

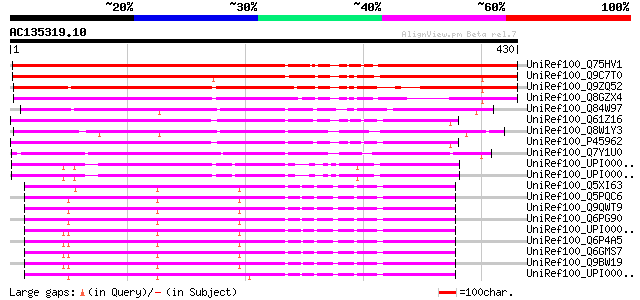
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.10 + phase: 1 /pseudo/partial
(430 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75HV1 Putative kinesin-related protein [Oryza sativa] 426 e-118
UniRef100_Q9C7T0 Kinesin, putative; 56847-62063 [Arabidopsis tha... 362 9e-99
UniRef100_Q9ZQ52 Putative kinesin heavy chain [Arabidopsis thali... 320 4e-86
UniRef100_Q8GZX4 Hypothetical protein OSJNBa0090O10.9 [Oryza sat... 299 9e-80
UniRef100_Q84W97 Putative kinesin [Arabidopsis thaliana] 229 9e-59
UniRef100_Q61Z16 Hypothetical protein CBG03312 [Caenorhabditis b... 209 1e-52
UniRef100_Q8W1Y3 Kinesin-like protein heavy chain [Arabidopsis t... 208 2e-52
UniRef100_P45962 Kinesin-like protein klp-3 [Caenorhabditis eleg... 208 3e-52
UniRef100_Q7Y1U0 Kinesin-like calmodulin binding protein [Gossyp... 185 2e-45
UniRef100_UPI0000499918 UPI0000499918 UniRef100 entry 184 3e-45
UniRef100_UPI00004989DF UPI00004989DF UniRef100 entry 184 3e-45
UniRef100_Q5XI63 Kinesin family member C1 [Rattus norvegicus] 182 1e-44
UniRef100_Q5PQC6 Hypothetical protein [Xenopus laevis] 182 2e-44
UniRef100_Q9QWT9 KIFC1 [Mus musculus] 182 2e-44
UniRef100_Q6PG90 Kinesin family member C5A [Mus musculus] 182 2e-44
UniRef100_UPI000036D6C9 UPI000036D6C9 UniRef100 entry 182 2e-44
UniRef100_Q6P4A5 KIFC1 protein [Homo sapiens] 181 3e-44
UniRef100_Q6GMS7 KIFC1 protein [Homo sapiens] 181 3e-44
UniRef100_Q9BW19 Kinesin-like protein KIFC1 [Homo sapiens] 181 3e-44
UniRef100_UPI000024D8A6 UPI000024D8A6 UniRef100 entry 181 4e-44
>UniRef100_Q75HV1 Putative kinesin-related protein [Oryza sativa]
Length = 840
Score = 426 bits (1096), Expect = e-118
Identities = 238/428 (55%), Positives = 296/428 (68%), Gaps = 18/428 (4%)
Query: 3 YEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEE 62
Y+ LK+KY + ERRRL NE+IEL+GNIRVFCRCRPL+ EI+NG + V S+ E
Sbjct: 150 YDGLKKKYADECAERRRLYNELIELRGNIRVFCRCRPLSTAEISNGCSSIVQIDPSHETE 209
Query: 63 LQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFT 122
LQ V SD +K FKFDHVF P DNQE VFA++ P+V SV+DG NVCIFAYGQTGTGKTFT
Sbjct: 210 LQFVPSDKDRKAFKFDHVFGPSDNQETVFAESLPVVRSVMDGFNVCIFAYGQTGTGKTFT 269
Query: 123 MEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKL 182
MEG PE RGVNYR LEELFR+SEER ++ Y VS+LEVYNEKI+DLL +S + +KL
Sbjct: 270 MEGIPEDRGVNYRALEELFRLSEERSSSVAYTFAVSILEVYNEKIRDLLDESSEQTGRKL 329
Query: 183 EVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIV 242
++KQ ADGTQEV GL+E +Y DGVWE LK G + RSVG+TSANELSSRSH + + V
Sbjct: 330 DIKQTADGTQEVAGLIEAPIYTIDGVWEKLKVGAKNRSVGATSANELSSRSH-SLVKVTV 388
Query: 243 DDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*IS 302
+ +V W +W + +A ++V V G +LKE N S +S
Sbjct: 389 RSEHLVTGQKWRS-HIW--LVDLAGSERVN-------KTEVEGD-RLKESQFINKS--LS 435
Query: 303 LYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCS 362
++ L ++ ++ RNSKLTH+LQSSLGGDCKTLMFVQISPSS D ETLCS
Sbjct: 436 ALGDVI----SALASKNAHIPYRNSKLTHLLQSSLGGDCKTLMFVQISPSSADSGETLCS 491
Query: 363 LNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHDEKEARKLQDNLQSVQMRLATREF 422
LNFA+RVR I+ GPARKQ D E K KQM EK +H+EKE KL ++LQ Q++ A+RE
Sbjct: 492 LNFASRVRAIDHGPARKQADPAETFKLKQMTEKIRHEEKENAKLLESLQLTQLKYASREN 551
Query: 423 MCRNLQDK 430
+ + LQ+K
Sbjct: 552 VIKTLQEK 559
>UniRef100_Q9C7T0 Kinesin, putative; 56847-62063 [Arabidopsis thaliana]
Length = 1195
Score = 362 bits (930), Expect = 9e-99
Identities = 205/434 (47%), Positives = 284/434 (65%), Gaps = 24/434 (5%)
Query: 3 YEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEE 62
+E LK K++ ER+ L N+++ELKGNIRVFCRCRPLN E G ++ + + + E
Sbjct: 464 HENLKVKFVAGEKERKELYNKILELKGNIRVFCRCRPLNFEETEAGVSMGIDVESTKNGE 523
Query: 63 LQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFT 122
+ V+ + KK FKFD VF P +Q VF T P SV+DG+NVCIFAYGQTGTGKTFT
Sbjct: 524 VIVMSNGFPKKSFKFDSVFGPNASQADVFEDTAPFATSVIDGYNVCIFAYGQTGTGKTFT 583
Query: 123 MEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLL--AGNSSEATK 180
MEGT RGVNYRTLE LFR+ + R+ YE+ VS+LEVYNE+I+DLL A S+ A K
Sbjct: 584 MEGTQHDRGVNYRTLENLFRIIKAREHRYNYEISVSVLEVYNEQIRDLLVPASQSASAPK 643
Query: 181 KLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYI 240
+ E++Q ++G VPGLVE V + VW++LK+G+ R+VG T+ANE SSRSH +
Sbjct: 644 RFEIRQLSEGNHHVPGLVEAPVKSIEEVWDVLKTGSNARAVGKTTANEHSSRSH--CIHC 701
Query: 241 IVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS* 300
++ + +L+ LW + +A ++V V G+ +LKE +N++
Sbjct: 702 VMVKGENLLNGECTKSKLW--LVDLAGSERVA-------KTEVQGE-RLKET--QNINKS 749
Query: 301 ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETL 360
+S ++ L N+ +++ RNSKLTH+LQ SLGGD KTLMFVQISP+ D +ETL
Sbjct: 750 LSALGDVIF----ALANKSSHIPFRNSKLTHLLQDSLGGDSKTLMFVQISPNENDQSETL 805
Query: 361 CSLNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMR 416
CSLNFA+RVRGIE GPA+KQ+D TELLKYKQM EK K D +++ RK+++ + ++ +
Sbjct: 806 CSLNFASRVRGIELGPAKKQLDNTELLKYKQMVEKWKQDMKGKDEQIRKMEETMYGLEAK 865
Query: 417 LATREFMCRNLQDK 430
+ R+ + LQDK
Sbjct: 866 IKERDTKNKTLQDK 879
>UniRef100_Q9ZQ52 Putative kinesin heavy chain [Arabidopsis thaliana]
Length = 1068
Score = 320 bits (821), Expect = 4e-86
Identities = 191/432 (44%), Positives = 273/432 (62%), Gaps = 51/432 (11%)
Query: 4 EILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSE-E 62
E LK+KY E +R+ L N + E KGNIRVFCRCRPLN E + SA ++V+F+ + E
Sbjct: 401 EDLKQKYSEEQAKRKELYNHIQETKGNIRVFCRCRPLNTEETSTKSA-TIVDFDGAKDGE 459
Query: 63 LQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFT 122
L V+ ++SKK FKFD V+ P+D Q VFA P+V SVLDG+NVCIFAYGQTGTGKTFT
Sbjct: 460 LGVITGNNSKKSFKFDRVYTPKDGQVDVFADASPMVVSVLDGYNVCIFAYGQTGTGKTFT 519
Query: 123 MEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKL 182
MEGTP++RGVNYRT+E+LF V+ ER+ TI Y + VS+LEVYNE+I+DLLA +S +KKL
Sbjct: 520 MEGTPQNRGVNYRTVEQLFEVARERRETISYNISVSVLEVYNEQIRDLLA--TSPGSKKL 577
Query: 183 EVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIV 242
E+KQ++DG+ VPGLVE +V + VW +L++G+ RSVGS + NE SSRSH + ++
Sbjct: 578 EIKQSSDGSHHVPGLVEANVENINEVWNVLQAGSNARSVGSNNVNEHSSRSHCMLSIMV- 636
Query: 243 DDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*IS 302
K +++ LW + +A +++ V G+ +LKE +N++ +S
Sbjct: 637 -KAKNLMNGDCTKSKLW--LVDLAGSERLA-------KTDVQGE-RLKEA--QNINRSLS 683
Query: 303 LYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCS 362
++ L +K +HI SPS D++ETL S
Sbjct: 684 ALGDVIYALA--------------TKSSHI---------------PYSPSEHDVSETLSS 714
Query: 363 LNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMRLA 418
LNFATRVRG+E GPARKQVD E+ K K M EK++ + ++ +K+++N+Q+++ +
Sbjct: 715 LNFATRVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNLEGKNK 774
Query: 419 TREFMCRNLQDK 430
R+ R+LQ+K
Sbjct: 775 GRDNSYRSLQEK 786
>UniRef100_Q8GZX4 Hypothetical protein OSJNBa0090O10.9 [Oryza sativa]
Length = 1045
Score = 299 bits (766), Expect = 9e-80
Identities = 180/431 (41%), Positives = 261/431 (59%), Gaps = 59/431 (13%)
Query: 4 EILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEEL 63
E LK KY E +R++L+N V E KGNIRVFCRCRPL+++E ++G +V + ++
Sbjct: 381 EDLKLKYYEEMAKRKKLHNIVEETKGNIRVFCRCRPLSKDETSSGYKCAVDFDGAKDGDI 440
Query: 64 QVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTM 123
+V ++KK FKFD V+ P DNQ V+A P+V SVLDG+NVCIFAYGQTGTGKTFTM
Sbjct: 441 AIVNGGAAKKTFKFDRVYMPTDNQADVYADASPLVTSVLDGYNVCIFAYGQTGTGKTFTM 500
Query: 124 EGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLE 183
EGT +RGVNYRTLEELF+++EER+ T+ Y + VS+LEVYNE+I+DLLA SS ++KKLE
Sbjct: 501 EGTERNRGVNYRTLEELFKIAEERKETVTYSISVSVLEVYNEQIRDLLA--SSPSSKKLE 558
Query: 184 VKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVD 243
+KQA++G+ VPG+VE V VW++L++G+ R+VGS + NE SSRSH + ++
Sbjct: 559 IKQASEGSHHVPGIVEAKVENIKEVWDVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVRA 618
Query: 244 DKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISL 303
+ +++ LW + +A +++ V G+ +LKE +N++ +S
Sbjct: 619 EN--LMNGECTRSKLW--LVDLAGSERLA-------KTDVQGE-RLKEA--QNINRSLSA 664
Query: 304 YHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSL 363
++ L + +++ RNSKLTH+LQ SL
Sbjct: 665 LGDVI----SALATKNSHIPYRNSKLTHLLQDSL-------------------------- 694
Query: 364 NFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMRLAT 419
E GPA+KQVD EL K KQM E++K D + RKL+DN Q+++ +
Sbjct: 695 ---------ELGPAKKQVDTAELQKVKQMLERAKQDIRLKDDSLRKLEDNCQNLENKAKG 745
Query: 420 REFMCRNLQDK 430
+E +NLQ+K
Sbjct: 746 KEQFYKNLQEK 756
>UniRef100_Q84W97 Putative kinesin [Arabidopsis thaliana]
Length = 983
Score = 229 bits (585), Expect = 9e-59
Identities = 152/408 (37%), Positives = 232/408 (56%), Gaps = 29/408 (7%)
Query: 10 YLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSD 69
Y V E R+L N+V +LKG+IRV+CR RP + + S + N E ++ +
Sbjct: 379 YHRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQSSFSSTIG--NMEDDTIGINTASRH 436
Query: 70 S-SKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG--- 125
S K F F+ VF P QE VF+ +P++ SVLDG+NVCIFAYGQTG+GKTFTM G
Sbjct: 437 GKSLKSFTFNKVFGPSATQEEVFSDMQPLIRSVLDGYNVCIFAYGQTGSGKTFTMSGPRD 496
Query: 126 -TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEV 184
T + +GVNYR L +LF ++E+R+ T +Y++ V M+E+YNE+++DLL + S K+LE+
Sbjct: 497 LTEKSQGVNYRALGDLFLLAEQRKDTFRYDIAVQMIEIYNEQVRDLLVTDGS--NKRLEI 554
Query: 185 KQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDD 244
+ ++ VP V V +++K+G++ R+VGST+ N+ SSRSH + V
Sbjct: 555 RNSSQKGLSVPDASLVPVSSTFDVIDLMKTGHKNRAVGSTALNDRSSRSH-SCLTVHVQG 613
Query: 245 KKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLY 304
+ + AV G + + +A ++V +T +LKE ++++ +S
Sbjct: 614 RDLTSGAVLRGCM---HLVDLAGSERVDKSEVT--------GDRLKEA--QHINRSLSAL 660
Query: 305 HHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLN 364
++ L P+ P RNSKLT +LQ SLGG KTLMFV ISP + + ET+ +L
Sbjct: 661 GDVIASLAHKNPHVPY----RNSKLTQLLQDSLGGQAKTLMFVHISPEADAVGETISTLK 716
Query: 365 FATRVRGIESGPARKQVDLTELLKYKQMAE--KSKHDEKEARKLQDNL 410
FA RV +E G AR D +++ + K+ K+ KEA Q+N+
Sbjct: 717 FAERVATVELGAARVNNDTSDVKELKEQIATLKAALARKEAESQQNNI 764
>UniRef100_Q61Z16 Hypothetical protein CBG03312 [Caenorhabditis briggsae]
Length = 598
Score = 209 bits (532), Expect = 1e-52
Identities = 137/382 (35%), Positives = 212/382 (54%), Gaps = 24/382 (6%)
Query: 1 DEYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNS 60
D + +L +Y +R++L+N+++EL GNIRVF R RP +E N V V++ N
Sbjct: 216 DFFRVLIERYKAEMEKRKQLHNQLVELNGNIRVFYRIRPQLASESDNQKPVVVIDEMDNG 275
Query: 61 EELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKT 120
S S K D V +QE +F + PI+ S +DG+NVCIFAYG TG+GKT
Sbjct: 276 VVHVSNTSGSRKTSAGADKVIPTGFSQEQIFNEVSPIITSCIDGYNVCIFAYGHTGSGKT 335
Query: 121 FTMEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATK 180
+TM+G E G+N R + +LF ++ER G IKY++ V+M+E+YNEKI+DLL + +
Sbjct: 336 YTMDGPVELPGINQRAIMQLFETAKERTGDIKYDIKVAMMEIYNEKIRDLL----NTSNT 391
Query: 181 KLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYI 240
L ++Q +G +PGL E V A V E L G + ++V +T AN SSRSH I +
Sbjct: 392 NLTIRQTEEGKGSIPGLEEVTVSSAQEVTETLARGRKNKAVAATEANIESSRSH-VIVRV 450
Query: 241 IVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS* 300
+V ++ G L + +A ++V+ + G+L LKE N S
Sbjct: 451 LVSATNLITKVTTVGRL---NLVDLAGSERVS-------QTNATGQL-LKEAQAINKS-- 497
Query: 301 ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETL 360
+S ++V+ L+ ++ RN +LT IL+ SL GD KTL+ V +SP S L E++
Sbjct: 498 LSELGNVVLALR----QNQKHIPFRNCQLTRILEDSLNGDSKTLVIVHLSPDSKSLNESI 553
Query: 361 CSLNFATRVRGI--ESGPARKQ 380
S+NFA ++ + +SG +++
Sbjct: 554 SSVNFAEKIGQVFTKSGTMKRE 575
>UniRef100_Q8W1Y3 Kinesin-like protein heavy chain [Arabidopsis thaliana]
Length = 1087
Score = 208 bits (530), Expect = 2e-52
Identities = 148/429 (34%), Positives = 231/429 (53%), Gaps = 40/429 (9%)
Query: 4 EILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEEL 63
E+ Y +V E R L NEV +LKG IRV+CR RP + + S V + N
Sbjct: 351 EVTSSSYHKVLEENRLLYNEVQDLKGTIRVYCRVRPFFQEQKDMQSTVDYIGENGN---- 406
Query: 64 QVVCSDSSKKQ------FKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGT 117
++ ++ K++ F F+ F +QE ++ T+P++ SVLDG NVCIFAYGQTG+
Sbjct: 407 -IIINNPFKQEKDARKIFSFNKAFGQTVSQEQIYIDTQPVIRSVLDGFNVCIFAYGQTGS 465
Query: 118 GKTFTMEG----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAG 173
GKT+TM G T GVNYR L +LF++S R + YE+ V M+E+YNE+++DLL
Sbjct: 466 GKTYTMSGPDLMTETTWGVNYRALRDLFQLSNARTHVVTYEIGVQMIEIYNEQVRDLLVS 525
Query: 174 NSSEATKKLEVKQAAD-GTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSR 232
+ S +++L+++ + VP V V ++++ G + R+VG+T+ NE SSR
Sbjct: 526 DGS--SRRLDIRNNSQLNGLNVPDANLIPVSNTRDVLDLMRIGQKNRAVGATALNERSSR 583
Query: 233 SHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEK 292
SH + + V K++ ++ G L + R +K + +V +LK +
Sbjct: 584 SH-SVLTVHVQGKELASGSILRGCLHLVDLAGSERVEK---------SEAVGERLKEAQH 633
Query: 293 G*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPS 352
++LS+ + + L + +++ RNSKLT +LQ SLGG KTLMFV I+P
Sbjct: 634 INKSLSALGDVIY--------ALAQKSSHVPYRNSKLTQVLQDSLGGQAKTLMFVHINPE 685
Query: 353 SVDLTETLCSLNFATRVRGIESGPARKQVDLTEL--LKYKQMAEKSKHDEKEARKLQDNL 410
+ ET+ +L FA RV IE G AR + E+ LK + + KS ++KEA + L
Sbjct: 686 VNAVGETISTLKFAQRVASIELGAARSNKETGEIRDLKDEISSLKSAMEKKEAE--LEQL 743
Query: 411 QSVQMRLAT 419
+S +R T
Sbjct: 744 RSGSIRNTT 752
>UniRef100_P45962 Kinesin-like protein klp-3 [Caenorhabditis elegans]
Length = 598
Score = 208 bits (529), Expect = 3e-52
Identities = 133/382 (34%), Positives = 214/382 (55%), Gaps = 24/382 (6%)
Query: 1 DEYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNS 60
D + +L +Y +R++L+N+++EL GNIRVF R RP +E N V V++ N
Sbjct: 216 DFFRVLIERYKAEMEKRKQLHNQLVELNGNIRVFYRIRPQLASETDNQKPVVVIDEMDNG 275
Query: 61 EELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKT 120
+ + K D V + +Q+ +F + PI+ S +DG+NVCIFAYG TG+GKT
Sbjct: 276 VVHVSNTTGTRKTSAGADKVIPTDFSQDQIFNEVSPIITSCIDGYNVCIFAYGHTGSGKT 335
Query: 121 FTMEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATK 180
+TM+G G+N R + +LF ++ER G IKY++ V+M+E+YNEKI+DLL + +
Sbjct: 336 YTMDGPVTMPGINQRAIMQLFETAKERTGDIKYDIKVAMMEIYNEKIRDLL----NTSNT 391
Query: 181 KLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYI 240
L ++Q +G +PGL E V A+ V E L G + ++V +T AN SSRSH I +
Sbjct: 392 NLAIRQTEEGRSSIPGLEEVSVNSAEEVTETLARGRKNKAVAATEANIESSRSH-VIVRV 450
Query: 241 IVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS* 300
+V ++ A G L + +A ++V+ + G+L LKE N S
Sbjct: 451 LVSATNLITKATTVGRL---NLVDLAGSERVS-------QTNATGQL-LKEAQAINKS-- 497
Query: 301 ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETL 360
+S ++V+ L+ ++ RN +LT IL+ SL GD KTL+ V +SP + L E++
Sbjct: 498 LSELGNVVLALR----QNQKHIPFRNCQLTRILEDSLNGDSKTLVIVHLSPDAKSLNESI 553
Query: 361 CSLNFATRVRGI--ESGPARKQ 380
S+NFA ++ + +SG +++
Sbjct: 554 SSVNFAEKIGQVFTKSGTMKRE 575
>UniRef100_Q7Y1U0 Kinesin-like calmodulin binding protein [Gossypium hirsutum]
Length = 1209
Score = 185 bits (470), Expect = 2e-45
Identities = 129/413 (31%), Positives = 213/413 (51%), Gaps = 31/413 (7%)
Query: 2 EYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSE 61
E E+L Y E + R+R N + ++KG +RVFCR RPLNE E+ ++ + +
Sbjct: 808 ELEVL---YKEEQILRKRYFNTIEDMKGKVRVFCRLRPLNEKEMLEKERKVLMGLDEFTV 864
Query: 62 ELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTF 121
E D KQ +D VF QE +F T+ +V S +DG+NVCIFAYGQTG+GKTF
Sbjct: 865 EHP--WKDDKAKQHMYDRVFDDSATQEDIFEDTRYLVQSAVDGYNVCIFAYGQTGSGKTF 922
Query: 122 TMEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKK 181
T+ G+ + G+ R + ELF++ + L M+E+Y + + DLL +++ K
Sbjct: 923 TIYGSDNNPGLTPRAIAELFKILRRDSNKFSFSLKAYMVELYQDTLVDLLLPKNAKRL-K 981
Query: 182 LEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYII 241
L++K+ + G V + + + I++ G+ R + T NE SSRSH I ++
Sbjct: 982 LDIKKDSKGMVAVENATVIPISTFEELKSIIQRGSERRHISGTQMNEESSRSH-LILSVV 1040
Query: 242 VDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*I 301
++ + +V G L + + R +K + SV +LK + ++LS+
Sbjct: 1041 IESTNLQTQSVARGKLSFVDLAGSERVKK---------SGSVGDQLKEAQSINKSLSA-- 1089
Query: 302 SLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLC 361
+ + L + ++ RN KLT ++ SLGG+ KTLMFV +SP+ +L ET
Sbjct: 1090 ------LGDVISALSSGSQHIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPAESNLDETYN 1143
Query: 362 SLNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKH------DEKEARKLQD 408
SL +A+RVR I + A K + E+++ K++ K DE++ +Q+
Sbjct: 1144 SLTYASRVRSIVN-DASKNISSKEVVRLKKLVAYWKEQAGRRGDEEDYEDIQE 1195
>UniRef100_UPI0000499918 UPI0000499918 UniRef100 entry
Length = 567
Score = 184 bits (468), Expect = 3e-45
Identities = 140/396 (35%), Positives = 207/396 (51%), Gaps = 55/396 (13%)
Query: 2 EYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENE-----IANGSAVSV--V 54
E E + K + ERR+L+NEV+ELKGN+RVFCR RP +NE + +AV V +
Sbjct: 210 ELEKTEDKLIHSEKERRKLHNEVMELKGNVRVFCRVRPPLKNEGISVGVTGDNAVIVNSI 269
Query: 55 NFESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQ 114
NF E+++ F FD F + Q+ VF + +V S LDG+ CIFAYGQ
Sbjct: 270 NFSGKKEKIK----------FGFDRAFDSDSTQQDVFEEISQLVQSSLDGYQTCIFAYGQ 319
Query: 115 TGTGKTFTMEGTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDLLAG 173
TG+GKT+TMEGT + G+ T+ ++F EE + ++++ V +E+YN I DLL
Sbjct: 320 TGSGKTYTMEGTNDKPGMIPLTVHKIFTAIEELKTLGWQFKISVKYVEIYNNNIFDLLV- 378
Query: 174 NSSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRS 233
+ E +KKL++K +P V A+ V ++ R R+V +T N SSRS
Sbjct: 379 -NEEESKKLQIKYNGP-LVILPEANVIEVSEAEEVDHLINIATRNRAVAATKCNAQSSRS 436
Query: 234 HWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG 293
H IF + + CG + + EQ+ FG LT V + G +L E G
Sbjct: 437 H-SIFMMDL-----------CGRNIG------SNEQR--FGGLT--LVDLAGSERLDESG 474
Query: 294 --------*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLM 345
+N++ +S +++ + N+ +++ RNSKLT +LQ+ LG D KTLM
Sbjct: 475 AKGERLEETKNINKSLSALGDVIV----AIANKDSHIPYRNSKLTELLQNCLGSDSKTLM 530
Query: 346 FVQISPSSVDLTETLCSLNFATRVRGIESGPARKQV 381
FV IS D ET+ SL FAT+V G A++ V
Sbjct: 531 FVNISSDQQDTLETISSLRFATKVNTCVIGTAKRHV 566
>UniRef100_UPI00004989DF UPI00004989DF UniRef100 entry
Length = 567
Score = 184 bits (468), Expect = 3e-45
Identities = 140/396 (35%), Positives = 207/396 (51%), Gaps = 55/396 (13%)
Query: 2 EYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENE-----IANGSAVSV--V 54
E E + K + ERR+L+NEV+ELKGN+RVFCR RP +NE + +AV V +
Sbjct: 210 ELEKTEDKLIHSEKERRKLHNEVMELKGNVRVFCRVRPPLKNEGISVGVTGDNAVIVNSI 269
Query: 55 NFESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQ 114
NF E+++ F FD F + Q+ VF + +V S LDG+ CIFAYGQ
Sbjct: 270 NFSGKKEKIK----------FGFDRAFDSDSTQQDVFEEISQLVQSSLDGYQTCIFAYGQ 319
Query: 115 TGTGKTFTMEGTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDLLAG 173
TG+GKT+TMEGT + G+ T+ ++F EE + ++++ V +E+YN I DLL
Sbjct: 320 TGSGKTYTMEGTNDKPGMIPLTVHKIFTAIEELKTLGWQFKISVKYVEIYNNNIFDLLV- 378
Query: 174 NSSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRS 233
+ E +KKL++K +P V A+ V ++ R R+V +T N SSRS
Sbjct: 379 -NEEESKKLQIKYNGP-LVILPEANVIEVSEAEEVDHLINIATRNRAVAATKCNAQSSRS 436
Query: 234 HWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG 293
H IF + + CG + + EQ+ FG LT V + G +L E G
Sbjct: 437 H-SIFMMDL-----------CGRNIG------SNEQR--FGGLT--LVDLAGSERLDESG 474
Query: 294 --------*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLM 345
+N++ +S +++ + N+ +++ RNSKLT +LQ+ LG D KTLM
Sbjct: 475 AKGERLEETKNINKSLSALGDVIV----AIANKDSHIPYRNSKLTELLQNCLGSDSKTLM 530
Query: 346 FVQISPSSVDLTETLCSLNFATRVRGIESGPARKQV 381
FV IS D ET+ SL FAT+V G A++ V
Sbjct: 531 FVNISSDQQDTLETISSLRFATKVNTCVIGTAKRHV 566
>UniRef100_Q5XI63 Kinesin family member C1 [Rattus norvegicus]
Length = 693
Score = 182 bits (463), Expect = 1e-44
Identities = 136/391 (34%), Positives = 204/391 (51%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVV-----------------N 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E E V
Sbjct: 313 LEMERRRLHNQLQELKGNIRVFCRVRPVLEGESTPSPGFLVFPPGPAGPSDPPTRLCLSR 372
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + +++ F FD VF P QE VF + +V S LDG+ VCIFAYGQT
Sbjct: 373 SDDRRSTLTRAPAAATRHDFSFDRVFPPGSKQEEVFEEISMLVQSALDGYPVCIFAYGQT 432
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R + LF V++E G Y + S +E+YNE ++DL
Sbjct: 433 GSGKTFTMEGGPRGDPQLEGLIPRAMRHLFSVAQEMSGQGWTYSFVASYVEIYNETVRDL 492
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G E+++A G++E V V V +L + R+V T+ N
Sbjct: 493 LATGTRKGQGGDCEIRRAGPGSEELTVTNARYVPVSCEKEVEALLHLAQQNRAVARTAQN 552
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH +F + + + + CG L + +A +++ G L + +
Sbjct: 553 ERSSRSH-SVFQLQISGEH-AARGLQCGAPL--NLVDLAGSERLDPG----LTLGPGERD 604
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 605 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 658
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 659 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 689
>UniRef100_Q5PQC6 Hypothetical protein [Xenopus laevis]
Length = 716
Score = 182 bits (462), Expect = 2e-44
Identities = 135/391 (34%), Positives = 209/391 (52%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANG-----------------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E E + +S+
Sbjct: 336 LEMERRRLHNQLQELKGNIRVFCRVRPVLEGESTPSPGFLVFPPGPAGPSDPPTGLSLSR 395
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + + + F FD VF P QE VF + +V S LDG+ VCIFAYGQT
Sbjct: 396 SDDRRSTLTGAPAPTVRHDFSFDRVFPPGSKQEEVFEEIAMLVQSALDGYPVCIFAYGQT 455
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R + LF V++E G Y + S +E+YNE ++DL
Sbjct: 456 GSGKTFTMEGGPRGDPQLEGLIPRAMRHLFSVAQEMSGQGWTYSFVASYVEIYNETVRDL 515
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A+ G++E V V V +L ++ R+V T+ N
Sbjct: 516 LATGPRKGQGGECEIRRASPGSEELTVTNARYVPVSCEKEVEALLHLAHQNRAVAHTAQN 575
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
+ SSRSH +F + + + + CG L + +A +++ G L + +
Sbjct: 576 KRSSRSH-SVFQLQISGEH-AARGLQCGAPL--NLVDLAGSERLDPG----LHLGPGERD 627
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 628 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 681
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 682 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 712
>UniRef100_Q9QWT9 KIFC1 [Mus musculus]
Length = 631
Score = 182 bits (462), Expect = 2e-44
Identities = 135/391 (34%), Positives = 209/391 (52%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANG-----------------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E E + +S+
Sbjct: 251 LEMERRRLHNQLQELKGNIRVFCRVRPVLEGESTPSPGFLVFPPGPAGPSDPPTGLSLSR 310
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + + + F FD VF P QE VF + +V S LDG+ VCIFAYGQT
Sbjct: 311 SDDRRSTLTGAPAPTVRHDFSFDRVFPPGSKQEEVFEEIAMLVQSALDGYPVCIFAYGQT 370
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R + LF V++E G Y + S +E+YNE ++DL
Sbjct: 371 GSGKTFTMEGGPRGDPQLEGLIPRAMRHLFSVAQEMSGQGWTYSFVASYVEIYNETVRDL 430
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A+ G++E V V V +L ++ R+V T+ N
Sbjct: 431 LATGPRKGQGGECEIRRASPGSEELTVTNARYVPVSCEKEVEALLHLAHQNRAVAHTAQN 490
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
+ SSRSH +F + + + + CG L + +A +++ G L + +
Sbjct: 491 KRSSRSH-SVFQLQISGEH-AARGLQCGAPL--NLVDLAGSERLDPG----LHLGPGERD 542
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 543 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 596
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 597 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 627
>UniRef100_Q6PG90 Kinesin family member C5A [Mus musculus]
Length = 674
Score = 182 bits (462), Expect = 2e-44
Identities = 135/391 (34%), Positives = 209/391 (52%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANG-----------------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E E + +S+
Sbjct: 294 LEMERRRLHNQLQELKGNIRVFCRVRPVLEGESTPSPGFLVFPPGPAGPSDPPTGLSLSR 353
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + + + F FD VF P QE VF + +V S LDG+ VCIFAYGQT
Sbjct: 354 SDDRRSTLTGAPAPTVRHDFSFDRVFPPGSKQEEVFEEIAMLVQSALDGYPVCIFAYGQT 413
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R + LF V++E G Y + S +E+YNE ++DL
Sbjct: 414 GSGKTFTMEGGPRGDPQLEGLIPRAMRHLFSVAQEMSGQGWTYSFVASYVEIYNETVRDL 473
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A+ G++E V V V +L ++ R+V T+ N
Sbjct: 474 LATGPRKGQGGECEIRRASPGSEELTVTNARYVPVSCEKEVEALLHLAHQNRAVAHTAQN 533
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
+ SSRSH +F + + + + CG L + +A +++ G L + +
Sbjct: 534 KRSSRSH-SVFQLQISGEH-AARGLQCGAPL--NLVDLAGSERLDPG----LHLGPGERD 585
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 586 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 639
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 640 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 670
>UniRef100_UPI000036D6C9 UPI000036D6C9 UniRef100 entry
Length = 673
Score = 182 bits (461), Expect = 2e-44
Identities = 137/391 (35%), Positives = 209/391 (53%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENE---------IANG--------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E +G + +S+
Sbjct: 293 LEMERRRLHNQLQELKGNIRVFCRVRPVLPGEPTPPPGLLLFPSGPGGPSDPPTRLSLSR 352
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + ++ F FD VF P Q+ VF + +V S LDG+ VCIFAYGQT
Sbjct: 353 SDERRGTLSGAPAPPTRHDFSFDRVFPPGSGQDEVFEEIAMLVQSALDGYPVCIFAYGQT 412
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R L LF V++E G Y + S +E+YNE ++DL
Sbjct: 413 GSGKTFTMEGGPGGDPQLEGLIPRALRHLFSVAQELSGQGWTYSFVASYVEIYNETVRDL 472
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A G++E V V V +L + R+V T+ N
Sbjct: 473 LATGTRKGQGGECEIRRAGPGSEELTVTNARYVPVSCEKEVEALLHLARQNRAVARTAQN 532
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH +F + + + + CG L + +A +++ G LA+ +
Sbjct: 533 ERSSRSH-SVFQLQISGEHS-SRGLQCGAPL--SLVDLAGSERLDPG----LALGPGERE 584
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 585 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 638
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 639 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 669
>UniRef100_Q6P4A5 KIFC1 protein [Homo sapiens]
Length = 709
Score = 181 bits (460), Expect = 3e-44
Identities = 137/391 (35%), Positives = 209/391 (53%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENE---------IANG--------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E +G + +S+
Sbjct: 329 LEMERRRLHNQLQELKGNIRVFCRVRPVLPGEPTPPPGLLLFPSGPGGPSDPPTRLSLSR 388
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + ++ F FD VF P Q+ VF + +V S LDG+ VCIFAYGQT
Sbjct: 389 SDERRGTLSGAPAPPTRHDFSFDRVFPPGSGQDEVFEEIAMLVQSALDGYPVCIFAYGQT 448
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R L LF V++E G Y + S +E+YNE ++DL
Sbjct: 449 GSGKTFTMEGGPGGDPQLEGLIPRALRHLFSVAQELSGQGWTYSFVASYVEIYNETVRDL 508
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A G++E V V V +L + R+V T+ N
Sbjct: 509 LATGTRKGQGGECEIRRAGPGSEELTVTNARYVPVSCEKEVDALLHLARQNRAVARTAQN 568
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH +F + + + + CG L + +A +++ G LA+ +
Sbjct: 569 ERSSRSH-SVFQLQISGEHS-SRGLQCGAPL--SLVDLAGSERLDPG----LALGPGERE 620
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 621 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 674
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 675 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 705
>UniRef100_Q6GMS7 KIFC1 protein [Homo sapiens]
Length = 720
Score = 181 bits (460), Expect = 3e-44
Identities = 137/391 (35%), Positives = 209/391 (53%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENE---------IANG--------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E +G + +S+
Sbjct: 340 LEMERRRLHNQLQELKGNIRVFCRVRPVLPGEPTPPPGLLLFPSGPGGPSDPPTRLSLSR 399
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + ++ F FD VF P Q+ VF + +V S LDG+ VCIFAYGQT
Sbjct: 400 SDERRGTLSGAPAPPTRHDFSFDRVFPPGSGQDEVFEEIAMLVQSALDGYPVCIFAYGQT 459
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R L LF V++E G Y + S +E+YNE ++DL
Sbjct: 460 GSGKTFTMEGGPGGDPQLEGLIPRALRHLFSVAQELSGQGWTYSFVASYVEIYNETVRDL 519
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A G++E V V V +L + R+V T+ N
Sbjct: 520 LATGTRKGQGGECEIRRAGPGSEELTVTNARYVPVSCEKEVDALLHLARQNRAVARTAQN 579
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH +F + + + + CG L + +A +++ G LA+ +
Sbjct: 580 ERSSRSH-SVFQLQISGEHS-SRGLQCGAPL--SLVDLAGSERLDPG----LALGPGERE 631
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 632 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 685
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 686 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 716
>UniRef100_Q9BW19 Kinesin-like protein KIFC1 [Homo sapiens]
Length = 673
Score = 181 bits (460), Expect = 3e-44
Identities = 137/391 (35%), Positives = 209/391 (53%), Gaps = 39/391 (9%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENE---------IANG--------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E +G + +S+
Sbjct: 293 LEMERRRLHNQLQELKGNIRVFCRVRPVLPGEPTPPPGLLLFPSGPGGPSDPPTRLSLSR 352
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + ++ F FD VF P Q+ VF + +V S LDG+ VCIFAYGQT
Sbjct: 353 SDERRGTLSGAPAPPTRHDFSFDRVFPPGSGQDEVFEEIAMLVQSALDGYPVCIFAYGQT 412
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R L LF V++E G Y + S +E+YNE ++DL
Sbjct: 413 GSGKTFTMEGGPGGDPQLEGLIPRALRHLFSVAQELSGQGWTYSFVASYVEIYNETVRDL 472
Query: 171 LA-GNSSEATKKLEVKQAADGTQE--VPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
LA G + E+++A G++E V V V +L + R+V T+ N
Sbjct: 473 LATGTRKGQGGECEIRRAGPGSEELTVTNARYVPVSCEKEVDALLHLARQNRAVARTAQN 532
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH +F + + + + CG L + +A +++ G LA+ +
Sbjct: 533 ERSSRSH-SVFQLQISGEHS-SRGLQCGAPL--SLVDLAGSERLDPG----LALGPGERE 584
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMFV
Sbjct: 585 RLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMFV 638
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPAR 378
ISP +++E+L SL FA++V G A+
Sbjct: 639 NISPLEENVSESLNSLRFASKVNQCVIGTAQ 669
>UniRef100_UPI000024D8A6 UPI000024D8A6 UniRef100 entry
Length = 663
Score = 181 bits (459), Expect = 4e-44
Identities = 133/392 (33%), Positives = 210/392 (52%), Gaps = 40/392 (10%)
Query: 13 VSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANG-----------------SAVSVVN 55
+ +ERRRL+N++ ELKGNIRVFCR RP+ E E + +S+
Sbjct: 282 LEMERRRLHNQLQELKGNIRVFCRVRPVLEGESTPSPGFLVFPPGPAGPSDRPTGLSLSR 341
Query: 56 FESNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQT 115
+ L + + + F FD VF P QE VF + +V S LDG+ VCIFAYGQT
Sbjct: 342 SDDRRSTLTGAPAPTVRHDFSFDRVFPPGSKQEEVFEEIAMLVQSALDGYPVCIFAYGQT 401
Query: 116 GTGKTFTME----GTPEHRGVNYRTLEELFRVSEERQGT-IKYELLVSMLEVYNEKIKDL 170
G+GKTFTME G P+ G+ R + LF V++E G Y + S +E+YNE ++DL
Sbjct: 402 GSGKTFTMEGGPRGDPQLAGLIPRAMRHLFSVAQEMSGQGWTYSFVASYVEIYNETVRDL 461
Query: 171 LA-GNSSEATKKLEVKQAADGTQEVPGLVETHV---YGADGVWEILKSGNRVRSVGSTSA 226
LA G + E+++A+ G++E+ +V + +L ++ R+V T+
Sbjct: 462 LATGPRKGQGGECEIRRASPGSEELTVTNARYVPVSCEKEASEALLHLAHQNRAVAHTAQ 521
Query: 227 NELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GK 286
N+ SSRSH +F + + + + CG L + +A +++ G L + +
Sbjct: 522 NKRSSRSH-SVFQLQISGEH-AARGLQCGAPL--NLVDLAGSERLDPG----LPLGPGER 573
Query: 287 LKLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMF 346
+L+E + ++S +S ++M L N+ +++ RNSKLT++LQ+SLGG K LMF
Sbjct: 574 DRLRET--QAINSSLSTLGLVIMALS----NKESHVPYRNSKLTYLLQNSLGGSAKMLMF 627
Query: 347 VQISPSSVDLTETLCSLNFATRVRGIESGPAR 378
V ISP +++E+L SL FA++V G A+
Sbjct: 628 VNISPLEENVSESLNSLRFASKVNQCVIGTAQ 659
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 644,964,274
Number of Sequences: 2790947
Number of extensions: 25305190
Number of successful extensions: 107198
Number of sequences better than 10.0: 1510
Number of HSP's better than 10.0 without gapping: 1404
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 101520
Number of HSP's gapped (non-prelim): 3111
length of query: 430
length of database: 848,049,833
effective HSP length: 130
effective length of query: 300
effective length of database: 485,226,723
effective search space: 145568016900
effective search space used: 145568016900
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC135319.10


