V EP W+ + WG P + D I I LP + L+ P
Sbjct: 274 TGTEV-EEPKWMAFNGRWGKPYDLTDKDISILYQISRLPAGGKVVDSLLWRFIPYSKKNI 332
Query: 543 EGPTGPKEKDNW 554
G K KD++
Sbjct: 333 NQANGIKSKDDF 344
>UniRef100_UPI000042E5AF UPI000042E5AF UniRef100 entry
Length = 442
Score = 63.9 bits (154), Expect = 1e-08
Identities = 61/206 (29%), Positives = 88/206 (42%), Gaps = 35/206 (16%)
Query: 262 YGPTVYFHPDEKYMPSSVSWF-------FKNGAILYTAGNAKGKAIDYHGTNLPGG---- 310
Y P V+ + +E+Y P V F ++NG+I G K +D NLP
Sbjct: 87 YAPLVHLYSEERYFPYDVKKFVTNFHVTWRNGSIY--PGTEKNMNLDKLA-NLPNSTDLF 143
Query: 311 -----GYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFN 365
++ WI T + +L G I+ A + V G + D + F FN
Sbjct: 144 LTANSDFDSDPEWI---TGLKNKPSLINGEIKDAPATLIVVDK-GNGWVDAFWFYFYSFN 199
Query: 366 -GPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLE- 423
GP + G HVGDWEH +R + GE V+ S HSGG +LE
Sbjct: 200 LGPYVMG-------QGPYGNHVGDWEHSLVRY--YKGEPVIVWMSAHSGGGGYYYDNLEK 250
Query: 424 -FIKENKPIVYSSRHGHASYPHAGTY 448
+ N PI++S+R HA+YP G +
Sbjct: 251 YSLDPNHPIIFSARGTHANYPSVGQH 276
>UniRef100_Q6BUX3 Similar to CA0982|IPF7397 Candida albicans [Debaryomyces hansenii]
Length = 436
Score = 58.5 bits (140), Expect = 5e-07
Identities = 69/274 (25%), Positives = 106/274 (38%), Gaps = 53/274 (19%)
Query: 262 YGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGN-AKGKAIDYHGT------NLPG----- 309
Y P V+ + +EKY+P + + N + Y G+ KG + + + N+P
Sbjct: 79 YAPLVHLYTEEKYLPYDIKDYVTNFFVTYENGSVVKGTEEEMNLSKLSKLPNVPDIFLLS 138
Query: 310 -GGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFN-GP 367
++ W+ T + +L G I++A + V G + D + F FN GP
Sbjct: 139 RTDFDKDPEWV---TGAKNKPSLIDGEIKNAPATLIVVDK-GNGWVDAFWFYFYSFNLGP 194
Query: 368 ATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLE--FI 425
+ G HVGDWEH +R N G V+ S H GG +LE I
Sbjct: 195 FVM-------GSGPYGNHVGDWEHSLVRFYN--GVPVIVWMSAHGGGGAYFYHNLEKWEI 245
Query: 426 KENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLG 485
+ PI++S+R HA+Y G Q L G+ +D + + + Y Y
Sbjct: 246 DDTHPIIFSARGTHANYVSVG---QHPHDLPYGILSDFTDRGPLWNPTKNY----LAYSY 298
Query: 486 DGVITEPC-----------------WLQYMREWG 502
DGV P WL + WG
Sbjct: 299 DGVFVYPAENNTNSRHRGREVEYGNWLSFTGHWG 332
>UniRef100_Q6C6U8 Similar to sp|P53285 Saccharomyces cerevisiae YGR141w [Yarrowia
lipolytica]
Length = 405
Score = 54.7 bits (130), Expect = 7e-06
Identities = 67/291 (23%), Positives = 106/291 (36%), Gaps = 55/291 (18%)
Query: 251 NLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGG 310
N Q+ + + P V+ + +EKY+P + + N I +G + L
Sbjct: 29 NPGQVPDYVLRHCPGVHLYSEEKYLPGDPADYVTNFKIDTASGKTVLNGSESDPVTLARL 88
Query: 311 G------------------YNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGA 352
G +N WI T E +L G I+ A+ + V G
Sbjct: 89 GQYSFEKHSSRTFMTSLTDFNADPDWI---TGEGNIPHLVSGKIDHAKSVLIVVDK-GDG 144
Query: 353 FTDIAMWVFCPFN-GPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEH 411
D + F PFN GP + G HVGDWEH +R + ++ ++ S H
Sbjct: 145 LVDAFWFYFYPFNLGPFVMGGG-------PYGNHVGDWEHSLVRFQDGVPQI--LWMSAH 195
Query: 412 SGGK--WVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFI 469
GG + NA + + +++S+R HA+Y G Q S L + +D +
Sbjct: 196 GGGNAYYYNAAEKMDNDPERRVLFSARGTHANYGSVG---QHSHDLPFYMLSDFTDRGAL 252
Query: 470 LD---SSFRYKIVAAE---------------YLGDGVITEPCWLQYMREWG 502
D + + Y + E DG P WL Y+ WG
Sbjct: 253 WDPVQNYYGYTLTHRERGRTGADRIWFDGRVLTSDGQSPHPSWLNYLGRWG 303
>UniRef100_O23558 Hypothetical protein AT4g17130 [Arabidopsis thaliana]
Length = 747
Score = 52.0 bits (123), Expect = 5e-05
Identities = 38/149 (25%), Positives = 56/149 (37%), Gaps = 28/149 (18%)
Query: 54 NKFEKVWRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQ 113
++F+K+ N + + F+RP P GF+ LG Y D+P VLV +
Sbjct: 525 SEFDKIGTIRNPCTDQI-YAFWRP-HPPPGFASLGDYLTPLDKPPTKGVLVVNTNLMR-- 580
Query: 114 ADCSESESPALKKPLNYSLIWC--------------MDSHDECVYFWLPNPPKGYKAVGI 159
+K+PL++ LIW D D W P PKGY A+
Sbjct: 581 ----------VKRPLSFKLIWSPLASGGLGGSSMDDKDERDSSCSIWFPEAPKGYVALSC 630
Query: 160 VVTTNPDEPKAEEVRCVRTDLTEVCETSD 188
VV++ P C+ C D
Sbjct: 631 VVSSGSTPPSLASTFCILASSVSPCSLRD 659
>UniRef100_Q6FQ07 Similar to sp|P53285 Saccharomyces cerevisiae YGR141w [Candida
glabrata]
Length = 446
Score = 52.0 bits (123), Expect = 5e-05
Identities = 56/219 (25%), Positives = 88/219 (39%), Gaps = 46/219 (21%)
Query: 264 PTVYFHPDEKYMPSSVSWFFKNGAILYTAGNA--------KGKAIDYHGTNLPGGGYNDG 315
P ++ + DEKY PS+ S + + +L G A K Y + G Y
Sbjct: 71 PLIHLYSDEKYWPSNFSEYVSHFKLLDDEGEAILDQLNSIKELKQSYVRYDEDGASYEVD 130
Query: 316 AFWIDLPTDEDARSN---LKKGNIESAELYVHVKPAL------GGAFTDIAMWVFCPFN- 365
+ I + + +D + +K + Y PAL G + D + F PFN
Sbjct: 131 SEDIFMTSIDDFDKDPGWMKGFKPKYGTGYAAQAPALVFVVDKGNGWVDAFWFYFYPFNW 190
Query: 366 GPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFI 425
GP + G H+GDWEH +R N G ++ S H GG +A+ E I
Sbjct: 191 GPFIMGYGPW-------GNHLGDWEHTLVRFYN--GVPKYIWMSAHGGG---SAYTYEAI 238
Query: 426 KENK----------------PIVYSSRHGHASYPHAGTY 448
++ K P+++S+R HA+YP G +
Sbjct: 239 EKKKRLRRIEGRITNELIEKPLLFSARGTHANYPSVGQH 277
>UniRef100_P53285 Hypothetical 54.5 kDa protein in CBF2-SKN1 intergenic region
[Saccharomyces cerevisiae]
Length = 467
Score = 52.0 bits (123), Expect = 5e-05
Identities = 52/217 (23%), Positives = 82/217 (36%), Gaps = 41/217 (18%)
Query: 264 PTVYFHPDEKYMPSSVSWFFKNGAILYTAGNA--------KGKAIDYHGTNLPGGGY--- 312
P ++ + +EKY P+ V F K + +G +Y+ L G +
Sbjct: 92 PLLHLYSEEKYWPADVKDFVKRFQLRDHSGEKIINEHLRDLSDLQEYYSVELENGTWGRV 151
Query: 313 -NDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPAL------GGAFTDIAMWVFCPFN 365
++G + L + L E ++ PA+ G + D + F PFN
Sbjct: 152 SSEGTYMTSLDDFDKGPDWLLGEQPEYGTGHIKKAPAVLFVVDKGNGWVDAFWFYFYPFN 211
Query: 366 -GPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEF 424
GP + G HVGDWEH +R + GE ++ S H GG +E
Sbjct: 212 WGPYIMG-------SGPWGNHVGDWEHSLVRF--YKGEPQYLWMSAHGGGSAYKFEAIEK 262
Query: 425 IKE-------------NKPIVYSSRHGHASYPHAGTY 448
IK KP+++S+R HA Y G +
Sbjct: 263 IKRLRRVDGKLTNEVIKKPLIFSARGTHAHYASVGQH 299
>UniRef100_UPI0000235C44 UPI0000235C44 UniRef100 entry
Length = 641
Score = 51.6 bits (122), Expect = 6e-05
Identities = 45/161 (27%), Positives = 70/161 (42%), Gaps = 22/161 (13%)
Query: 350 GGAFTDIAMWVFCPFNGPATLKVSLMNIEMN-KIGEHVGDWEHFTLRVSNFTGELWSVFF 408
G D + F FN L N+ +N + G H+GDWEH +R + G ++FF
Sbjct: 421 GNGIVDAFWFYFYSFN--------LGNVVLNVRFGNHIGDWEHCLVRFHH--GIPKALFF 470
Query: 409 SEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNF 468
S HSGG+ + LE I +P++YS+ HA Y G + S L G+ +D
Sbjct: 471 SAHSGGEAYSYEALEKI-GRRPVIYSATGTHAMYATPGVH---SYILPWGLLHDQTDRGP 526
Query: 469 ILDSSFRYKIVAAEYLGDGV------ITEPC-WLQYMREWG 502
+ D + +++ D + T P W + WG
Sbjct: 527 LWDPLLNSHMYTYDHVSDTLRASTLSPTAPTEWFYFNGHWG 567
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.139 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,084,803,022
Number of Sequences: 2790947
Number of extensions: 49263079
Number of successful extensions: 96429
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 96263
Number of HSP's gapped (non-prelim): 62
length of query: 560
length of database: 848,049,833
effective HSP length: 133
effective length of query: 427
effective length of database: 476,853,882
effective search space: 203616607614
effective search space used: 203616607614
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135461.13


