

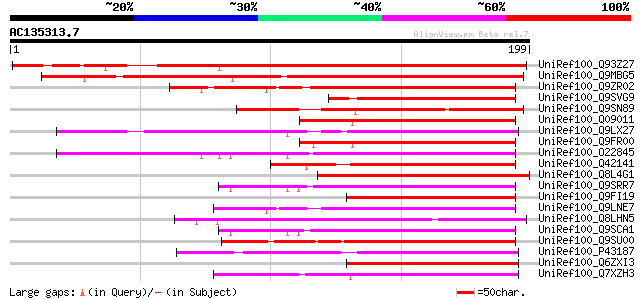
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135313.7 - phase: 0
(199 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93Z27 AT5g39670/MIJ24_140 [Arabidopsis thaliana] 166 4e-40
UniRef100_Q9MBG5 Similarity to calmodulin [Arabidopsis thaliana] 145 6e-34
UniRef100_Q9ZR02 Putative calmodulin [Arabidopsis thaliana] 83 4e-15
UniRef100_Q9SVG9 Putative calcium-binding protein [Arabidopsis t... 82 6e-15
UniRef100_Q9SN89 Putative calcium binding protein [Arabidopsis t... 80 4e-14
UniRef100_Q09011 Calcium-binding protein CAST [Solanum tuberosum] 80 4e-14
UniRef100_Q9LX27 Calmodulin-like protein [Arabidopsis thaliana] 79 5e-14
UniRef100_Q9FR00 Avr9/Cf-9 rapidly elicited protein 31 [Nicotian... 79 5e-14
UniRef100_O22845 Putative calcium binding protein [Arabidopsis t... 79 9e-14
UniRef100_Q42141 Calcium-binding protein [Arabidopsis thaliana] 78 1e-13
UniRef100_Q8L4G1 Hypothetical protein OJ1118_B03.113 [Oryza sativa] 77 3e-13
UniRef100_Q9SRR7 Putative calmodulin [Arabidopsis thaliana] 77 3e-13
UniRef100_Q9FI19 Calmodulin-like protein [Arabidopsis thaliana] 76 4e-13
UniRef100_Q9LNE7 T21E18.4 protein [Arabidopsis thaliana] 76 6e-13
UniRef100_Q8LHN5 Hypothetical protein P0519E12.112 [Oryza sativa] 76 6e-13
UniRef100_Q9SCA1 Calcium-binding protein [Lotus japonicus] 74 2e-12
UniRef100_Q9SU00 Putative calmodulin [Arabidopsis thaliana] 74 2e-12
UniRef100_P43187 Calcium-binding allergen Bet v 3 [Betula verruc... 72 6e-12
UniRef100_Q6ZXI3 Putative calmodulin-like protein [Populus euram... 71 1e-11
UniRef100_Q7XZH3 Hypothetical protein OSJNBb0033J23.19 [Oryza sa... 71 1e-11
>UniRef100_Q93Z27 AT5g39670/MIJ24_140 [Arabidopsis thaliana]
Length = 204
Score = 166 bits (419), Expect = 4e-40
Identities = 97/205 (47%), Positives = 131/205 (63%), Gaps = 23/205 (11%)
Query: 2 ALPQISQSNSISNSTSPLFGLIDLFLYLTFFNKI---HNFFSSIWFFLLCQIHSGSSEVR 58
+L + QS+S+S PLFGLI+ FL + FF + FFS W V+
Sbjct: 15 SLSKCKQSSSLS---FPLFGLINFFL-IGFFRWVSFAQLFFSRFWPL-----------VQ 59
Query: 59 EEKKVSESKCSSQENESNIGR-----DNGDMIERDEVKMVMEKMGFFCSSESEELEEKYG 113
++ VSE K E +++I D+ D + R++V MVM+ +G E+E L+++Y
Sbjct: 60 HQQCVSEKKSKDLEFQTSIKHEEYRDDDDDGLCREDVGMVMKSLGLSTDQENEGLQKQYS 119
Query: 114 SKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMI 173
SKE+ +FEE EPSLEE+KQAFDVFDEN+DGFID +LQRVL ILGLKQGS ENC++MI
Sbjct: 120 SKEVSNLFEEKEPSLEEVKQAFDVFDENRDGFIDPIDLQRVLTILGLKQGSNLENCRRMI 179
Query: 174 TIFDENQDGRIDFIEFVNIMKNHFC 198
FD ++DGRIDF FV M+N+FC
Sbjct: 180 RSFDGSKDGRIDFYGFVKFMENNFC 204
>UniRef100_Q9MBG5 Similarity to calmodulin [Arabidopsis thaliana]
Length = 194
Score = 145 bits (366), Expect = 6e-34
Identities = 85/192 (44%), Positives = 116/192 (60%), Gaps = 11/192 (5%)
Query: 13 SNSTSPLFGLIDLFL--YLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCSS 70
S+S+ PLF L + FL + + + F S F L Q H + + K E+
Sbjct: 6 SSSSLPLFALFNFFLISFCRWVSSTRIFLSR--FVPLLQHHQRVFDKKNNKDQQETLTKQ 63
Query: 71 QENESNIGRDNGDM-----IERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEENE 125
++++ + D+ D I R+E +MVM +G F + + +L+E+Y +KE+ +FEE E
Sbjct: 64 EDDDDDDDDDDDDDDDDIDISREEAEMVMRSLGLFYNDD--QLQEQYSAKEVSSLFEEKE 121
Query: 126 PSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRID 185
SLEE+KQAFDVFDENKDGFIDA ELQRVL ILG KQGS +NC MI D N+DG+ID
Sbjct: 122 ASLEEVKQAFDVFDENKDGFIDAIELQRVLTILGFKQGSYLDNCLVMIRSLDGNKDGKID 181
Query: 186 FIEFVNIMKNHF 197
F EFV M+ F
Sbjct: 182 FNEFVKFMETSF 193
>UniRef100_Q9ZR02 Putative calmodulin [Arabidopsis thaliana]
Length = 154
Score = 83.2 bits (204), Expect = 4e-15
Identities = 55/137 (40%), Positives = 83/137 (60%), Gaps = 10/137 (7%)
Query: 62 KVSESKCSSQE-NESNIGRDNGDMIERDEVKMVMEKM---GFFCSSESEELEEKYGSKEL 117
K + K +++E NES ++ G +I DE+ +++K+ G C + EE E Y +
Sbjct: 15 KDGDGKITTKELNESF--KNLGIIIPEDELTQIIQKIDVNGDGCV-DIEEFGELY---KT 68
Query: 118 CEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFD 177
V +E+E E++K+AF+VFD N DGFI EL+ VL LGLKQG E C+KMI D
Sbjct: 69 IMVEDEDEVGEEDMKEAFNVFDRNGDGFITVDELKAVLSSLGLKQGKTLEECRKMIMQVD 128
Query: 178 ENQDGRIDFIEFVNIMK 194
+ DGR++++EF +MK
Sbjct: 129 VDGDGRVNYMEFRQMMK 145
Score = 34.7 bits (78), Expect = 1.5
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 2/65 (3%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
EL + F +FD++ DG I KEL LG+ + ++I D N DG +D EF
Sbjct: 5 ELNRVFQMFDKDGDGKITTKELNESFKNLGIIIPE--DELTQIIQKIDVNGDGCVDIEEF 62
Query: 190 VNIMK 194
+ K
Sbjct: 63 GELYK 67
>UniRef100_Q9SVG9 Putative calcium-binding protein [Arabidopsis thaliana]
Length = 191
Score = 82.4 bits (202), Expect = 6e-15
Identities = 43/72 (59%), Positives = 49/72 (67%), Gaps = 3/72 (4%)
Query: 123 ENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDG 182
ENE L E AF VFDEN DGFI A+ELQ VL LGL +G E E +KMI D NQDG
Sbjct: 116 ENESDLAE---AFKVFDENGDGFISARELQTVLKKLGLPEGGEMERVEKMIVSVDRNQDG 172
Query: 183 RIDFIEFVNIMK 194
R+DF EF N+M+
Sbjct: 173 RVDFFEFKNMMR 184
Score = 40.0 bits (92), Expect = 0.035
Identities = 27/93 (29%), Positives = 50/93 (53%), Gaps = 5/93 (5%)
Query: 104 ESEELEEKYGSKELCEVFEENEPSLEELK--QAFDVFDENKDGFIDAKELQRVLVILGLK 161
ES E+K +++ F PSL L+ + FD+FD+N DGFI +EL + L LGL
Sbjct: 2 ESNNNEKKKVARQSSS-FRLRSPSLNALRLQRIFDLFDKNGDGFITVEELSQALTRLGL- 59
Query: 162 QGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
++ + + + + + + ++F +F ++ K
Sbjct: 60 -NADLSDLKSTVESYIQPGNTGLNFDDFSSLHK 91
>UniRef100_Q9SN89 Putative calcium binding protein [Arabidopsis thaliana]
Length = 183
Score = 79.7 bits (195), Expect = 4e-14
Identities = 45/111 (40%), Positives = 70/111 (62%), Gaps = 10/111 (9%)
Query: 88 DEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEENEPSLEEL-KQAFDVFDENKDGFI 146
+EVK +++ EE EE + E+NE +E+ K+AF +FDEN+DGFI
Sbjct: 81 EEVKAIIDDSEALYECLIEEGEEY--------LLEKNEMMGKEIVKEAFRLFDENQDGFI 132
Query: 147 DAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMKNHF 197
D EL+ VL +LG + ++ E C+KM+ ++DEN+DG+IDF EFV +++ F
Sbjct: 133 DENELKHVLSLLGYDECTKME-CRKMVKVYDENRDGKIDFYEFVKLIEKSF 182
>UniRef100_Q09011 Calcium-binding protein CAST [Solanum tuberosum]
Length = 199
Score = 79.7 bits (195), Expect = 4e-14
Identities = 42/85 (49%), Positives = 60/85 (70%), Gaps = 2/85 (2%)
Query: 112 YGSKELCEVFEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENC 169
+GSK ++ +P+ +E LK+AFDVFDEN DGFI AKELQ VL LGL +GSE +
Sbjct: 109 FGSKCEDKLGLNPDPAQDESDLKEAFDVFDENGDGFISAKELQVVLEKLGLPEGSEIDRV 168
Query: 170 QKMITIFDENQDGRIDFIEFVNIMK 194
+ MI+ +++ DGR+DF EF ++M+
Sbjct: 169 EMMISSVEQDHDGRVDFFEFKDMMR 193
Score = 37.0 bits (84), Expect = 0.30
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 121 FEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
F PSL L++ FDVFD N D I +EL + L +LGL ++ + M+ + +
Sbjct: 29 FRLRSPSLNSIRLRRIFDVFDRNHDCLISVEELSQALNLLGL--DADLSEIESMVKLHIK 86
Query: 179 NQDGRIDFIEFVNIMKN 195
++ + F +F + ++
Sbjct: 87 PENTGLRFEDFETLHRS 103
>UniRef100_Q9LX27 Calmodulin-like protein [Arabidopsis thaliana]
Length = 195
Score = 79.3 bits (194), Expect = 5e-14
Identities = 54/195 (27%), Positives = 89/195 (44%), Gaps = 31/195 (15%)
Query: 19 LFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCSSQENESNIG 78
L+ L + FL K+ FF W+ + ++ +ES +
Sbjct: 7 LYNLFNSFLLCLVPKKLRVFFPPSWYI------DDKNPPPPDESETESPVDLKRVFQMFD 60
Query: 79 RDNGDMIERDEVKMVMEKMGFFCSSES------------------EELEEKYGSKELCEV 120
++ I ++E+ +E +G F + E E YGS +
Sbjct: 61 KNGDGRITKEELNDSLENLGIFMPDKDLIQMIQKMDANGDGCVDINEFESLYGS-----I 115
Query: 121 FEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQ 180
EE E +++ AF+VFD++ DGFI +EL V+ LGLKQG E C++MI DE+
Sbjct: 116 VEEKEEG--DMRDAFNVFDQDGDGFITVEELNSVMTSLGLKQGKTLECCKEMIMQVDEDG 173
Query: 181 DGRIDFIEFVNIMKN 195
DGR+++ EF+ +MK+
Sbjct: 174 DGRVNYKEFLQMMKS 188
>UniRef100_Q9FR00 Avr9/Cf-9 rapidly elicited protein 31 [Nicotiana tabacum]
Length = 205
Score = 79.3 bits (194), Expect = 5e-14
Identities = 41/86 (47%), Positives = 61/86 (70%), Gaps = 3/86 (3%)
Query: 112 YGSK-ELCEVFEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFEN 168
+GSK E ++ + +P +E LK AF+VFDEN DGFI AKELQ VL LGL +G+E +
Sbjct: 113 FGSKYEEDKIVLDQDPDQDEVDLKDAFNVFDENGDGFISAKELQAVLEKLGLPEGNEIDR 172
Query: 169 CQKMITIFDENQDGRIDFIEFVNIMK 194
+ MI+ D++ DG++DF+EF ++M+
Sbjct: 173 VEMMISSVDQDHDGQVDFVEFKDMMR 198
Score = 36.2 bits (82), Expect = 0.51
Identities = 22/71 (30%), Positives = 36/71 (49%), Gaps = 4/71 (5%)
Query: 121 FEENEPSLEE--LKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
F PSL L++ FDVFD N D I EL + L +LGL ++ + M+ + +
Sbjct: 33 FRLRSPSLNSIRLRRIFDVFDRNHDSLISVDELSQALNLLGL--DADQSEIESMVRSYIK 90
Query: 179 NQDGRIDFIEF 189
+ + + F +F
Sbjct: 91 SGNNGLRFEDF 101
>UniRef100_O22845 Putative calcium binding protein [Arabidopsis thaliana]
Length = 215
Score = 78.6 bits (192), Expect = 9e-14
Identities = 58/202 (28%), Positives = 94/202 (45%), Gaps = 27/202 (13%)
Query: 19 LFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREEKKVSESKCSSQE-NESNI 77
L+ +++ FL K+ F WF +S S S S ++ + S +
Sbjct: 7 LYNILNSFLLSLVPKKLRTLFPLSWFDKTLHKNSPPSPSTMLPSPSSSSAPTKRIDPSEL 66
Query: 78 GR------DNGD-MIERDEVKMVMEKMGFFCSSES------------------EELEEKY 112
R NGD I ++E+ +E +G + + +E E Y
Sbjct: 67 KRVFQMFDKNGDGRITKEELNDSLENLGIYIPDKDLTQMIHKIDANGDGCVDIDEFESLY 126
Query: 113 GSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKM 172
S + E + E E++K AF+VFD++ DGFI +EL+ V+ LGLKQG + C+KM
Sbjct: 127 SSI-VDEHHNDGETEEEDMKDAFNVFDQDGDGFITVEELKSVMASLGLKQGKTLDGCKKM 185
Query: 173 ITIFDENQDGRIDFIEFVNIMK 194
I D + DGR+++ EF+ +MK
Sbjct: 186 IMQVDADGDGRVNYKEFLQMMK 207
>UniRef100_Q42141 Calcium-binding protein [Arabidopsis thaliana]
Length = 101
Score = 78.2 bits (191), Expect = 1e-13
Identities = 42/96 (43%), Positives = 59/96 (60%), Gaps = 7/96 (7%)
Query: 101 CSSESEELEEKY--GSKELCEVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVIL 158
C + ++ L+E + G C+ E++ L+ AF+VFDE+ DGFI A ELQ+VL L
Sbjct: 5 CCTPTKTLDESFFGGEGSCCDGSPESD-----LEXAFNVFDEDGDGFISAVELQKVLKKL 59
Query: 159 GLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
GL + E E +KMI D N DGR+DF EF N+M+
Sbjct: 60 GLPEAGEIEQVEKMIVSVDSNHDGRVDFFEFKNMMQ 95
>UniRef100_Q8L4G1 Hypothetical protein OJ1118_B03.113 [Oryza sativa]
Length = 218
Score = 77.0 bits (188), Expect = 3e-13
Identities = 38/81 (46%), Positives = 53/81 (64%)
Query: 119 EVFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
E+ + S ELK AF VFD N+DGFI A EL V+ LG K+G +E+C +MI FDE
Sbjct: 111 ELLDRKMASEGELKDAFYVFDRNEDGFICASELWSVMRRLGFKEGQRYEDCMRMIHTFDE 170
Query: 179 NQDGRIDFIEFVNIMKNHFCC 199
++DGRI ++EF +M++ C
Sbjct: 171 DRDGRISYLEFRRMMEDADGC 191
>UniRef100_Q9SRR7 Putative calmodulin [Arabidopsis thaliana]
Length = 153
Score = 76.6 bits (187), Expect = 3e-13
Identities = 45/129 (34%), Positives = 74/129 (56%), Gaps = 17/129 (13%)
Query: 81 NGD-MIERDEVKMVMEKMGFFCSSES-----EELE---------EKYGSKELCEVFEENE 125
NGD I + E+ +E +G + + E+++ E++G L + E
Sbjct: 16 NGDGKITKQELNDSLENLGIYIPDKDLVQMIEKIDLNGDGYVDIEEFGG--LYQTIMEER 73
Query: 126 PSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRID 185
E++++AF+VFD+N+DGFI +EL+ VL LGLKQG E+C++MI+ D + DG ++
Sbjct: 74 DEEEDMREAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCKRMISKVDVDGDGMVN 133
Query: 186 FIEFVNIMK 194
F EF +MK
Sbjct: 134 FKEFKQMMK 142
Score = 34.7 bits (78), Expect = 1.5
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
EL + F +FD N DG I +EL L LG+ ++ +MI D N DG +D EF
Sbjct: 5 ELARIFQMFDRNGDGKITKQELNDSLENLGIYIPD--KDLVQMIEKIDLNGDGYVDIEEF 62
>UniRef100_Q9FI19 Calmodulin-like protein [Arabidopsis thaliana]
Length = 181
Score = 76.3 bits (186), Expect = 4e-13
Identities = 36/65 (55%), Positives = 47/65 (71%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
+L++AF+VFDE+ DGFI A ELQ+VL LGL + E E +KMI D N DGR+DF EF
Sbjct: 111 DLEEAFNVFDEDGDGFISAVELQKVLKKLGLPEAGEIEQVEKMIVSVDSNHDGRVDFFEF 170
Query: 190 VNIMK 194
N+M+
Sbjct: 171 KNMMQ 175
Score = 40.8 bits (94), Expect = 0.021
Identities = 25/76 (32%), Positives = 40/76 (51%), Gaps = 4/76 (5%)
Query: 121 FEENEPSLEELK--QAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
F PSL L+ + FD+FD+N DGFI +EL + L LGL ++F + + + F +
Sbjct: 17 FRLRSPSLNALRLHRVFDLFDKNNDGFITVEELSQALSRLGL--DADFSDLKSTVDSFIK 74
Query: 179 NQDGRIDFIEFVNIMK 194
+ F +F + K
Sbjct: 75 PDKTGLRFDDFAALHK 90
>UniRef100_Q9LNE7 T21E18.4 protein [Arabidopsis thaliana]
Length = 150
Score = 75.9 bits (185), Expect = 6e-13
Identities = 46/119 (38%), Positives = 70/119 (58%), Gaps = 9/119 (7%)
Query: 79 RDNGDMIERDEVKMVMEKM---GFFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAF 135
R G I E+ ++EK+ G C + +E E Y + + +E + E++K+AF
Sbjct: 31 RSLGIYIPDKELTQMIEKIDVNGDGCV-DIDEFGELYKT-----IMDEEDEEEEDMKEAF 84
Query: 136 DVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
+VFD+N DGFI EL+ VL LGLKQG ++C+KMI D + DGR+++ EF +MK
Sbjct: 85 NVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCKKMIKKVDVDGDGRVNYKEFRQMMK 143
Score = 42.7 bits (99), Expect = 0.005
Identities = 26/65 (40%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
ELK+ F +FD+N DG I KEL L LG+ + +MI D N DG +D EF
Sbjct: 5 ELKRVFQMFDKNGDGTITGKELSETLRSLGIYIPD--KELTQMIEKIDVNGDGCVDIDEF 62
Query: 190 VNIMK 194
+ K
Sbjct: 63 GELYK 67
>UniRef100_Q8LHN5 Hypothetical protein P0519E12.112 [Oryza sativa]
Length = 213
Score = 75.9 bits (185), Expect = 6e-13
Identities = 44/139 (31%), Positives = 75/139 (53%), Gaps = 6/139 (4%)
Query: 64 SESKCSS--QENESNIG--RDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCE 119
S+ CSS E+E + + + + EV+ +ME++G E L+ + G E+
Sbjct: 77 SDEPCSSCGGEDEDVVAAVEEEEAHLSKVEVEEIMERIGLGVGGHGEGLKARMGRDEVSR 136
Query: 120 VFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDEN 179
+F+ +EPS E+++AF VFD N DGFIDA +L+ L LG ++ + C+ MI +
Sbjct: 137 LFDADEPSFAEVRRAFAVFDGNADGFIDADDLRAALARLGFRE--DAAACRAMIAASCGS 194
Query: 180 QDGRIDFIEFVNIMKNHFC 198
D R++ +FV ++ C
Sbjct: 195 VDARMNLFQFVKFLETGLC 213
>UniRef100_Q9SCA1 Calcium-binding protein [Lotus japonicus]
Length = 230
Score = 74.3 bits (181), Expect = 2e-12
Identities = 45/129 (34%), Positives = 72/129 (54%), Gaps = 17/129 (13%)
Query: 81 NGD-MIERDEVKMVMEKMGFFCSSES-----EELE---------EKYGSKELCEVFEENE 125
NGD I + E+ +E +G F + E ++ +++G EL + +
Sbjct: 96 NGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNGDGCVDIDEFG--ELYQSIMDER 153
Query: 126 PSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRID 185
E++++AF+VFD+N DGFI +EL+ VL LG+KQG E+C+KMI D + DG +D
Sbjct: 154 DEEEDMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVD 213
Query: 186 FIEFVNIMK 194
+ EF +MK
Sbjct: 214 YKEFKQMMK 222
Score = 38.5 bits (88), Expect = 0.10
Identities = 25/60 (41%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
ELK+ F +FD N DG I KEL L LG+ + +MI D N DG +D EF
Sbjct: 85 ELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPD--KELTQMIERIDVNGDGCVDIDEF 142
>UniRef100_Q9SU00 Putative calmodulin [Arabidopsis thaliana]
Length = 152
Score = 73.9 bits (180), Expect = 2e-12
Identities = 43/113 (38%), Positives = 70/113 (61%), Gaps = 4/113 (3%)
Query: 82 GDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFDEN 141
G M+ +E+ ++ KM + + +++GS E+ EE E E++++AF VFD+N
Sbjct: 34 GIMVPENEINEMIAKMDV--NGDGAMDIDEFGSLYQ-EMVEEKEEE-EDMREAFRVFDQN 89
Query: 142 KDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
DGFI +EL+ VL +GLKQG E+C+KMI+ D + DG ++F EF +M+
Sbjct: 90 GDGFITDEELRSVLASMGLKQGRTLEDCKKMISKVDVDGDGMVNFKEFKQMMR 142
>UniRef100_P43187 Calcium-binding allergen Bet v 3 [Betula verrucosa]
Length = 205
Score = 72.4 bits (176), Expect = 6e-12
Identities = 47/131 (35%), Positives = 65/131 (48%), Gaps = 9/131 (6%)
Query: 65 ESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEEN 124
ES S E NIG D I + + F E E+ E+ K + E +
Sbjct: 78 ESTVKSFTREGNIGLQFEDFIS---LHQSLNDSYFAYGGEDEDDNEEDMRKSILSQEEAD 134
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
F VFDE+ DG+I A+ELQ VL LG +GSE + +KMI D N+DGR+
Sbjct: 135 SFG------GFKVFDEDGDGYISARELQMVLGKLGFSEGSEIDRVEKMIVSVDSNRDGRV 188
Query: 185 DFIEFVNIMKN 195
DF EF ++M++
Sbjct: 189 DFFEFKDMMRS 199
Score = 40.0 bits (92), Expect = 0.035
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 8/65 (12%)
Query: 131 LKQAFDVFDENKDGFIDAKELQRVLVILGLKQG-SEFENCQKMITIFDENQDGRI--DFI 187
L++ FD+FD+N DG I EL R L +LGL+ SE E+ K T ++G I F
Sbjct: 41 LRRIFDLFDKNSDGIITVDELSRALNLLGLETDLSELESTVKSFT-----REGNIGLQFE 95
Query: 188 EFVNI 192
+F+++
Sbjct: 96 DFISL 100
>UniRef100_Q6ZXI3 Putative calmodulin-like protein [Populus euramericana]
Length = 193
Score = 71.2 bits (173), Expect = 1e-11
Identities = 34/66 (51%), Positives = 46/66 (69%)
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
+L +AF VFDE+ DG+I A ELQ VL LGL + E + +MIT D NQDGR+DF EF
Sbjct: 123 DLSEAFKVFDEDGDGYISAHELQVVLRKLGLPEAKEIDRIHQMITSVDRNQDGRVDFFEF 182
Query: 190 VNIMKN 195
++M++
Sbjct: 183 KDMMRS 188
Score = 37.0 bits (84), Expect = 0.30
Identities = 22/69 (31%), Positives = 39/69 (55%), Gaps = 4/69 (5%)
Query: 126 PSLE--ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGR 183
PSL L++ FD+FD+N DG I +E+ + L +LGL ++F + + I +
Sbjct: 21 PSLNFLRLRRIFDLFDKNGDGMITIEEISQALSLLGL--DADFSDLEFTIKSHIKPGSSG 78
Query: 184 IDFIEFVNI 192
+ F +FV++
Sbjct: 79 LSFEDFVSL 87
>UniRef100_Q7XZH3 Hypothetical protein OSJNBb0033J23.19 [Oryza sativa]
Length = 213
Score = 71.2 bits (173), Expect = 1e-11
Identities = 40/124 (32%), Positives = 70/124 (56%), Gaps = 9/124 (7%)
Query: 79 RDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEENEPSLE-------EL 131
R G + R +V V+ +G + E ++ +++ + +CE E E EL
Sbjct: 90 RRGGAGLSRHDVAAVVASLGMVAAGEDDDDDDE--ACGVCEAVAAVEEMAEGKVAGEGEL 147
Query: 132 KQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVN 191
++AF VFD ++DG++ A EL V+ LG+++G+ + +C +MI +D + DGRI F EF
Sbjct: 148 REAFYVFDRDEDGYVSAAELWNVMRRLGIEEGARYGDCVRMIAAYDGDGDGRISFQEFRA 207
Query: 192 IMKN 195
+M+N
Sbjct: 208 MMEN 211
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 334,854,532
Number of Sequences: 2790947
Number of extensions: 14293184
Number of successful extensions: 65363
Number of sequences better than 10.0: 2375
Number of HSP's better than 10.0 without gapping: 787
Number of HSP's successfully gapped in prelim test: 1599
Number of HSP's that attempted gapping in prelim test: 60668
Number of HSP's gapped (non-prelim): 4482
length of query: 199
length of database: 848,049,833
effective HSP length: 121
effective length of query: 78
effective length of database: 510,345,246
effective search space: 39806929188
effective search space used: 39806929188
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC135313.7


