

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.10 - phase: 0
(548 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
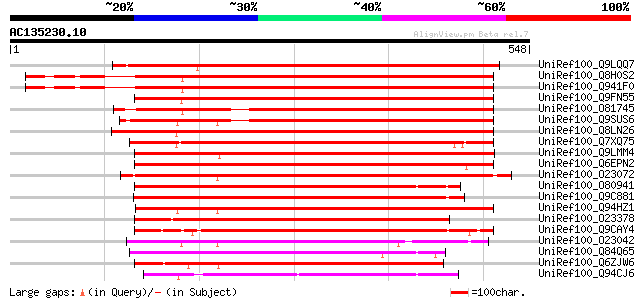
UniRef100_Q9LQQ7 F24B9.5 protein [Arabidopsis thaliana] 603 e-171
UniRef100_Q8H0S2 At4g23490/F16G20_190 [Arabidopsis thaliana] 561 e-158
UniRef100_Q941F0 AT4g23490/F16G20_190 [Arabidopsis thaliana] 558 e-157
UniRef100_Q9FN55 Arabidopsis thaliana genomic DNA, chromosome 5,... 556 e-157
UniRef100_O81745 Hypothetical protein F16G20.190 [Arabidopsis th... 521 e-146
UniRef100_Q9SUS6 Hypothetical protein AT4g11350 [Arabidopsis tha... 506 e-142
UniRef100_Q8LN26 Hypothetical protein OSJNBa0053C23.29 [Oryza sa... 503 e-141
UniRef100_Q7XQ75 OSJNBa0011J08.14 protein [Oryza sativa] 501 e-140
UniRef100_Q9LMM4 F22L4.11 protein [Arabidopsis thaliana] 476 e-133
UniRef100_Q6EPN2 Fringe-related protein-like [Oryza sativa] 473 e-132
UniRef100_O23072 A_IG005I10.16 protein [Arabidopsis thaliana] 455 e-126
UniRef100_O80941 Putative zinc finger protein [Arabidopsis thali... 376 e-102
UniRef100_Q9C881 Hypothetical protein T16O9.11 [Arabidopsis thal... 342 2e-92
UniRef100_Q94HZ1 Hypothetical protein OSJNBa0076F20.12 [Oryza sa... 333 8e-90
UniRef100_O23378 Hypothetical protein dl3665c [Arabidopsis thali... 330 7e-89
UniRef100_Q9CAY4 Hypothetical protein F24K9.9 [Arabidopsis thali... 321 3e-86
UniRef100_O23042 YUP8H12.11 protein [Arabidopsis thaliana] 312 2e-83
UniRef100_Q84Q65 Hypothetical protein OSJNBa0071M09.15 [Oryza sa... 294 4e-78
UniRef100_Q6ZJW6 Putative fringe-related protein [Oryza sativa] 279 2e-73
UniRef100_Q94CJ6 Hypothetical protein At5g12460 [Arabidopsis tha... 274 6e-72
>UniRef100_Q9LQQ7 F24B9.5 protein [Arabidopsis thaliana]
Length = 560
Score = 603 bits (1554), Expect = e-171
Identities = 294/435 (67%), Positives = 343/435 (78%), Gaps = 27/435 (6%)
Query: 109 EEQENKSPIVVRLTPEQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVV 168
EE ++ P V L P Q + +T L HIVFGIAASS LW TRKEYIK WWRP +TRGVV
Sbjct: 101 EEDDDVPPRVPALYP-QRPRMFNTTLDHIVFGIAASSVLWETRKEYIKSWWRPGKTRGVV 159
Query: 169 WLDQRVSTQRNEGLPDIRISDDTSKFRY-------------------------TNRQGQR 203
W+D+RV T RN+ LP+IRIS DTS+FRY T+ G R
Sbjct: 160 WIDKRVRTYRNDPLPEIRISQDTSRFRYLLISTISDVFYKKSLIQIKKSCFRYTHPVGDR 219
Query: 204 SALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDHRHFYYVGSSSESHV 263
SA+RISRVVTETL+LG K VRWFVMGDDDTVFVVDNVV +LSKYDH FYYVGSSSE+HV
Sbjct: 220 SAVRISRVVTETLRLGKKGVRWFVMGDDDTVFVVDNVVNVLSKYDHTQFYYVGSSSEAHV 279
Query: 264 QNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDRMQACMAELGVPLTK 323
QNI FSY+MA+GGGGFAISY LA+EL MQDRCIQRYP LYGSDDR+QACM ELGVPLTK
Sbjct: 280 QNIFFSYSMAFGGGGFAISYALALELLRMQDRCIQRYPGLYGSDDRIQACMTELGVPLTK 339
Query: 324 EAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQSIRHLMESINQDSSG 383
E GFHQYDVYGDLLGLLGAHPVAPLVSLHH+DVVQPIFP M R++++RHLM S D +
Sbjct: 340 EPGFHQYDVYGDLLGLLGAHPVAPLVSLHHIDVVQPIFPKMKRSRALRHLMSSAVLDPAS 399
Query: 384 IMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTR 443
I QQSICYD+NRFWSISVSWGF+VQI+RG++SPRELEMPSRTFLNW+R+ADY YAFNTR
Sbjct: 400 IFQQSICYDQNRFWSISVSWGFVVQIIRGIISPRELEMPSRTFLNWFRKADYIGYAFNTR 459
Query: 444 PVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTKS-PFCRWRMESPEKITSIVVTKR 502
PV++HPCQ+PFV+Y++ +D RQ++G Y+ D+T+ P CRWR++SP KI S+VV KR
Sbjct: 460 PVSRHPCQRPFVFYLNSAKYDEGRRQVIGYYNLDKTRRIPGCRWRLDSPGKIDSVVVLKR 519
Query: 503 RDPLRWKKVRLLIIF 517
DPLRW KV ++F
Sbjct: 520 PDPLRWHKVGPHLLF 534
>UniRef100_Q8H0S2 At4g23490/F16G20_190 [Arabidopsis thaliana]
Length = 526
Score = 561 bits (1447), Expect = e-158
Identities = 276/500 (55%), Positives = 351/500 (70%), Gaps = 49/500 (9%)
Query: 17 ITTWLILITIVLYILYSCNILLFNNEQECPMTTTTLDDTTQ-DDVHMTSYNISSTSDKNI 75
+ WLI + YI+Y ++ T+ + DD+T V S N+SS N+
Sbjct: 36 LMVWLICFIVFTYIIYMLKLV---------STSRSCDDSTSFTTVSALSTNVSS----NV 82
Query: 76 EHKVLPIENKEQMQDETEKETEQDEDDFREGEGEEQENKSPIVVRLTPEQMAQRQDTELK 135
+ ++ + +E E++T D+ T+L
Sbjct: 83 SSLSTSLASRRRNWEEEEEDTVVDKL------------------------------TDLN 112
Query: 136 HIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNEG-----LPDIRISDD 190
H+VFGIAASS LW RKEYIKIW++PK+ RG VWLD+ V ++ LP ++IS
Sbjct: 113 HVVFGIAASSKLWKQRKEYIKIWYKPKRMRGYVWLDKEVKKSLSDDDDEKLLPPVKISGG 172
Query: 191 TSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDHR 250
T+ F YTN+QGQRSALRISR+V+ETL+LG K+VRWFVMGDDDTVFV+DN++R+L KYDH
Sbjct: 173 TASFPYTNKQGQRSALRISRIVSETLRLGPKNVRWFVMGDDDTVFVIDNLIRVLRKYDHE 232
Query: 251 HFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDRM 310
YY+GS SESH+QNI FSY MAYGGGGFAISYPLA L+ MQDRCIQRYPALYGSDDRM
Sbjct: 233 QMYYIGSLSESHLQNIFFSYGMAYGGGGFAISYPLAKALSKMQDRCIQRYPALYGSDDRM 292
Query: 311 QACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQSI 370
QACMAELGVPLTKE GFHQYDVYG+L GLL AHPV P VS+HHLDVV+PIFP+M+R +++
Sbjct: 293 QACMAELGVPLTKELGFHQYDVYGNLFGLLAAHPVTPFVSMHHLDVVEPIFPNMTRVRAL 352
Query: 371 RHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNWY 430
+ + E + DS+G++QQSICYDK++ W+ISVSWG+ VQI RG+ SPRE+EMPSRTFLNWY
Sbjct: 353 KKITEPMKLDSAGLLQQSICYDKHKSWTISVSWGYAVQIFRGIFSPREMEMPSRTFLNWY 412
Query: 431 RRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTKSPFCRWRMES 490
+RADYTAYAFNTRPV+++PCQKPFV+YMS T FD V Y+ + P CRW+M +
Sbjct: 413 KRADYTAYAFNTRPVSRNPCQKPFVFYMSSTKFDQQLNTTVSEYTIHRVSHPSCRWKMTN 472
Query: 491 PEKITSIVVTKRRDPLRWKK 510
P +I +IVV K+ DP W++
Sbjct: 473 PAEINTIVVYKKPDPHLWER 492
>UniRef100_Q941F0 AT4g23490/F16G20_190 [Arabidopsis thaliana]
Length = 526
Score = 558 bits (1439), Expect = e-157
Identities = 275/500 (55%), Positives = 350/500 (70%), Gaps = 49/500 (9%)
Query: 17 ITTWLILITIVLYILYSCNILLFNNEQECPMTTTTLDDTTQ-DDVHMTSYNISSTSDKNI 75
+ WLI + YI+Y ++ T+ + DD+T V S N+SS N+
Sbjct: 36 LMVWLICFIVFTYIIYMLKLV---------STSRSCDDSTSFTTVSALSTNVSS----NV 82
Query: 76 EHKVLPIENKEQMQDETEKETEQDEDDFREGEGEEQENKSPIVVRLTPEQMAQRQDTELK 135
+ ++ + +E E++T D+ T+L
Sbjct: 83 SSLSTSLASRRRNWEEEEEDTVVDKL------------------------------TDLN 112
Query: 136 HIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNEG-----LPDIRISDD 190
H+VFGIAASS LW RKEYIKIW++ K+ RG VWLD+ V ++ LP ++IS
Sbjct: 113 HVVFGIAASSKLWKQRKEYIKIWYKQKRMRGYVWLDKEVKKSLSDDDDEKLLPPVKISGG 172
Query: 191 TSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDHR 250
T+ F YTN+QGQRSALRISR+V+ETL+LG K+VRWFVMGDDDTVFV+DN++R+L KYDH
Sbjct: 173 TASFPYTNKQGQRSALRISRIVSETLRLGPKNVRWFVMGDDDTVFVIDNLIRVLRKYDHE 232
Query: 251 HFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDRM 310
YY+GS SESH+QNI FSY MAYGGGGFAISYPLA L+ MQDRCIQRYPALYGSDDRM
Sbjct: 233 QMYYIGSLSESHLQNIFFSYGMAYGGGGFAISYPLAKALSKMQDRCIQRYPALYGSDDRM 292
Query: 311 QACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQSI 370
QACMAELGVPLTKE GFHQYDVYG+L GLL AHPV P VS+HHLDVV+PIFP+M+R +++
Sbjct: 293 QACMAELGVPLTKELGFHQYDVYGNLFGLLAAHPVTPFVSMHHLDVVEPIFPNMTRVRAL 352
Query: 371 RHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNWY 430
+ + E + DS+G++QQSICYDK++ W+ISVSWG+ VQI RG+ SPRE+EMPSRTFLNWY
Sbjct: 353 KKITEPMKLDSAGLLQQSICYDKHKSWTISVSWGYAVQIFRGIFSPREMEMPSRTFLNWY 412
Query: 431 RRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTKSPFCRWRMES 490
+RADYTAYAFNTRPV+++PCQKPFV+YMS T FD V Y+ + P CRW+M +
Sbjct: 413 KRADYTAYAFNTRPVSRNPCQKPFVFYMSSTKFDQQLNTTVSEYTIHRVSHPSCRWKMTN 472
Query: 491 PEKITSIVVTKRRDPLRWKK 510
P +I +IVV K+ DP W++
Sbjct: 473 PAEINTIVVYKKPDPHLWER 492
>UniRef100_Q9FN55 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MYC6
[Arabidopsis thaliana]
Length = 524
Score = 556 bits (1434), Expect = e-157
Identities = 257/382 (67%), Positives = 307/382 (80%), Gaps = 3/382 (0%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNE---GLPDIRIS 188
T +H+VFGIAAS+ LW RKEYIKIW++P Q R VWL++ V+ + E LP ++IS
Sbjct: 110 TGFQHVVFGIAASARLWKQRKEYIKIWYKPNQMRSYVWLEKPVTEEDEEDEISLPPVKIS 169
Query: 189 DDTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYD 248
DTSKF Y N+QG RSA+RISR+VTETLKLGLKDVRWFVMGDDDTVFV +N++R+L KYD
Sbjct: 170 GDTSKFPYKNKQGHRSAIRISRIVTETLKLGLKDVRWFVMGDDDTVFVAENLIRVLRKYD 229
Query: 249 HRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDD 308
H YY+GS SESH+QNI+FSY MAYGGGGFAISYPLAV L+ MQDRCI+RYPALYGSDD
Sbjct: 230 HNQMYYIGSLSESHLQNIYFSYGMAYGGGGFAISYPLAVALSKMQDRCIKRYPALYGSDD 289
Query: 309 RMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQ 368
RMQACMAELGVPLTKE GFHQYDVYG+L GLL AHPVAPLV+LHHLDVV+PIFP+M+R
Sbjct: 290 RMQACMAELGVPLTKELGFHQYDVYGNLFGLLAAHPVAPLVTLHHLDVVEPIFPNMTRVD 349
Query: 369 SIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLN 428
+++HL DS+G+MQQSICYDK R W++SVSWGF VQI RG+ S RE+EMPSRTFLN
Sbjct: 350 ALKHLQVPAKLDSAGLMQQSICYDKRRKWTVSVSWGFAVQIFRGIFSAREIEMPSRTFLN 409
Query: 429 WYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTKSPFCRWRM 488
WYRRADYTAYAFNTRPV++HPCQKPFV+YM+ T + V Y + P CRW+M
Sbjct: 410 WYRRADYTAYAFNTRPVSRHPCQKPFVFYMTSTRVHRVTNMTVSRYEIHRVAHPECRWKM 469
Query: 489 ESPEKITSIVVTKRRDPLRWKK 510
+P I +++V K+ DP W +
Sbjct: 470 ANPSDIKTVIVYKKPDPHLWDR 491
>UniRef100_O81745 Hypothetical protein F16G20.190 [Arabidopsis thaliana]
Length = 614
Score = 521 bits (1341), Expect = e-146
Identities = 250/407 (61%), Positives = 308/407 (75%), Gaps = 33/407 (8%)
Query: 110 EQENKSPIVVRLTPEQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVW 169
E+E + +V +LT +L H+VFGIAASS LW RKEYIKIW++PK+ RG VW
Sbjct: 206 EEEEEDTVVDKLT----------DLNHVVFGIAASSKLWKQRKEYIKIWYKPKRMRGYVW 255
Query: 170 LDQRVSTQRNEG-----LPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVR 224
LD+ V ++ LP ++IS T+ F YTN+QGQRSALRISR+V+ETL+LG K+VR
Sbjct: 256 LDKEVKKSLSDDDDEKLLPPVKISGGTASFPYTNKQGQRSALRISRIVSETLRLGPKNVR 315
Query: 225 WFVMGDDDTVFVVDNVVRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYP 284
WFVMGDDDT+ YY+GS SESH+QNI FSY MAYGGGGFAISYP
Sbjct: 316 WFVMGDDDTM------------------YYIGSLSESHLQNIFFSYGMAYGGGGFAISYP 357
Query: 285 LAVELATMQDRCIQRYPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHP 344
LA L+ MQDRCIQRYPALYGSDDRMQACMAELGVPLTKE GFHQYDVYG+L GLL AHP
Sbjct: 358 LAKALSKMQDRCIQRYPALYGSDDRMQACMAELGVPLTKELGFHQYDVYGNLFGLLAAHP 417
Query: 345 VAPLVSLHHLDVVQPIFPSMSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWG 404
V P VS+HHLDVV+PIFP+M+R ++++ + E + DS+G++QQSICYDK++ W+ISVSWG
Sbjct: 418 VTPFVSMHHLDVVEPIFPNMTRVRALKKITEPMKLDSAGLLQQSICYDKHKSWTISVSWG 477
Query: 405 FMVQILRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFD 464
+ VQI RG+ SPRE+EMPSRTFLNWY+RADYTAYAFNTRPV+++PCQKPFV+YMS T FD
Sbjct: 478 YAVQIFRGIFSPREMEMPSRTFLNWYKRADYTAYAFNTRPVSRNPCQKPFVFYMSSTKFD 537
Query: 465 TASRQIVGVYSRDQTKSPFCRWRMESPEKITSIVVTKRRDPLRWKKV 511
V Y+ + P CRW+M +P +I +IVV K+ DP W++V
Sbjct: 538 QQLNTTVSEYTIHRVSHPSCRWKMTNPAEINTIVVYKKPDPHLWERV 584
>UniRef100_Q9SUS6 Hypothetical protein AT4g11350 [Arabidopsis thaliana]
Length = 489
Score = 506 bits (1303), Expect = e-142
Identities = 246/402 (61%), Positives = 300/402 (74%), Gaps = 29/402 (7%)
Query: 117 IVVRLTPEQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVS- 175
+ V+ P A+++ T+L H+VFGIAASS LW RKEYIKIW++PK+ RG VWLD+ V
Sbjct: 75 VTVKAVP---AEQEATDLNHVVFGIAASSKLWKQRKEYIKIWYKPKKMRGYVWLDEEVKI 131
Query: 176 ---TQRNEGLPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKL----GLKDVRWFVM 228
T E LP +RIS DTS F YTN+QG RSA+RISR+V+ETL K+VRWFVM
Sbjct: 132 KSETGDQESLPSVRISGDTSSFPYTNKQGHRSAIRISRIVSETLMSLDSESKKNVRWFVM 191
Query: 229 GDDDTVFVVDNVVRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVE 288
GDDDT+ YY+GS SESH+QNI FSY MAYGGGGFAISYPLAV
Sbjct: 192 GDDDTM------------------YYIGSLSESHLQNIIFSYGMAYGGGGFAISYPLAVA 233
Query: 289 LATMQDRCIQRYPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPL 348
L+ MQD+CIQRYPALYGSDDRMQACMAELGVPLTKE GFHQYDV+G+L GLL AHP+ P
Sbjct: 234 LSKMQDQCIQRYPALYGSDDRMQACMAELGVPLTKEIGFHQYDVHGNLFGLLAAHPITPF 293
Query: 349 VSLHHLDVVQPIFPSMSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQ 408
VS+HHLDVV+PIFP+M+R ++I+ L + DS+ ++QQSICYDK++ W+ISVSWGF VQ
Sbjct: 294 VSMHHLDVVEPIFPNMTRVRAIKKLTTPMKIDSAALLQQSICYDKHKSWTISVSWGFAVQ 353
Query: 409 ILRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASR 468
+ RG SPRE+EMPSRTFLNWY+RADYTAYAFNTRPV+++ CQKPFV++MS FD
Sbjct: 354 VFRGSFSPREMEMPSRTFLNWYKRADYTAYAFNTRPVSRNHCQKPFVFHMSSAKFDPQLN 413
Query: 469 QIVGVYSRDQTKSPFCRWRMESPEKITSIVVTKRRDPLRWKK 510
V Y+R + P CRW M +PE+I +IVV K+ DP W +
Sbjct: 414 TTVSEYTRHRVPQPACRWDMANPEEINTIVVYKKPDPHLWNR 455
>UniRef100_Q8LN26 Hypothetical protein OSJNBa0053C23.29 [Oryza sativa]
Length = 527
Score = 503 bits (1295), Expect = e-141
Identities = 242/410 (59%), Positives = 300/410 (73%), Gaps = 7/410 (1%)
Query: 108 GEEQENKSPIVVRLTPEQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGV 167
G + SP + P + A T L+HI FGI ASS LW +RKEYIK+WWRP + RG
Sbjct: 81 GSSPSSSSPPPTPVRPMRSADEAPTGLRHIAFGIGASSALWKSRKEYIKLWWRPGRMRGF 140
Query: 168 VWLDQRV----STQRNEGLPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKLGLKDV 223
VW+D+ V S GLP I +S DTSKF YT+ G RSALRISR+V+ET +LGL V
Sbjct: 141 VWMDRPVEEFYSKSSRTGLPPIMVSSDTSKFPYTHGAGSRSALRISRIVSETFRLGLPGV 200
Query: 224 RWFVMGDDDTVFVVDNVVRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISY 283
RWFVMGDDDTVF+ +N+V +LS+YDHR YY+GS SESH+QN+ FSY MA+GGGGFAIS
Sbjct: 201 RWFVMGDDDTVFLPENLVHVLSQYDHRQPYYIGSPSESHIQNLIFSYGMAFGGGGFAISR 260
Query: 284 PLAVELATMQDRCIQRYPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAH 343
LA ELA MQD C+ RYPALYGSDDR+ ACM+ELGVPLT+ GFHQ D++GD+LGLLGAH
Sbjct: 261 ALAEELAKMQDGCLHRYPALYGSDDRIHACMSELGVPLTRHPGFHQCDLWGDVLGLLGAH 320
Query: 344 PVAPLVSLHHLDVVQPIFPSM-SRAQSIRHLMES-INQDSSGIMQQSICYDKNRFWSISV 401
PVAPLV+LHHLD ++P+FP+ SRA ++R L + + DS+ + QQS+CYD+ W++SV
Sbjct: 321 PVAPLVTLHHLDFLEPVFPTTPSRAGALRKLFDGPVRLDSAAVAQQSVCYDREHHWTVSV 380
Query: 402 SWGFMVQILRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKT 461
SWGF V ++RGVLSPRE+E P R+FLNWY+RADYTAY+FNTRPVA+ PCQKP VYYM +
Sbjct: 381 SWGFAVMVVRGVLSPREMETPMRSFLNWYKRADYTAYSFNTRPVARQPCQKPRVYYMRDS 440
Query: 462 HFDTASRQIVGVYSRDQTKSPFCRWRMESPEKITS-IVVTKRRDPLRWKK 510
D V Y R + K P CRWR+ P + IVV K+ DP WK+
Sbjct: 441 RMDRRRNVTVTEYDRHRGKQPDCRWRIPDPAALVDHIVVLKKPDPDLWKR 490
>UniRef100_Q7XQ75 OSJNBa0011J08.14 protein [Oryza sativa]
Length = 524
Score = 501 bits (1291), Expect = e-140
Identities = 255/400 (63%), Positives = 304/400 (75%), Gaps = 18/400 (4%)
Query: 127 AQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQT-RGVVWLDQRV-----STQRNE 180
A +T L+H+VFGIAASS W+ RKEYIK+WWRP+ RG VWLD+ V ST R
Sbjct: 87 AASTETTLQHVVFGIAASSRFWDKRKEYIKVWWRPRGAMRGYVWLDREVRESNMSTART- 145
Query: 181 GLPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNV 240
GLP IRIS DTS F YT+R+G RSA+RISR+V+ET +LGL VRWFVMGDDDTVF DN+
Sbjct: 146 GLPAIRISSDTSGFPYTHRRGHRSAIRISRIVSETFRLGLPGVRWFVMGDDDTVFFPDNL 205
Query: 241 VRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRY 300
+ +L+K+DHR YY+GS SESH+QNI+FSY MAYGGGGFAIS PLA LA MQD CI+RY
Sbjct: 206 LTVLNKFDHRQPYYIGSLSESHLQNIYFSYGMAYGGGGFAISRPLAEALARMQDGCIRRY 265
Query: 301 PALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPI 360
PALYGSDDR+QACMAELGVPLTK GFHQYDVYGDLLGLL AHPVAP+V+LHHLDVVQP+
Sbjct: 266 PALYGSDDRIQACMAELGVPLTKHPGFHQYDVYGDLLGLLAAHPVAPIVTLHHLDVVQPL 325
Query: 361 FP-SMSRAQSIRHLMES-INQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRE 418
FP + SR ++R L + I D +GIMQQSICYD W++SV+WGF V + RGV+SPRE
Sbjct: 326 FPNAKSRPAAVRRLFDGPIELDPAGIMQQSICYDGGNRWTVSVAWGFAVLVSRGVMSPRE 385
Query: 419 LEMPSRTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASR---QIVGVYS 475
+EMP+RTFLNWYRRADYTAYAFNTRP+A+ PCQKP VYY+S A R V Y
Sbjct: 386 MEMPARTFLNWYRRADYTAYAFNTRPLARSPCQKPAVYYLSSARRAAALRGGDTTVTRYE 445
Query: 476 R----DQTKSPFCRWRMESPE-KITSIVVTKRRDPLRWKK 510
R ++T+ P CRW + P+ + IVV K+ DP W +
Sbjct: 446 RWRRANETR-PACRWNIADPDAHLDHIVVLKKPDPGLWDR 484
>UniRef100_Q9LMM4 F22L4.11 protein [Arabidopsis thaliana]
Length = 509
Score = 476 bits (1226), Expect = e-133
Identities = 229/390 (58%), Positives = 288/390 (73%), Gaps = 9/390 (2%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPK-QTRGVVWLDQRVSTQRN--EGLPDIRIS 188
TELKH+VFGIAAS+ W RK+Y+K+WW+P + GVVWLDQ ++ N + LP IRIS
Sbjct: 84 TELKHVVFGIAASAKFWKHRKDYVKLWWKPNGEMNGVVWLDQHINQNDNVSKTLPPIRIS 143
Query: 189 DDTSKFRYTNRQGQRSALRISRVVTETLKL--GL---KDVRWFVMGDDDTVFVVDNVVRI 243
DTS+F+Y +G RSA+RI+R+V+ET++L G K+VRW VMGDDDTVF +N+V++
Sbjct: 144 SDTSRFQYRYPKGLRSAIRITRIVSETVRLLNGTELEKNVRWIVMGDDDTVFFPENLVKV 203
Query: 244 LSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPAL 303
L KYDH FYY+GSSSESH+QN+ FSY MAYGGGGFAISYPLA L MQDRCIQRY L
Sbjct: 204 LRKYDHNQFYYIGSSSESHIQNLKFSYGMAYGGGGFAISYPLAKALEKMQDRCIQRYSEL 263
Query: 304 YGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPS 363
YGSDDR+ ACM+ELGVPLTKE GFHQ D+YG LLGLL AHP+APLVS+HHLD+V P+FP+
Sbjct: 264 YGSDDRIHACMSELGVPLTKEVGFHQIDLYGKLLGLLSAHPLAPLVSIHHLDLVDPVFPN 323
Query: 364 MSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPS 423
M R ++R M DS + QQSICYD + W++SVSWG+ VQI+RGVLS RE+ +P+
Sbjct: 324 MGRVNAMRRFMVPAKLDSPSLAQQSICYDADHRWTVSVSWGYTVQIIRGVLSAREMVIPT 383
Query: 424 RTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSR-DQTKSP 482
RTF++WY++AD +YAFNTRP+AK CQ+P VYY+S D A R+ Y R P
Sbjct: 384 RTFIDWYKQADERSYAFNTRPIAKSACQRPRVYYLSNALPDLALRRTASEYVRWYDMWEP 443
Query: 483 FCRWRMESPEKITSIVVTKRRDPLRWKKVR 512
C W M P + ++V K+ DP RW K R
Sbjct: 444 ECDWDMSDPSEFERVIVYKKPDPDRWNKHR 473
>UniRef100_Q6EPN2 Fringe-related protein-like [Oryza sativa]
Length = 516
Score = 473 bits (1218), Expect = e-132
Identities = 233/389 (59%), Positives = 289/389 (73%), Gaps = 10/389 (2%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQ-TRGVVWLDQRVSTQR-NEGLPDIRISD 189
T L+H+VFGIAAS+ LW RK+YIKIWWRP RG VW+DQ V +GLP I+IS
Sbjct: 91 TTLQHVVFGIAASARLWEKRKDYIKIWWRPNAGMRGFVWMDQPVRESGVPDGLPPIKISS 150
Query: 190 DTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDH 249
+TS F Y NR+G RSA+RISR+V+ET +LGL VRW+VMGDDDTVF+ DN+V +L K DH
Sbjct: 151 NTSGFPYKNRRGHRSAIRISRIVSETFRLGLSGVRWYVMGDDDTVFLPDNLVAVLQKLDH 210
Query: 250 RHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDR 309
R YY+G SESH+QNI FSY MA+GGGGFAIS PLA L MQD CI RYP+LYGSDDR
Sbjct: 211 RQPYYIGYPSESHLQNIFFSYGMAFGGGGFAISQPLAARLERMQDACIHRYPSLYGSDDR 270
Query: 310 MQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFP-SMSRAQ 368
+ ACMAELGVPLT+ GFHQYDVYGDLLGLL AHPVAPLVSLHHLDVV+P+FP + SR
Sbjct: 271 IHACMAELGVPLTRHPGFHQYDVYGDLLGLLAAHPVAPLVSLHHLDVVRPLFPNARSRPA 330
Query: 369 SIRHLMES-INQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFL 427
++R L E + DS+G +QQSICYD W++SVSWGF+V RG++S RE+E+P+RTFL
Sbjct: 331 ALRRLFEGPVALDSAGAVQQSICYDARNRWTVSVSWGFVVMASRGMISAREMELPARTFL 390
Query: 428 NWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTA--SRQIVGVYSRDQTKS---P 482
NWY+RADY A+AFNTRP+A+ PC+KP YY+S A V Y R + ++ P
Sbjct: 391 NWYKRADYKAHAFNTRPLARRPCEKPSFYYLSSARRTVARDGETTVTTYQRWRHRNDMRP 450
Query: 483 FCRWRMESPEK-ITSIVVTKRRDPLRWKK 510
CRW++ P+ + ++VV K+ DP W +
Sbjct: 451 PCRWKIADPDALLDTVVVLKKPDPGLWDR 479
>UniRef100_O23072 A_IG005I10.16 protein [Arabidopsis thaliana]
Length = 785
Score = 455 bits (1171), Expect = e-126
Identities = 228/426 (53%), Positives = 297/426 (69%), Gaps = 17/426 (3%)
Query: 118 VVRLTPEQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQT-RGVVWLDQRVS- 175
V+RL Q + Q TELKHIVFGIAASS+LW R+EY+K WW+P G VWLD+ ++
Sbjct: 65 VIRLGSGQTPEEQ-TELKHIVFGIAASSDLWKHRREYVKTWWKPNGVMNGAVWLDKPIND 123
Query: 176 -TQRNEGLPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKL-----GLKDVRWFVMG 229
+ LP IRIS DTS F+Y R G RSA+RI+R+V+ET+++ ++VRW VMG
Sbjct: 124 TVSSSSALPQIRISSDTSSFKYRYRNGHRSAIRITRIVSETVRMLNGTEAERNVRWVVMG 183
Query: 230 DDDTVFVVDNVVRILSKYDHRHFYYVGSSSESHVQNIH-FSYAMAYGGGGFAISYPLAVE 288
DDDTVF +N+VR+L KYDH+ FYY+G+ SESH+QN+H FSY MAYGGGGFAISYPLA
Sbjct: 184 DDDTVFFTENLVRVLRKYDHKQFYYIGAPSESHLQNLHQFSYGMAYGGGGFAISYPLAKV 243
Query: 289 LATMQDRCIQRYPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPL 348
L MQDRCI+RY LYGSDDR+ ACMAELGVPLTKE GFHQ+DVYG+LLGLL HP AP+
Sbjct: 244 LEKMQDRCIERYSDLYGSDDRIHACMAELGVPLTKEVGFHQFDVYGNLLGLLSVHPQAPI 303
Query: 349 VSLHHLDVVQPIFPSMSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQ 408
VS+HHLDVV PIFP +R +++ LM DS+ ++QQS+CYDK+ W++S+SWG+ VQ
Sbjct: 304 VSIHHLDVVDPIFPKTNRVNALKKLMIPAKLDSASLVQQSVCYDKSHQWTMSISWGYTVQ 363
Query: 409 ILRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASR 468
I R + R +E+P+RTF +W+ R+D+T AFNTRPV CQ+P V+Y S +++S
Sbjct: 364 ITRTYMPARMMEVPTRTFNDWHLRSDFTNLAFNTRPVTWTDCQRPRVFYFSHAFSNSSSS 423
Query: 469 Q--IVGVYSRDQTKSPFCRWRMESPEKITSIVVTKRRDPLRWKKVRLLIIFFI--QRSNF 524
+ Y R P C W + P +I I V K+ P RW KV +I F+ + SNF
Sbjct: 424 DTTTISQYLRHDEWYPKCEWGIADPSEINQIFVYKKPTPDRWSKV---VIRFVPSKDSNF 480
Query: 525 NTIHTI 530
++ I
Sbjct: 481 AFVNAI 486
>UniRef100_O80941 Putative zinc finger protein [Arabidopsis thaliana]
Length = 532
Score = 376 bits (965), Expect = e-102
Identities = 173/347 (49%), Positives = 240/347 (68%), Gaps = 4/347 (1%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRN--EGLPDIRISD 189
T++ HI FGI S W R Y ++WWRP TRG +WLD+ P ++S
Sbjct: 96 TDISHIAFGIGGSIQTWRDRSRYSELWWRPNVTRGFIWLDEEPPLNMTWLSTSPPYQVSA 155
Query: 190 DTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDH 249
DTS+F YT G RSA+R++R++ ET +LGL DVRWF+MGDDDTVF VDN++ +L+KYDH
Sbjct: 156 DTSRFSYTCWYGSRSAIRMARIIKETFELGLTDVRWFIMGDDDTVFFVDNLITVLNKYDH 215
Query: 250 RHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDR 309
YY+G +SES Q+I SYAMAYGGGG AISYPLAVEL + D CI RY +LYGSD +
Sbjct: 216 NQMYYIGGNSESVEQDIVHSYAMAYGGGGIAISYPLAVELVKLLDGCIDRYASLYGSDQK 275
Query: 310 MQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQS 369
++AC++E+GVPLTKE GFHQ D+ G+ GLL AHPVAPLV+LHHLD V PIFP ++ +
Sbjct: 276 IEACLSEIGVPLTKELGFHQVDIRGNPYGLLAAHPVAPLVTLHHLDYVDPIFPGTTQIDA 335
Query: 370 IRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNW 429
+R L+ + D S I+Q S C+D+ R W +SVSWG+ +QI +++ +ELE P TF +W
Sbjct: 336 LRRLVSAYKTDPSRIIQHSFCHDQTRNWYVSVSWGYTIQIYPTLVTAKELETPFLTFKSW 395
Query: 430 YRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSR 476
R + ++F+TRP+++ PC++P VY++ + ++ S Q + Y +
Sbjct: 396 -RTSSSEPFSFDTRPISEDPCERPLVYFLDRV-YEVGSGQTLTTYRK 440
>UniRef100_Q9C881 Hypothetical protein T16O9.11 [Arabidopsis thaliana]
Length = 548
Score = 342 bits (877), Expect = 2e-92
Identities = 156/351 (44%), Positives = 234/351 (66%), Gaps = 3/351 (0%)
Query: 131 DTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQR-NEGLPDIRISD 189
D + H++FGIA SS LW RKE +++WW+P Q RG VWL+++VS + ++ LP I +S+
Sbjct: 128 DLSMNHLMFGIAGSSQLWERRKELVRLWWKPSQMRGHVWLEEQVSPEEGDDSLPPIIVSE 187
Query: 190 DTSKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDH 249
D+S+FRYTN G S LRISR+ E+ +L L +VRWFV+GDDDT+F V N++ +LSKYD
Sbjct: 188 DSSRFRYTNPTGHPSGLRISRIAMESFRLSLPNVRWFVLGDDDTIFNVHNLLAVLSKYDP 247
Query: 250 RHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDR 309
Y+G+ SESH N +FS+ MA+GGGG AISYPLA L+ + D C+ RYP LYGSDDR
Sbjct: 248 SEMVYIGNPSESHSANSYFSHNMAFGGGGIAISYPLAEALSRIHDDCLDRYPKLYGSDDR 307
Query: 310 MQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQS 369
+ AC+ ELGVPL++E GFHQ+D+ G+ GLL +HP+AP VS+HH++ V P++P +S S
Sbjct: 308 LHACITELGVPLSREPGFHQWDIKGNAHGLLSSHPIAPFVSIHHVEAVNPLYPGLSTLDS 367
Query: 370 IRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNW 429
++ L +++ D ++Q+SICYD + ++S G++VQ+ +L PR+LE +F W
Sbjct: 368 LKLLTRAMSLDPRSVLQRSICYDHTHKLTFAISLGYVVQVFPSILLPRDLERAELSFSAW 427
Query: 430 YRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTK 480
+ + + + + C+KP ++++ + +G YSR K
Sbjct: 428 NGISQPSEFDLDIKLPISSLCKKPILFFLKEV--GQEGNATLGTYSRSLVK 476
>UniRef100_Q94HZ1 Hypothetical protein OSJNBa0076F20.12 [Oryza sativa]
Length = 516
Score = 333 bits (854), Expect = 8e-90
Identities = 170/390 (43%), Positives = 242/390 (61%), Gaps = 12/390 (3%)
Query: 134 LKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVS-----TQRNEG--LPDIR 186
L HIVFGIA S++LW R+EY+++WW P RG VWLD + EG LP IR
Sbjct: 93 LGHIVFGIAGSAHLWPRRREYVRMWWDPAAMRGHVWLDAGAPAAPGPSASGEGSLLPPIR 152
Query: 187 ISDDTSKFRYTNRQGQRSALRISRVVTETLKL---GLKDVRWFVMGDDDTVFVVDNVVRI 243
+S+DTS+FRYTN G S LRI+R+ E ++L G RW V+ DDDTV DN+V +
Sbjct: 153 VSEDTSRFRYTNPTGHPSGLRIARIAAEAVRLVGGGGGGARWVVLVDDDTVVSADNLVAV 212
Query: 244 LSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPAL 303
L KYD R YVG+ SESH N +FS++MA+GGGG A+S PLA LA D CI+RYP L
Sbjct: 213 LGKYDWREMVYVGAPSESHSANTYFSHSMAFGGGGVALSLPLATALARTLDVCIERYPKL 272
Query: 304 YGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPS 363
YGSDDR+ AC+ ELGVPL++E GFHQ+D+ G+ G+L AHP+AP +S+HHL++V PI+P
Sbjct: 273 YGSDDRLHACITELGVPLSREYGFHQWDIRGNAHGILAAHPIAPFISIHHLELVDPIYPG 332
Query: 364 MSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPS 423
++ +S+ +++ + +Q+SICYDK + ++ +VS G++VQ+ VL PRELE
Sbjct: 333 LNSLESLELFTKAMKTEPMSFLQRSICYDKRQKYTFAVSLGYVVQVYPYVLLPRELERSE 392
Query: 424 RTFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFD--TASRQIVGVYSRDQTKS 481
RT++ + R + T + F+T+ + K C+KP ++++ D + RD K+
Sbjct: 393 RTYIAYNRMSQRTEFDFDTKDIQKSLCKKPILFFLKDVWKDGNITRGSYIRASVRDDLKN 452
Query: 482 PFCRWRMESPEKITSIVVTKRRDPLRWKKV 511
+R I I V+ RW V
Sbjct: 453 KVFCFRSPPLPDIDEIQVSASPLSKRWHLV 482
>UniRef100_O23378 Hypothetical protein dl3665c [Arabidopsis thaliana]
Length = 520
Score = 330 bits (846), Expect = 7e-89
Identities = 163/334 (48%), Positives = 224/334 (66%), Gaps = 2/334 (0%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNEGLPDIRISDDT 191
T +H++F IAAS + W R Y+++W+ P+ TR VV+LD R + + LP + +S D
Sbjct: 78 TRRRHLLFSIAASHDSWLRRSSYVRLWYSPESTRAVVFLD-RGGLESDLTLPPVIVSKDV 136
Query: 192 SKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDHRH 251
S+F Y G RSA+R++RVV ET+ G KDVRWFV GDDDTVF VDN+V +LSKYDHR
Sbjct: 137 SRFPYNFPGGLRSAIRVARVVKETVDRGDKDVRWFVFGDDDTVFFVDNLVTVLSKYDHRK 196
Query: 252 FYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDRMQ 311
++YVGS+SE + QN+ +S+ MA+GGGGFAIS LA LA + D C+ RY +YGSD R+
Sbjct: 197 WFYVGSNSEFYDQNVRYSFDMAFGGGGFAISASLAKVLAKVLDSCLMRYSHMYGSDSRIF 256
Query: 312 ACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQSIR 371
+C+AELGV LT E GFHQ DV G++ GLL AHP++PLVSLHHLD V P FP +R +S+
Sbjct: 257 SCVAELGVTLTHEPGFHQIDVRGNIFGLLCAHPLSPLVSLHHLDAVDPFFPKRNRTESVA 316
Query: 372 HLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNWYR 431
HL+ + + DS I+QQS+CYD ++SV WG+ VQ+ G +L +TF W R
Sbjct: 317 HLIGAASFDSGRILQQSVCYDSLNTVTVSVVWGYAVQVYEGNKLLPDLLTLQKTFSTWRR 376
Query: 432 RADYTA-YAFNTRPVAKHPCQKPFVYYMSKTHFD 464
+ + Y F+TR + PC +P V+++ D
Sbjct: 377 GSGVQSNYMFSTREYPRDPCGRPLVFFLDSVGSD 410
>UniRef100_Q9CAY4 Hypothetical protein F24K9.9 [Arabidopsis thaliana]
Length = 505
Score = 321 bits (823), Expect = 3e-86
Identities = 165/390 (42%), Positives = 237/390 (60%), Gaps = 21/390 (5%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNEGLPDIRISDDT 191
T + HI F IA ++ W R +YI +WWR TRG VWLD+ V N D+R S T
Sbjct: 91 TNISHIFFSIAGAAETWIDRSQYISLWWR-NTTRGFVWLDEPVKIPENHS--DVRFSIPT 147
Query: 192 -------SKFRYTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRIL 244
++F++++ R+A+RI+R++ ++ +L L +VRWFVMGDDDTVF +N+V++L
Sbjct: 148 RVSDPGWTRFKFSS---SRAAVRIARIIWDSYRLNLPNVRWFVMGDDDTVFFTENLVKVL 204
Query: 245 SKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALY 304
SKYDH +Y+G +SES Q++ +Y MA+GGGGFA+S PLA LA D C+QRY Y
Sbjct: 205 SKYDHEQMWYIGGNSESVEQDVMHAYDMAFGGGGFALSRPLAARLAAAMDDCLQRYFYFY 264
Query: 305 GSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSM 364
GSD R+ +C++E+GVP T+E GFHQ D+ GD G L AHP+APLVSLHHL + P+FP+
Sbjct: 265 GSDQRIASCISEIGVPFTEERGFHQLDIRGDPYGFLAAHPLAPLVSLHHLVYLDPMFPNK 324
Query: 365 SRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSR 424
+ +S++ LM+ D + I+QQ C+D+ R WSIS+SWG+ +QI L+ EL P +
Sbjct: 325 NPIESLQTLMKPYTLDPNRILQQINCHDRKRQWSISISWGYTIQIYTYFLTATELTTPLQ 384
Query: 425 TFLNWYRRADYTAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVYSRDQTKSPFC 484
TF W +D + FNTRP+ PC++P Y+M D YS C
Sbjct: 385 TFKTWRSSSD-GPFVFNTRPLKPDPCERPVTYFMDGAE-DVRDSGTKTWYSIGDKNYGHC 442
Query: 485 ----RWRMESPEKITSIVVTKRRDPLRWKK 510
R+ ++I +V + + DP W K
Sbjct: 443 GKIEHTRLTKVKRI--LVTSMKTDPEYWNK 470
>UniRef100_O23042 YUP8H12.11 protein [Arabidopsis thaliana]
Length = 479
Score = 312 bits (799), Expect = 2e-83
Identities = 168/415 (40%), Positives = 244/415 (58%), Gaps = 41/415 (9%)
Query: 124 EQMAQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNE--- 180
+Q + T+++HIVFGI +S+ W R+EY+K+WW ++ RG V++++ + + +N
Sbjct: 60 DQDQSQSPTKIEHIVFGIGSSAISWRARREYVKLWWDAQKMRGCVFVERPLPSSQNHTDS 119
Query: 181 -----GLPDIRISDDTSKFRYTNRQGQRSALRISRVVTETLKL---GLKDVRWFVMGDDD 232
G ++TS +RYT R G R+A+RI+R V ET++L ++VRW+V GDDD
Sbjct: 120 YLLPPGTVKSNRPNETSVYRYTWRGGDRNAIRIARCVLETVRLFNTSSEEVRWYVFGDDD 179
Query: 233 TVFVVDNVVRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATM 292
T+F+ +N+ R LSKYDH +YY+GS+SE + QN F + MA+GGGG+A+S LA LA
Sbjct: 180 TIFIPENLARTLSKYDHTSWYYIGSTSEIYHQNSMFGHDMAFGGGGYALSSSLANVLARN 239
Query: 293 QDRCIQRYPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLH 352
D CI+RYP LYG D R+ AC+ ELGV L+KE GFHQ+DV G+ LG+L +H PLVSLH
Sbjct: 240 FDSCIERYPHLYGGDSRVYACVLELGVGLSKEPGFHQFDVRGNALGILTSHSTRPLVSLH 299
Query: 353 HLDVVQPIFPSMSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQI--- 409
H+ + PIFP+ + ++RHL ++ D I Q S+CYD+ W+ISVSWG+ VQ+
Sbjct: 300 HMAHIDPIFPNSTTFSAVRHLFSAVQLDPLRIFQLSVCYDRWYSWTISVSWGYTVQVNTF 359
Query: 410 -----------------LRGVLSPRELEMPSRTFLNWYRRADY-TAYAFNTRPVAKHPCQ 451
LR VL +E TF W + + Y FNTR + + PCQ
Sbjct: 360 LPFFVQDSNNIDGRHLFLRDVLRAQE------TFRPWQKSGGLASVYTFNTREIHRDPCQ 413
Query: 452 KPFVYYMSKTHFDTASRQIVGVYSRDQTKSPFCRWRMESPEKITSI-VVTKRRDP 505
+P +YM + I VY + + + SP KI I V ++R DP
Sbjct: 414 RPVTFYMQHVSSSSHDGTIKSVYKQAYENCTYD--PVTSPRKIHEIRVFSRRLDP 466
>UniRef100_Q84Q65 Hypothetical protein OSJNBa0071M09.15 [Oryza sativa]
Length = 676
Score = 294 bits (753), Expect = 4e-78
Identities = 164/357 (45%), Positives = 212/357 (58%), Gaps = 24/357 (6%)
Query: 127 AQRQDTELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVSTQRNEGLPDIR 186
A R T L HIVF I AS+ W R+ Y +WWRP RG VWLD S Q P R
Sbjct: 68 AARSPTTLAHIVFVIGASNATWAKRRVYTGLWWRPGAMRGHVWLDDEPSGQWRPSWPPYR 127
Query: 187 I-SDDTSKFRYTNRQGQRSALRISRV--VTETLKLGLKDVRWFVMGDDDTVFVVDNVVRI 243
+ D ++F + R A ++ E + G +VRW VMGDDDTVF +N+V +
Sbjct: 128 VLRPDEARFGKEHAAAARMAWAVAEAFQAAEAGREGDGEVRWLVMGDDDTVFFPENLVAV 187
Query: 244 LSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPAL 303
L KYDHR YYVGS+SES QN+ SY+MA+GGGG+AISYP A LA + D C+ RY
Sbjct: 188 LDKYDHREMYYVGSTSESVGQNVVHSYSMAFGGGGYAISYPAAAALAGIMDGCLDRYNEF 247
Query: 304 YGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPS 363
YGSD R+QAC+AELGVPLT E GFHQ D+ G + GLL AHPVAPLVSLHHLD + PI P+
Sbjct: 248 YGSDHRVQACLAELGVPLTTEPGFHQLDLKGHVYGLLAAHPVAPLVSLHHLDRLNPISPN 307
Query: 364 -MSRAQSIRHLMESINQDSSGIMQQSICYD------------KNRF-WSISVSWGFMVQI 409
+ R ++R L+ + D S +QQ+ICY + +F S+SVSWG+MV +
Sbjct: 308 WLKRLPAVRSLVGASRHDPSRTLQQAICYHHDARGGGRRRRRRRQFTLSVSVSWGYMVHL 367
Query: 410 LRGVLSPRELEMPSRTFLNWYRRADYTAYAFNTRPVAKH-----PC-QKPFVYYMSK 460
+ P EL+ P RTF W + + NTRP A PC +KP ++Y+ +
Sbjct: 368 YPAAVPPHELQTPLRTFRAW-SGSPAGPFTVNTRPEATPNATALPCHRKPIMFYLDR 423
>UniRef100_Q6ZJW6 Putative fringe-related protein [Oryza sativa]
Length = 493
Score = 279 bits (713), Expect = 2e-73
Identities = 144/340 (42%), Positives = 216/340 (63%), Gaps = 14/340 (4%)
Query: 132 TELKHIVFGIAASSNLWNTRKEYIKIWWRPKQTRGVVWLD--QRVSTQRNEGLP-DIRI- 187
T L H+VFGIA+S R +++W RP R ++LD + +E LP ++R
Sbjct: 72 TTLAHVVFGIASSRRTLPLRLPLLRLWLRPP-ARAFLFLDGPAPAAAAASEPLPPNLRFC 130
Query: 188 --SDDTSKFRYTNRQGQRSALRISRVVTETLKLG------LKDVRWFVMGDDDTVFVVDN 239
S D S+F YT+ +G SA+R++R+ E L+L RW V+ DDDT FV+ N
Sbjct: 131 VSSTDASRFPYTHPRGLPSAVRVARIAKELLQLDDHHHATPPPPRWLVLADDDTAFVLPN 190
Query: 240 VVRILSKYDHRHFYYVGSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQR 299
++ LS+YD R +Y+G+ SES QN +AMAYGGGG A+S+PLA LA + D C+ R
Sbjct: 191 LLHTLSRYDWREPWYLGARSESAAQNAWHGFAMAYGGGGIAVSWPLAARLARVLDSCLLR 250
Query: 300 YPALYGSDDRMQACMAELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQP 359
YP LYGSD R+ AC+AELGV LT E GFHQ D++GD+ GLL AHP+ PLVSLHHLD V P
Sbjct: 251 YPHLYGSDARIHACLAELGVELTHEPGFHQIDLHGDISGLLRAHPLTPLVSLHHLDHVYP 310
Query: 360 IFPSMSRAQSIRHLMESINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPREL 419
++P M RA +++H + N D + I+QQ++CYD ++ ++S++WG+ VQ+ +G + +L
Sbjct: 311 LYPGMDRATAVKHFFRAANADPARILQQTVCYDHSKAITVSIAWGYSVQVYKGNVLLPDL 370
Query: 420 EMPSRTFLNWYRRADYT-AYAFNTRPVAKHPCQKPFVYYM 458
+TF+ W R + T + F+T+ + C++ ++++
Sbjct: 371 LAVQKTFVPWKRGRNATDVFMFDTKHYPRDECKRAALFFL 410
>UniRef100_Q94CJ6 Hypothetical protein At5g12460 [Arabidopsis thaliana]
Length = 415
Score = 274 bits (700), Expect = 6e-72
Identities = 137/339 (40%), Positives = 202/339 (59%), Gaps = 16/339 (4%)
Query: 142 AASSNLWNTRKEYIKIWWRPKQTRGVVWLDQRVST------QRNEGLPDIRISDDTSKFR 195
++S+ W R+ YI+ WWRP T+G V+L++ Q++ + S T+KF+
Sbjct: 20 SSSTKTWRYRRGYIEPWWRPNITKGYVFLERPPGPDLLPWPQQSPPFSVNKESFITNKFK 79
Query: 196 YTNRQGQRSALRISRVVTETLKLGLKDVRWFVMGDDDTVFVVDNVVRILSKYDHRHFYYV 255
+ +R+ + E+ K K+ RWFV+GDDDT+F +DN+V+ L +Y+H+ YYV
Sbjct: 80 --------TQIRLFYSLQESFKKASKETRWFVIGDDDTLFFLDNLVKALDRYNHKKHYYV 131
Query: 256 GSSSESHVQNIHFSYAMAYGGGGFAISYPLAVELATMQDRCIQRYPALYGSDDRMQACMA 315
G +SE+ N F++ M YGGGG+A+SYP V L + + CI+RY +Y SD C+A
Sbjct: 132 GMNSENVWSNAIFAFDMGYGGGGYALSYPTVVTLLSNMEECIKRYLGVY-SDLLSFRCLA 190
Query: 316 ELGVPLTKEAGFHQYDVYGDLLGLLGAHPVAPLVSLHHLDVVQPIFPSMSRAQSIRHLME 375
+LG+ LT E G HQ D++GD+ GLL AHP +PL+SLHH DV+ PIFP M+R QS+ HLME
Sbjct: 191 DLGIDLTLEKGMHQNDLHGDISGLLSAHPQSPLISLHHFDVIDPIFPGMNRQQSVNHLME 250
Query: 376 SINQDSSGIMQQSICYDKNRFWSISVSWGFMVQILRGVLSPRELEMPSRTFLNWYRRADY 435
+ D S ++QQ+ICY + WS+SVSWG+ V I + + L+ P TF W +
Sbjct: 251 TAKTDQSRVLQQTICYQRGYNWSVSVSWGYSVHIYQSIYPRSHLKRPLETFRPW-KDVRI 309
Query: 436 TAYAFNTRPVAKHPCQKPFVYYMSKTHFDTASRQIVGVY 474
AY FNTR V PC+ P ++ D + +Y
Sbjct: 310 PAYGFNTRRVTNDPCEMPRQFFFDSVVEDKNQSLVTTIY 348
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 914,637,506
Number of Sequences: 2790947
Number of extensions: 38377064
Number of successful extensions: 187691
Number of sequences better than 10.0: 550
Number of HSP's better than 10.0 without gapping: 253
Number of HSP's successfully gapped in prelim test: 316
Number of HSP's that attempted gapping in prelim test: 180593
Number of HSP's gapped (non-prelim): 3158
length of query: 548
length of database: 848,049,833
effective HSP length: 132
effective length of query: 416
effective length of database: 479,644,829
effective search space: 199532248864
effective search space used: 199532248864
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135230.10


