

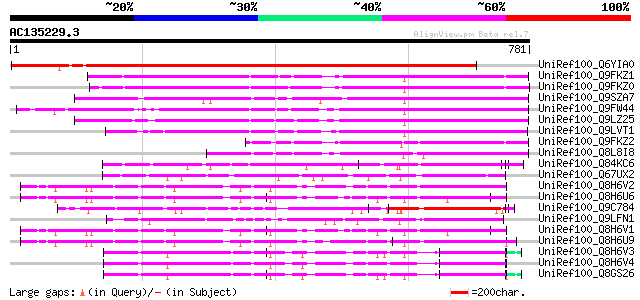
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135229.3 - phase: 0
(781 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6YIA0 Disease resistance protein-like protein MsR1 [M... 1181 0.0
UniRef100_Q9FKZ1 Probable disease resistance protein At5g66900 [... 450 e-125
UniRef100_Q9FKZ0 Probable disease resistance protein At5g66910 [... 423 e-117
UniRef100_Q9SZA7 Probable disease resistance protein At4g33300 [... 360 1e-97
UniRef100_Q9FW44 Disease resistance protein ADR1 [Arabidopsis th... 354 6e-96
UniRef100_Q9LZ25 Probable disease resistance protein At5g04720 [... 352 2e-95
UniRef100_Q9LVT1 Putative disease resistance protein At5g47280 [... 336 1e-90
UniRef100_Q9FKZ2 Probable disease resistance protein At5g66890 [... 259 2e-67
UniRef100_Q8L8I8 NBS/LRR [Pinus taeda] 229 2e-58
UniRef100_Q84KC6 NBS-LRR disease resistance protein homologue [H... 162 3e-38
UniRef100_Q67UX2 Putative NBS-LRR resistance protein RGH2 [Oryza... 145 6e-33
UniRef100_Q8H6V2 Putative rp3 protein [Zea mays] 132 3e-29
UniRef100_Q8H6U6 Putative rp3 protein [Zea mays] 132 3e-29
UniRef100_Q9C784 Disease resistance protein, putative [Arabidops... 132 5e-29
UniRef100_Q9LFN1 RPP1 disease resistance protein-like [Arabidops... 131 9e-29
UniRef100_Q8H6V1 Putative rp3 protein [Zea mays] 130 2e-28
UniRef100_Q8H6U9 Putative rp3 protein [Zea mays] 130 2e-28
UniRef100_Q8H6V3 Putative rp3 protein [Zea mays] 128 7e-28
UniRef100_Q8H6V4 Putative rp3 protein [Zea mays] 127 2e-27
UniRef100_Q8GS26 Putative rp3 protein [Zea mays] 127 2e-27
>UniRef100_Q6YIA0 Disease resistance protein-like protein MsR1 [Medicago sativa]
Length = 704
Score = 1181 bits (3055), Expect = 0.0
Identities = 609/707 (86%), Positives = 646/707 (91%), Gaps = 13/707 (1%)
Query: 3 SATTHIITLTTRF--NDVLNKISEIVETARKNQSSRLLFRLILKDLSPVVQDIKHYNEHL 60
S+ THIITL TRF +DVLNKIS+I E A++NQSSR RLIL++LSPVVQDIK YN+HL
Sbjct: 2 SSATHIITLATRFQFHDVLNKISQIAEKAQENQSSRRSLRLILRNLSPVVQDIKQYNDHL 61
Query: 61 DQPREEINSLIEE-----SACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNHE 115
D PREEINSLIEE SACK S ENDS VV++ + N+ E + ++ E
Sbjct: 62 DHPREEINSLIEENDAEESACKCSSENDSNVVEK----LKNETFEAGPPFKCPFDV--PE 115
Query: 116 NPKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIF 175
NPKFTVGLDIPFSKLKMELLR GSSTLVLTGLGGLGKTTLATKLCWD+EVNGKF ENIIF
Sbjct: 116 NPKFTVGLDIPFSKLKMELLRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFMENIIF 175
Query: 176 VTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSS 235
VTFSKTPMLKTIVERIH+HCGY VPEFQ+DEDAVN+LGLLLKKVEGSPLLLVLDDVWPSS
Sbjct: 176 VTFSKTPMLKTIVERIHEHCGYPVPEFQNDEDAVNRLGLLLKKVEGSPLLLVLDDVWPSS 235
Query: 236 ESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIID 295
ESLVEKLQFQ++ FKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA MEKNSSDIID
Sbjct: 236 ESLVEKLQFQISDFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYALMEKNSSDIID 295
Query: 296 KNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKI 355
KNLVEKVVRSC GLPLTIKVIATSLKNRPHDLW KIVKELSQGHSILDSNTELLTRLQKI
Sbjct: 296 KNLVEKVVRSCQGLPLTIKVIATSLKNRPHDLWRKIVKELSQGHSILDSNTELLTRLQKI 355
Query: 356 FDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNL 415
FDVLEDNP I+ECFMDLALFPEDHRIPVAALVDMWAELY+LDD GIQAMEIINKLGIMNL
Sbjct: 356 FDVLEDNPTIIECFMDLALFPEDHRIPVAALVDMWAELYRLDDTGIQAMEIINKLGIMNL 415
Query: 416 ANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLA 475
ANVIIPRK+ASDTDNNNYNNHFI+LHDILRELGIY+STKEPFEQRKRLIIDM+ NKSGL
Sbjct: 416 ANVIIPRKDASDTDNNNYNNHFIMLHDILRELGIYRSTKEPFEQRKRLIIDMHKNKSGLT 475
Query: 476 EKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHT 535
EKQQGLM RI SKFMRLCVKQNPQQLAARILSVSTDETCALDWS M+PAQVEVLILN+HT
Sbjct: 476 EKQQGLMIRILSKFMRLCVKQNPQQLAARILSVSTDETCALDWSQMEPAQVEVLILNLHT 535
Query: 536 KQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKN 595
KQYSLPEWI KMSKL+VLIITNY HPS+L N ELL SLQNLE+IRLERISVPSF T+KN
Sbjct: 536 KQYSLPEWIGKMSKLKVLIITNYTVHPSELTNFELLSSLQNLEKIRLERISVPSFATVKN 595
Query: 596 LKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTN 655
LKKLSLYMCNT LAFEKGSILISDAFPNLEELNIDYCKDL+VL TGICDIISLKKL+VTN
Sbjct: 596 LKKLSLYMCNTRLAFEKGSILISDAFPNLEELNIDYCKDLLVLPTGICDIISLKKLSVTN 655
Query: 656 CHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 702
CHK+SSLP+DIGKLENLELLSLSSCTDLEAIPTSI KLLNLKHLDIS
Sbjct: 656 CHKISSLPEDIGKLENLELLSLSSCTDLEAIPTSIEKLLNLKHLDIS 702
>UniRef100_Q9FKZ1 Probable disease resistance protein At5g66900 [Arabidopsis
thaliana]
Length = 809
Score = 450 bits (1157), Expect = e-125
Identities = 266/670 (39%), Positives = 390/670 (57%), Gaps = 36/670 (5%)
Query: 118 KFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVT 177
K VGLD P +LK LL TLV++ G GKTTL ++LC D ++ GKFK +I F
Sbjct: 166 KVIVGLDWPLGELKKRLLDDSVVTLVVSAPPGCGKTTLVSRLCDDPDIKGKFK-HIFFNV 224
Query: 178 FSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPLLLVLDDVWPSSE 236
S TP + IV+ + QH GY+ F++D A L LL+++ E P+LLVLDDVW ++
Sbjct: 225 VSNTPNFRVIVQNLLQHNGYNALTFENDSQAEVGLRKLLEELKENGPILLVLDDVWRGAD 284
Query: 237 SLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDK 296
S ++K Q ++ +KILVTSR FP F + LKPL +DA L H+A N+S +
Sbjct: 285 SFLQKFQIKLPNYKILVTSRFDFPSFDSNYRLKPLEDDDARALLIHWASRPCNTSPDEYE 344
Query: 297 NLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN-TELLTRLQKI 355
+L++K+++ CNG P+ I+V+ SLK R + W V+ S+G IL +L LQ
Sbjct: 345 DLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVESWSEGEKILGKPYPTVLECLQPS 404
Query: 356 FDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNL 415
FD L+ P + ECF+D+ F ED +I + ++DMW ELY + + + L NL
Sbjct: 405 FDALD--PNLKECFLDMGSFLEDQKIRASVIIDMWVELYGKGSSILYMY--LEDLASQNL 460
Query: 416 ANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLA 475
++ N + ++ YN+ + HDILREL I QS + +RKRL +++ N
Sbjct: 461 LKLVPLGTN--EHEDGFYNDFLVTQHDILRELAICQSEFKENLERKRLNLEILENT---- 514
Query: 476 EKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHT 535
F C+ + A +LS+STD+ + W M VE L+LN+ +
Sbjct: 515 -------------FPDWCLNT----INASLLSISTDDLFSSKWLEMDCPNVEALVLNLSS 557
Query: 536 KQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGT--- 592
Y+LP +I+ M KL+VL ITN+GF+P++L+N L SL NL+RIRLE++S+
Sbjct: 558 SDYALPSFISGMKKLKVLTITNHGFYPARLSNFSCLSSLPNLKRIRLEKVSITLLDIPQL 617
Query: 593 -LKNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKK 650
L +LKKLSL MC+ F + I++S+A L+E++IDYC DL L I +I+SLK
Sbjct: 618 QLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDELPYWISEIVSLKT 677
Query: 651 LNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 710
L++TNC+KLS LP+ IG L LE+L L S +L +P + L NL+ LDIS+C+ L L
Sbjct: 678 LSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELPEATEGLSNLRFLDISHCLGLRKL 737
Query: 711 PEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEV 770
P+E G L NLK + M C+ ELP SV NL+NL+ + CDEET WE + + N++++
Sbjct: 738 PQEIGKLQNLKKISMRKCSGCELPESVTNLENLE-VKCDEETGLLWERLKPKMRNLRVQE 796
Query: 771 LHVDVNLNWL 780
++ NLN L
Sbjct: 797 EEIEHNLNLL 806
>UniRef100_Q9FKZ0 Probable disease resistance protein At5g66910 [Arabidopsis
thaliana]
Length = 815
Score = 423 bits (1088), Expect = e-117
Identities = 259/669 (38%), Positives = 384/669 (56%), Gaps = 36/669 (5%)
Query: 121 VGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSK 180
VGLD P +LK +LL +S +V++G G GKTTL TKLC D E+ G+FK+ I + S
Sbjct: 173 VGLDWPLVELKKKLL--DNSVVVVSGPPGCGKTTLVTKLCDDPEIEGEFKK-IFYSVVSN 229
Query: 181 TPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPLLLVLDDVWPSSESLV 239
TP + IV+ + Q G F D A L LL+++ + +LLVLDDVW SE L+
Sbjct: 230 TPNFRAIVQNLLQDNGCGAITFDDDSQAETGLRDLLEELTKDGRILLVLDDVWQGSEFLL 289
Query: 240 EKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLV 299
K Q + +KILVTS+ F T L PL +E A +L +A ++S ++L+
Sbjct: 290 RKFQIDLPDYKILVTSQFDFTSLWPTYHLVPLKYEYARSLLIQWASPPLHTSPDEYEDLL 349
Query: 300 EKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSIL-DSNTELLTRLQKIFDV 358
+K+++ CNG PL I+V+ SLK + LW V+ S+G +IL ++N + RLQ F+V
Sbjct: 350 QKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGETILGNANPTVRQRLQPSFNV 409
Query: 359 LEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQA-MEIINKLGIMNLAN 417
L+ P + ECFMD+ F +D +I + ++D+W ELY + M +N+L NL
Sbjct: 410 LK--PHLKECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSSTNKFMLYLNELASQNLLK 467
Query: 418 VIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEK 477
++ + ++ YN + H+ILREL I+QS EP QRK+L +++ +
Sbjct: 468 LV--HLGTNKREDGFYNELLVTQHNILRELAIFQSELEPIMQRKKLNLEIREDN------ 519
Query: 478 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 537
F C+ Q + AR+LS+ TD+ + W M VE L+LNI +
Sbjct: 520 -----------FPDECLNQ---PINARLLSIYTDDLFSSKWLEMDCPNVEALVLNISSLD 565
Query: 538 YSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGT----L 593
Y+LP +IA+M KL+VL I N+GF+P++L+N L SL NL+RIR E++SV L
Sbjct: 566 YALPSFIAEMKKLKVLTIANHGFYPARLSNFSCLSSLPNLKRIRFEKVSVTLLDIPQLQL 625
Query: 594 KNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 652
+LKKLS +MC+ F + I +S A NL+E++IDYC DL L I +++SLK L+
Sbjct: 626 GSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWIPEVVSLKTLS 685
Query: 653 VTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 712
+TNC+KLS LP+ IG L LE+L + SC +L +P + +L NL+ LDIS+C+ L LP+
Sbjct: 686 ITNCNKLSQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDISHCLGLRKLPQ 745
Query: 713 EFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLH 772
E G L L+N+ M C+ ELP SV L+NL+ + CDE T WE + N+++
Sbjct: 746 EIGKLQKLENISMRKCSGCELPDSVRYLENLE-VKCDEVTGLLWERLMPEMRNLRVHTEE 804
Query: 773 VDVNLNWLL 781
+ NL LL
Sbjct: 805 TEHNLKLLL 813
>UniRef100_Q9SZA7 Probable disease resistance protein At4g33300 [Arabidopsis
thaliana]
Length = 855
Score = 360 bits (923), Expect = 1e-97
Identities = 230/719 (31%), Positives = 382/719 (52%), Gaps = 60/719 (8%)
Query: 98 VEETLYKAREILELLNHENPKFTVGLDIPFSKLKMELLRGGSSTLVLTGLGGLGKTTLAT 157
+ E + +A + N ++ KF VGL++ K+K + ++G+GG+GKTTLA
Sbjct: 159 ISEAMKRAEAMEIETNDDSEKFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAK 218
Query: 158 KLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK 217
+L D EV F+ I+F+T S++P+L+ + E I F S +A N +
Sbjct: 219 ELQRDHEVQCHFENRILFLTVSQSPLLEELRELIWG--------FLSGCEAGNPVPDCNF 270
Query: 218 KVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAV 277
+G+ L++LDDVW ++++L F+ G LV SR T ++ L+ ++A+
Sbjct: 271 PFDGARKLVILDDVW-TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAI 329
Query: 278 TLFHHYAQMEKN-----SSDIIDKNLVE-----------KVVRSCNGLPLTIKVIATSLK 321
+LF A +K+ D++ ++ ++ +V C GLPL +KV SL
Sbjct: 330 SLFCLCAFGQKSIPLGFCKDLVKQHNIQSFSILRVLCLAQVANECKGLPLALKVTGASLN 389
Query: 322 NRPHDLWHKIVKELSQGHSILDSN-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHR 380
+P W +++ LS+G DS+ + LL +++ D L+ K +CF+DL FPED +
Sbjct: 390 GKPEMYWKGVLQRLSKGEPADDSHESRLLRQMEASLDNLDQTTK--DCFLDLGAFPEDRK 447
Query: 381 IPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNH---F 437
IP+ L+++W EL+ +D+ A I+ L NL + + Y +H F
Sbjct: 448 IPLDVLINIWIELHDIDEGN--AFAILVDLSHKNLLTL-----GKDPRLGSLYASHYDIF 500
Query: 438 IILHDILRELGIYQSTKEPFEQRKRLII-----------DMNNNKSGLAEKQQGLMTRIF 486
+ HD+LR+L ++ S +RKRL++ + NN++ +A+ + F
Sbjct: 501 VTQHDVLRDLALHLSNAGKVNRRKRLLMPKRELDLPGDWERNNDEHYIAQIVSIHTGKCF 560
Query: 487 SKF-MRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIA 545
S + L + N Q + E + W M+ + E+LILN + +Y LP +I+
Sbjct: 561 STLQIFLVLSCNNHQ------DHNVGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFIS 614
Query: 546 KMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGT----LKNLKKLSL 601
KMS+L+VL+I N G P+ L++ + L L + LER+ VP LKNL K+SL
Sbjct: 615 KMSRLKVLVIINNGMSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSL 674
Query: 602 YMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSS 661
+C +F++ + ++D FP L +L ID+C DLV L + IC + SL L++TNC +L
Sbjct: 675 ILCKINKSFDQTGLDVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGE 734
Query: 662 LPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLK 721
LP+++ KL+ LE+L L +C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+
Sbjct: 735 LPKNLSKLQALEILRLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLE 794
Query: 722 NLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
+DM C + P S V+L++L+ + CD + A WE+ + + +KIE +L+WL
Sbjct: 795 KIDMRECCFSDRPSSAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 853
>UniRef100_Q9FW44 Disease resistance protein ADR1 [Arabidopsis thaliana]
Length = 787
Score = 354 bits (908), Expect = 6e-96
Identities = 240/784 (30%), Positives = 416/784 (52%), Gaps = 70/784 (8%)
Query: 11 LTTRFNDVLNKISEIVETARKNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQPRE----- 65
L+ L EI+E ARK L +L+ ++ + H N+ D ++
Sbjct: 58 LSNHHQTQLGVFYEILEKARK------LCEKVLRCNRWNLKHVYHANKMKDLEKQISRFL 111
Query: 66 --EINSLIEESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNHENPKFTVGL 123
+I + C D ++ N ++ + ++L ++E+ +P+ L
Sbjct: 112 NSQILLFVLAEVCHLRVNGDR--IERNMDRLLTERNDSLSFPETMMEIETVSDPEIQTVL 169
Query: 124 DIPFSKLKMELLRGGSSTLV-LTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTP 182
++ K+K + + + L ++G+ G GKTTLA +L D++V G FK ++F+T S++P
Sbjct: 170 ELGKKKVKEMMFKFTDTHLFGISGMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSP 229
Query: 183 MLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKL 242
+ + + C + EF D G+ +K L++LDDVW + ESL ++L
Sbjct: 230 NFENL-----ESC---IREFLYD-------GVHQRK------LVILDDVW-TRESL-DRL 266
Query: 243 QFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKV 302
++ G LV SR TT ++ L ++A++L A +K+ +K LV++V
Sbjct: 267 MSKIRGSTTLVVSRSKLADPRTTYNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQV 326
Query: 303 VRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN-TELLTRLQKIFDVLED 361
V C GLPL++KV+ SLKN+P W +VK L +G + +++ + + +++ + L+
Sbjct: 327 VDECKGLPLSLKVLGASLKNKPERYWEGVVKRLLRGEAADETHESRVFAHMEESLENLD- 385
Query: 362 NPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIP 421
PKI +CF+D+ FPED +IP+ L +W E + +D+ A + +L NL ++
Sbjct: 386 -PKIRDCFLDMGAFPEDKKIPLDLLTSVWVERHDIDEE--TAFSFVLRLADKNLLTIV-- 440
Query: 422 RKNASDTDNN-NYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQG 480
N D + Y + F+ HD+LR+L ++ S + +R+RL++ K +
Sbjct: 441 -NNPRFGDVHIGYYDVFVTQHDVLRDLALHMSNRVDVNRRERLLMP----------KTEP 489
Query: 481 LMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSL 540
++ R + K + A+I+S+ T E ++W M + EVLILN + Y L
Sbjct: 490 VLPREWEK-------NKDEPFDAKIVSLHTGEMDEMNWFDMDLPKAEVLILNFSSDNYVL 542
Query: 541 PEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGT----LKNL 596
P +I KMS+LRVL+I N G P++L+ + +L L + L+R+ VP + LKNL
Sbjct: 543 PPFIGKMSRLRVLVIINNGMSPARLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNL 602
Query: 597 KKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNC 656
K+ L C +F + S IS FP+L +L ID+C DL+ L++ I I SL L++TNC
Sbjct: 603 HKIHLIFCKVKNSFVQTSFDISKIFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNC 661
Query: 657 HKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 716
++ LP+++ +++LE L L +C +L ++P + +L LK++DIS C+SL SLPE+FG
Sbjct: 662 PRILELPKNLSNVQSLERLRLYACPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGK 721
Query: 717 LCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVN 776
L +L+ +DM C+ + LP SV L +L+ + CDEET++ WE + ++ + IEV
Sbjct: 722 LGSLEKIDMRECSLLGLPSSVAALVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFT 781
Query: 777 LNWL 780
++WL
Sbjct: 782 VDWL 785
>UniRef100_Q9LZ25 Probable disease resistance protein At5g04720 [Arabidopsis
thaliana]
Length = 811
Score = 352 bits (904), Expect = 2e-95
Identities = 222/691 (32%), Positives = 368/691 (53%), Gaps = 46/691 (6%)
Query: 98 VEETLYKAREILELLNHENPKFTVGLDIPFSKLKMELLRG--GSSTLVLTGLGGLGKTTL 155
+ E L A +E++ + VGLD+ K+K L + G + ++G+ G GKTTL
Sbjct: 157 LREALKTAEATVEMVTTDGADLGVGLDLGKRKVKEMLFKSIDGERLIGISGMSGSGKTTL 216
Query: 156 ATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLL 215
A +L DEEV G F ++F+T S++P L E + H + +++ A
Sbjct: 217 AKELARDEEVRGHFGNKVLFLTVSQSPNL----EELRAHIWGFLTSYEAGVGAT------ 266
Query: 216 LKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHED 275
+ S L++LDDVW + ESL + + + G LV SR T ++ L +
Sbjct: 267 ---LPESRKLVILDDVW-TRESLDQLMFENIPGTTTLVVSRSKLADSRVTYDVELLNEHE 322
Query: 276 AVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKEL 335
A LF +K ++LV++VV C GLPL++KVI SLK RP W V+ L
Sbjct: 323 ATALFCLSVFNQKLVPSGFSQSLVKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERL 382
Query: 336 SQGHSILDSN-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELY 394
S+G +++ + + +++ + L+ PK +CF+ L FPED +IP+ L+++ EL+
Sbjct: 383 SRGEPADETHESRVFAQIEATLENLD--PKTRDCFLVLGAFPEDKKIPLDVLINVLVELH 440
Query: 395 KLDDNGIQAMEIINKLGIMNLANVII-PRKNASDTDNNNYNNHFIILHDILRELGIYQST 453
L+D A +I L NL ++ PR T +Y + F+ HD+LR++ + S
Sbjct: 441 DLED--ATAFAVIVDLANRNLLTLVKDPRFGHMYT---SYYDIFVTQHDVLRDVALRLSN 495
Query: 454 KEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDET 513
R+RL++ K++ ++ R + + N + AR++S+ T E
Sbjct: 496 HGKVNNRERLLMP----------KRESMLPREWER-------NNDEPYKARVVSIHTGEM 538
Query: 514 CALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGS 573
+DW M+ + EVLIL+ + +Y LP +IAKM KL L+I N G P++L++ + +
Sbjct: 539 TQMDWFDMELPKAEVLILHFSSDKYVLPPFIAKMGKLTALVIINNGMSPARLHDFSIFTN 598
Query: 574 LQNLERIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNI 629
L L+ + L+R+ VP + L+NL KLSL C + ++ + I+ FP L +L I
Sbjct: 599 LAKLKSLWLQRVHVPELSSSTVPLQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTI 658
Query: 630 DYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTS 689
D+C DL+ L + IC I SL +++TNC ++ LP+++ KL+ L+LL L +C +L ++P
Sbjct: 659 DHCDDLLELPSTICGITSLNSISITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVE 718
Query: 690 IGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCD 749
I +L LK++DIS C+SLSSLPE+ G + L+ +D C+ +P SVV L +L+ + CD
Sbjct: 719 ICELPRLKYVDISQCVSLSSLPEKIGKVKTLEKIDTRECSLSSIPNSVVLLTSLRHVICD 778
Query: 750 EETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
E WE Q + +++E + +WL
Sbjct: 779 REALWMWEKVQKAVAGLRVEAAEKSFSRDWL 809
>UniRef100_Q9LVT1 Putative disease resistance protein At5g47280 [Arabidopsis
thaliana]
Length = 623
Score = 336 bits (862), Expect = 1e-90
Identities = 216/643 (33%), Positives = 355/643 (54%), Gaps = 43/643 (6%)
Query: 144 LTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQ 203
++G+ G GKT LA +L DEEV G F ++F+T S++P L+ + I +F
Sbjct: 14 ISGMIGSGKTILAKELARDEEVRGHFANRVLFLTVSQSPNLEELRSLIR--------DFL 65
Query: 204 SDEDAVNKLGLLLKKVEG-SPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRF 262
+ +A G L + G + L++LDDV + ESL ++L F + G LV S+
Sbjct: 66 TGHEA--GFGTALPESVGHTRKLVILDDV-RTRESL-DQLMFNIPGTTTLVVSQSKLVDP 121
Query: 263 STTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKN 322
TT ++ L DA +LF A +K+ K+LV++VV GLPL++KV+ SL +
Sbjct: 122 RTTYDVELLNEHDATSLFCLSAFNQKSVPSGFSKSLVKQVVGESKGLPLSLKVLGASLND 181
Query: 323 RPHDLWHKIVKELSQGHSILDSN-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRI 381
RP W V+ LS+G + +++ +++ +++ + L+ PK ECF+D+ FPE +I
Sbjct: 182 RPETYWAIAVERLSRGEPVDETHESKVFAQIEATLENLD--PKTKECFLDMGAFPEGKKI 239
Query: 382 PVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVII-PRKNASDTDNNNYNNHFIIL 440
PV L++M +++ L+D A +++ L NL ++ P A T +Y + F+
Sbjct: 240 PVDVLINMLVKIHDLEDAA--AFDVLVDLANRNLLTLVKDPTFVAMGT---SYYDIFVTQ 294
Query: 441 HDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQ 500
HD+LR++ ++ + + +R RL++ T + S++ R N +
Sbjct: 295 HDVLRDVALHLTNRGKVSRRDRLLMPKRE-------------TMLPSEWER----SNDEP 337
Query: 501 LAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGF 560
AR++S+ T E +DW M + EVLI+N + Y LP +IAKM LRV +I N G
Sbjct: 338 YNARVVSIHTGEMTEMDWFDMDFPKAEVLIVNFSSDNYVLPPFIAKMGMLRVFVIINNGT 397
Query: 561 HPSKLNNIELLGSLQNLERIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSIL 616
P+ L++ + SL NL + LER+ VP + LKNL KL L +C +F++ +I
Sbjct: 398 SPAHLHDFPIPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICKINNSFDQTAID 457
Query: 617 ISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLS 676
I+ FP L ++ IDYC DL L + IC I SL +++TNC + LP++I KL+ L+LL
Sbjct: 458 IAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLLR 517
Query: 677 LSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFS 736
L +C +L+++P I +L L ++DIS+C+SLSSLPE+ GN+ L+ +DM C+ +P S
Sbjct: 518 LYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMRECSLSSIPSS 577
Query: 737 VVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNW 779
V+L +L +TC E W++ + + ++IE N+ W
Sbjct: 578 AVSLTSLCYVTCYREALWMWKEVEKAVPGLRIEATEKWFNMTW 620
>UniRef100_Q9FKZ2 Probable disease resistance protein At5g66890 [Arabidopsis
thaliana]
Length = 415
Score = 259 bits (663), Expect = 2e-67
Identities = 164/435 (37%), Positives = 245/435 (55%), Gaps = 41/435 (9%)
Query: 356 FDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNL 415
FD L N + ECF+D+A F ED RI + ++D+W+ Y G + M + L NL
Sbjct: 9 FDALPHN--LRECFLDMASFLEDQRIIASTIIDLWSASY-----GKEGMNNLQDLASRNL 61
Query: 416 ANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFE--QRKRLIIDMNNNKSG 473
++ +N + ++ YN + ++LRE I Q KE +RKRL +++ +NK
Sbjct: 62 LKLLPIGRN--EYEDGFYNELLVKQDNVLREFAINQCLKESSSIFERKRLNLEIQDNK-- 117
Query: 474 LAEKQQGLMTRIFSKFMRLCVKQNPQQ---LAARILSVSTDETCALDWSHMQPAQVEVLI 530
F C+ NP+Q + A + S+STD++ A W M VE L+
Sbjct: 118 ---------------FPNWCL--NPKQPIVINASLFSISTDDSFASSWFEMDCPNVEALV 160
Query: 531 LNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISV--- 587
LNI + Y+LP +IA M +L+V+II N+G P+KL N+ L SL NL+RIR E++S+
Sbjct: 161 LNISSSNYALPNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRIRFEKVSISLL 220
Query: 588 --PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDI 645
P G LK+L+KLSL+ C+ + A + +S+ +L+E+ IDYC +L L I +
Sbjct: 221 DIPKLG-LKSLEKLSLWFCHVVDALNELED-VSETLQSLQEIEIDYCYNLDELPYWISQV 278
Query: 646 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCI 705
+SLKKL+VTNC+KL + + IG L +LE L LSSC L +P +I +L NL+ LD+S
Sbjct: 279 VSLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGF 338
Query: 706 SLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHN 765
L +LP E G L L+ + M C ELP SV NL+NL+ + CDE+TA W+ + + N
Sbjct: 339 QLKNLPLEIGKLKKLEKISMKDCYRCELPDSVKNLENLE-VKCDEDTAFLWKILKPEMKN 397
Query: 766 MKIEVLHVDVNLNWL 780
+ I + NLN L
Sbjct: 398 LTITEEKTEHNLNLL 412
>UniRef100_Q8L8I8 NBS/LRR [Pinus taeda]
Length = 479
Score = 229 bits (584), Expect = 2e-58
Identities = 156/500 (31%), Positives = 257/500 (51%), Gaps = 42/500 (8%)
Query: 296 KNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTE-LLTRLQK 354
++LV++V C GLPL +KVI +SL+ + +W ++LS+ SI + E LL RL+
Sbjct: 5 EDLVKQVAAECKGLPLALKVIGSSLRGKRRPIWINAERKLSKSESISEYYKESLLKRLET 64
Query: 355 IFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMN 414
DVL+D K +CF+DL FP+ + V L+D+W + +++ A E++ +L N
Sbjct: 65 SIDVLDDKHK--QCFLDLGAFPKGRKFSVETLLDIWVYVRQME--WTDAFEVLLELASRN 120
Query: 415 LANVI-IPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSG 473
L N+ P A D + + HD++R+L +Y ++++ KRL +K
Sbjct: 121 LLNLTGYPGSGA--IDYSCASELTFSQHDVMRDLALYLASQDNIISPKRLFTPSKEDK-- 176
Query: 474 LAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNI 533
+ T S Q A+ +S+ T DW + +VE L L
Sbjct: 177 -------IPTEWLSTL-------KDQASRAQFVSIYTGAMQEQDWCQIDFPEVEALALFF 222
Query: 534 HTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSF--- 590
QY LP ++ + KL+VLI+ NY + + + S + + L ++ VP+
Sbjct: 223 SANQYCLPTFLHRTPKLKVLIVYNYSSMRANIIGLPRFSSPIQIRSVFLHKLIVPASLYE 282
Query: 591 --GTLKNLKKLSLYMCNTILAFEKGSILISDA------FPNLEELNIDYCKDLVVLQTGI 642
+ + L+KLS+ +C + G+ + D FPN+ E+NID+C DL L +
Sbjct: 283 NCRSWERLEKLSVCLCEGL-----GNSSLVDMELEPLNFPNITEINIDHCSDLGELPLKL 337
Query: 643 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 702
C++ SL++L+VTNCH + +LP D+G+L++L +L LS+C L +P SI KL L++LDIS
Sbjct: 338 CNLTSLQRLSVTNCHLIQNLPDDMGRLKSLRVLRLSACPSLSRLPPSICKLGQLEYLDIS 397
Query: 703 NCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVV--NLQNLKTITCDEETAATWEDFQ 760
C L LP EF L NL+ LDM C+ ++ +V+ +L+ + D+E A
Sbjct: 398 LCRCLQDLPSEFDQLSNLETLDMRECSGLKKVPTVIQSSLKRVVISDSDKEYEAWXSIKA 457
Query: 761 HMLHNMKIEVLHVDVNLNWL 780
LH + I+V+ +L WL
Sbjct: 458 STLHTLTIDVVPEIFSLAWL 477
>UniRef100_Q84KC6 NBS-LRR disease resistance protein homologue [Hordeum vulgare]
Length = 1262
Score = 162 bits (411), Expect = 3e-38
Identities = 165/645 (25%), Positives = 294/645 (45%), Gaps = 55/645 (8%)
Query: 140 STLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSV 199
+ L + GLGG+GKTTLA + D + N + ++V S+ L I I
Sbjct: 188 TVLPICGLGGIGKTTLAQLVFNDAQFNDYHR---VWVYVSQVFDLNKIGNSIISQVSGKG 244
Query: 200 PEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKLQFQM---TGFKILVTSR 256
E ++K L ++ L+VLDD+W + +++L+ + T K+LVT+R
Sbjct: 245 SEHSHTLQHISKQ--LKDLLQDKKTLIVLDDLWETGYFQLDQLKLMLNVSTKMKVLVTTR 302
Query: 257 -VAFPRFSTTC-----ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVE----KVVRSC 306
+ R +L PL ++ + ++ + DK +E K+ R C
Sbjct: 303 SIDIARKMGNVGVEPYMLDPLDNDMCWRIIKQSSRFQSRP----DKEQLEPNGQKIARKC 358
Query: 307 NGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIM 366
GLPL + + L W I +S S++ +L L+ ++ L P +
Sbjct: 359 GGLPLAAQALGFLLSGMDLSEWEAIC--ISDIWDEPFSDSTVLPSLKLSYNTL--TPYMR 414
Query: 367 ECFMDLALFPEDHRIPVAALVDMWAELYKLD-DNGIQAMEIINKLGIMNLANVIIPRKNA 425
CF +FP+ H I L+ W L ++ N A+++ K L +
Sbjct: 415 LCFAYCGIFPKGHNISKDYLIHQWIALGFIEPSNKFSAIQLGGKYVRQFLGMSFLHHSKL 474
Query: 426 SDTDNNNYNNHFIILHDILR-----ELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQ- 479
+T N ++HD+ R EL ++ + + K I + +++ +
Sbjct: 475 PETFGNAMFTMHDLVHDLARSVITEELVVFDAEIVSDNRIKEYCIYASLTNCNISDHNKV 534
Query: 480 GLMTRIFSKFMRLCVKQNPQ--------QLAARILSVSTDETCALDWSHMQPAQVEVLIL 531
MT IF +R+ + + Q R+L +S + Q Q+EVLI
Sbjct: 535 RKMTTIFPPKLRVMHFSDCKLHGSAFSFQKCLRVLDLSGCSIKDFASALGQLKQLEVLIA 594
Query: 532 N-IHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISV--P 588
+ +Q+ PE I ++SKL L ++ +++ L SL +L+ + V
Sbjct: 595 QKLQDRQF--PESITRLSKLHYLNLSGSRGISEIPSSVGKLVSLVHLDLSYCTNVKVIPK 652
Query: 589 SFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDII 646
+ G L+NL+ L L C + + + GS+ NL+ LN+ C +L L + +
Sbjct: 653 ALGILRNLQTLDLSWCEKLESLPESLGSV------QNLQRLNLSNCFELEALPESLGSLK 706
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
++ L++++C+KL SLP+ +G L+N++ L LS C L ++P ++G+L NL+ +D+S C
Sbjct: 707 DVQTLDLSSCYKLESLPESLGSLKNVQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGCKK 766
Query: 707 LSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTITCDE 750
L + PE FG+L NL+ L+++ C +E LP S +L+NL+T+ E
Sbjct: 767 LETFPESFGSLENLQILNLSNCFELESLPESFGSLKNLQTLNLVE 811
Score = 129 bits (325), Expect = 3e-28
Identities = 89/257 (34%), Positives = 148/257 (56%), Gaps = 17/257 (6%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 585
++ L ++ K S+PE + ++ L+ L ++ + ++ ++ LGSL+NL+ + L
Sbjct: 828 LQTLDFSVCHKLESVPESLGGLNNLQTLKLS---VCDNLVSLLKSLGSLKNLQTLDLSGC 884
Query: 586 ----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQT 640
S+P S G+L+NL+ L+L C + + + NL+ LNI +C +LV L
Sbjct: 885 KKLESLPESLGSLENLQILNLSNCFKLESLPESL----GRLKNLQTLNISWCTELVFLPK 940
Query: 641 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLD 700
+ ++ +L +L+++ C KL SLP +G LENLE L+LS C LE++P S+G L NL+ LD
Sbjct: 941 NLGNLKNLPRLDLSGCMKLESLPDSLGSLENLETLNLSKCFKLESLPESLGGLQNLQTLD 1000
Query: 701 ISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTIT---CDE-ETAAT 755
+ C L SLPE G L NL+ L ++ C +E LP S+ L+NL+T+T CD+ E+
Sbjct: 1001 LLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQTLTLSVCDKLESLPE 1060
Query: 756 WEDFQHMLHNMKIEVLH 772
LH +K++V +
Sbjct: 1061 SLGSLKNLHTLKLQVCY 1077
Score = 127 bits (319), Expect = 1e-27
Identities = 86/228 (37%), Positives = 133/228 (57%), Gaps = 15/228 (6%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 584
+E L L+ K SLPE + + L+ L + KL ++ E LG L+NL+ ++L
Sbjct: 972 LETLNLSKCFKLESLPESLGGLQNLQTLDLLVC----HKLESLPESLGGLKNLQTLQLSF 1027
Query: 585 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 639
S+P S G LKNL+ L+L +C+ + + + + NL L + C L L
Sbjct: 1028 CHKLESLPESLGGLKNLQTLTLSVCDKLESLPESL----GSLKNLHTLKLQVCYKLKSLP 1083
Query: 640 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 699
+ I +L LN++ CH L S+P+ +G LENL++L+LS+C LE+IP S+G L NL+ L
Sbjct: 1084 ESLGSIKNLHTLNLSVCHNLESIPESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTL 1143
Query: 700 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
+S C L SLP+ GNL NL+ LD++ C +E LP S+ +L+NL+T+
Sbjct: 1144 ILSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTL 1191
Score = 120 bits (300), Expect = 2e-25
Identities = 82/230 (35%), Positives = 133/230 (57%), Gaps = 19/230 (8%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 584
++ L L + K SLPE + + L+ L ++ KL ++ E LG L+NL+ + L
Sbjct: 996 LQTLDLLVCHKLESLPESLGGLKNLQTLQLS----FCHKLESLPESLGGLKNLQTLTLSV 1051
Query: 585 I----SVP-SFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVV 637
S+P S G+LKNL L L +C + + + GSI NL LN+ C +L
Sbjct: 1052 CDKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSI------KNLHTLNLSVCHNLES 1105
Query: 638 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 697
+ + + +L+ LN++NC KL S+P+ +G L+NL+ L LS CT L ++P ++G L NL+
Sbjct: 1106 IPESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQ 1165
Query: 698 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
LD+S C L SLP+ G+L NL+ L+++ C +E LP + +L+ L+T+
Sbjct: 1166 TLDLSGCKKLESLPDSLGSLENLQTLNLSNCFKLESLPEILGSLKKLQTL 1215
Score = 119 bits (297), Expect = 5e-25
Identities = 79/228 (34%), Positives = 135/228 (58%), Gaps = 15/228 (6%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 584
++ L L+ K SLPE + + L+ L ++ KL ++ E LGSL+NL ++L+
Sbjct: 1020 LQTLQLSFCHKLESLPESLGGLKNLQTLTLSVC----DKLESLPESLGSLKNLHTLKLQV 1075
Query: 585 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 639
S+P S G++KNL L+L +C+ + + + + NL+ LN+ C L +
Sbjct: 1076 CYKLKSLPESLGSIKNLHTLNLSVCHNLESIPESV----GSLENLQILNLSNCFKLESIP 1131
Query: 640 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 699
+ + +L+ L ++ C +L SLP+++G L+NL+ L LS C LE++P S+G L NL+ L
Sbjct: 1132 KSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTL 1191
Query: 700 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
++SNC L SLPE G+L L+ L++ C +E LP S+ +L++L+T+
Sbjct: 1192 NLSNCFKLESLPEILGSLKKLQTLNLFRCGKLESLPESLGSLKHLQTL 1239
Score = 118 bits (296), Expect = 6e-25
Identities = 84/230 (36%), Positives = 130/230 (56%), Gaps = 19/230 (8%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRL-- 582
++ L L+ K SLPE + + L++L ++N KL ++ E LG L+NL+ + +
Sbjct: 876 LQTLDLSGCKKLESLPESLGSLENLQILNLSNC----FKLESLPESLGRLKNLQTLNISW 931
Query: 583 --ERISVP-SFGTLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVV 637
E + +P + G LKNL +L L C + L GS+ NLE LN+ C L
Sbjct: 932 CTELVFLPKNLGNLKNLPRLDLSGCMKLESLPDSLGSL------ENLETLNLSKCFKLES 985
Query: 638 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 697
L + + +L+ L++ CHKL SLP+ +G L+NL+ L LS C LE++P S+G L NL+
Sbjct: 986 LPESLGGLQNLQTLDLLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQ 1045
Query: 698 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
L +S C L SLPE G+L NL L + C ++ LP S+ +++NL T+
Sbjct: 1046 TLTLSVCDKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSIKNLHTL 1095
Score = 116 bits (290), Expect = 3e-24
Identities = 81/227 (35%), Positives = 124/227 (53%), Gaps = 16/227 (7%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRL-- 582
V+ L L+ K SLP+ + ++ LR + ++ KL E GSL+NL+ + L
Sbjct: 732 VQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGC----KKLETFPESFGSLENLQILNLSN 787
Query: 583 --ERISVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 639
E S+P SFG+LKNL+ L+L C + + + NL+ L+ C L +
Sbjct: 788 CFELESLPESFGSLKNLQTLNLVECKKLESLPESL----GGLKNLQTLDFSVCHKLESVP 843
Query: 640 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 699
+ + +L+ L ++ C L SL + +G L+NL+ L LS C LE++P S+G L NL+ L
Sbjct: 844 ESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQTLDLSGCKKLESLPESLGSLENLQIL 903
Query: 700 DISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTI 746
++SNC L SLPE G L NL+ L+++ C EL F NL NLK +
Sbjct: 904 NLSNCFKLESLPESLGRLKNLQTLNISWCT--ELVFLPKNLGNLKNL 948
Score = 116 bits (290), Expect = 3e-24
Identities = 79/225 (35%), Positives = 129/225 (57%), Gaps = 9/225 (4%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 585
V+ L L+ K SLPE + + ++ L ++ S N+ L +L+ ++ +++
Sbjct: 708 VQTLDLSSCYKLESLPESLGSLKNVQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGCKKL 767
Query: 586 SV--PSFGTLKNLKKLSLYMCNTILAFEKGSILIS-DAFPNLEELNIDYCKDLVVLQTGI 642
SFG+L+NL+ L+L C FE S+ S + NL+ LN+ CK L L +
Sbjct: 768 ETFPESFGSLENLQILNLSNC-----FELESLPESFGSLKNLQTLNLVECKKLESLPESL 822
Query: 643 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 702
+ +L+ L+ + CHKL S+P+ +G L NL+ L LS C +L ++ S+G L NL+ LD+S
Sbjct: 823 GGLKNLQTLDFSVCHKLESVPESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQTLDLS 882
Query: 703 NCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
C L SLPE G+L NL+ L+++ C +E LP S+ L+NL+T+
Sbjct: 883 GCKKLESLPESLGSLENLQILNLSNCFKLESLPESLGRLKNLQTL 927
Score = 110 bits (276), Expect = 1e-22
Identities = 77/222 (34%), Positives = 124/222 (55%), Gaps = 15/222 (6%)
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 584
++ L L++ K SLPE + + L L + KL ++ E LGS++NL + L
Sbjct: 1044 LQTLTLSVCDKLESLPESLGSLKNLHTLKLQVC----YKLKSLPESLGSIKNLHTLNLSV 1099
Query: 585 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 639
S+P S G+L+NL+ L+L C + + K + NL+ L + +C LV L
Sbjct: 1100 CHNLESIPESVGSLENLQILNLSNCFKLESIPKSL----GSLKNLQTLILSWCTRLVSLP 1155
Query: 640 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 699
+ ++ +L+ L+++ C KL SLP +G LENL+ L+LS+C LE++P +G L L+ L
Sbjct: 1156 KNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTLNLSNCFKLESLPEILGSLKKLQTL 1215
Query: 700 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNL 740
++ C L SLPE G+L +L+ L + C +E LP S+ NL
Sbjct: 1216 NLFRCGKLESLPESLGSLKHLQTLVLIDCPKLEYLPKSLENL 1257
>UniRef100_Q67UX2 Putative NBS-LRR resistance protein RGH2 [Oryza sativa]
Length = 1216
Score = 145 bits (365), Expect = 6e-33
Identities = 175/663 (26%), Positives = 294/663 (43%), Gaps = 88/663 (13%)
Query: 140 STLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSV 199
S + + GLGG+GKTTLA + D+E + + +V S L IV I H S
Sbjct: 195 SIIPVVGLGGMGKTTLAKAVYTDKETH--MFDVKAWVHVSMEFQLNKIVSGIISHVEGST 252
Query: 200 PEFQSDEDAVNKLGLLLKKVEGSPL-LLVLDDVWPSS----ESLVEKLQFQMTGFKILVT 254
P +D + L L ++ + L L++LDD+W E L+E LQ G KI+VT
Sbjct: 253 PANIAD---LQYLKSQLDRILCNKLYLIILDDLWEEGWSKLEKLMEMLQSGKKGSKIIVT 309
Query: 255 SR----------VAFPRFSTTCILKPLAHE-DAVTLFHHYAQMEK-NSSDIIDKNLVEKV 302
+R + F T +K + D ME SD++D + +++
Sbjct: 310 TRSEKVVNTLSTIRLSYFHTVDPIKLVGMSIDECWFIMKPRNMENCEFSDLVD--IGKEI 367
Query: 303 VRSCNGLPLTIKVIATSL-KNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIF-DVLE 360
+ C+G+PL K + + K+R + W +I + +ILD+ + L+ +
Sbjct: 368 AQRCSGVPLVAKALGYVMQKHRTREEWMEI-----KNSNILDTKDDEEGILKGLLLSYYH 422
Query: 361 DNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQ-----AMEIINKL-GIMN 414
P++ CFM ++FP H I L+ W L + D Q AME +N+L G+
Sbjct: 423 MPPQLKLCFMYCSMFPMSHVIDHDCLIQQWIALGFIQDTDGQPLQKVAMEYVNELLGMSF 482
Query: 415 LANVIIPRKNASDT-DNNNYNNHFIILHDILRELGIYQSTKE-----PFEQRKRLIIDMN 468
L P AS H +HD++ EL + + E E R ++N
Sbjct: 483 LTIFTSPTVLASRMLFKPTLKLH---MHDMVHELARHVAGNEFSHTNGAENRNTKRDNLN 539
Query: 469 N------NKSGLAEKQQGLMTRIFSKFMRLCVKQN-PQQLAARILSVSTDETCALDWSHM 521
N++ + + L T++ + R C K + P+Q + L + LD
Sbjct: 540 FHYHLLLNQNETSSAYKSLATKVRALHFRGCDKMHLPKQAFSHTLCLRV-----LDLGGR 594
Query: 522 QPAQVEVLILNIHTKQY---------SLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLG 572
Q +++ + + +Y S + + L+ LI++N N +G
Sbjct: 595 QVSELPSSVYKLKLLRYLDASSLRISSFSKSFNHLLNLQALILSNTYLKTLPTN----IG 650
Query: 573 SLQNLERIRLERIS-----VPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEEL 627
LQ L+ L + SFG L +L L+L C+ + A +F NL L
Sbjct: 651 CLQKLQYFDLSGCANLNELPTSFGDLSSLLFLNLASCHELEALPM-------SFGNLNRL 703
Query: 628 ---NIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLE 684
++ C L L C + L L++++C+ L LP I +L LE L+++SC+ ++
Sbjct: 704 QFLSLSDCYKLNSLPESCCQLHDLAHLDLSDCYNLGKLPDCIDQLSKLEYLNMTSCSKVQ 763
Query: 685 AIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNL 743
A+P S+ KL L+HL++S C+ L +LP G+L L++LD+ + +LP S+ N+ L
Sbjct: 764 ALPESLCKLTMLRHLNLSYCLRLENLPSCIGDL-QLQSLDIQGSFLLRDLPNSIFNMSTL 822
Query: 744 KTI 746
KT+
Sbjct: 823 KTV 825
Score = 47.4 bits (111), Expect = 0.002
Identities = 28/83 (33%), Positives = 42/83 (49%), Gaps = 1/83 (1%)
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN-LKHLDISNCI 705
S++ L + + L +LP+ I +L LS+ C +LE +P +G L+ + I C
Sbjct: 1110 SIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCP 1169
Query: 706 SLSSLPEEFGNLCNLKNLDMATC 728
LSSLPE L LK L + C
Sbjct: 1170 MLSSLPESIRRLTKLKKLRITNC 1192
Score = 39.7 bits (91), Expect = 0.35
Identities = 27/80 (33%), Positives = 41/80 (50%), Gaps = 2/80 (2%)
Query: 669 LENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN-LKNLDMAT 727
L ++E L+L S L A+P +I +L L I C L +LPE G+ L+ + + T
Sbjct: 1108 LSSIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDT 1167
Query: 728 CASI-ELPFSVVNLQNLKTI 746
C + LP S+ L LK +
Sbjct: 1168 CPMLSSLPESIRRLTKLKKL 1187
Score = 38.1 bits (87), Expect = 1.0
Identities = 31/96 (32%), Positives = 49/96 (50%), Gaps = 7/96 (7%)
Query: 565 LNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNL 624
L++IE L +L ++ +R ++ F +L +LS+ C + + + D F L
Sbjct: 1108 LSSIESL-TLMSIAGLRALPEAIQCF---TSLWRLSILGCGELETLPEW---LGDYFTCL 1160
Query: 625 EELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLS 660
EE++ID C L L I + LKKL +TNC LS
Sbjct: 1161 EEISIDTCPMLSSLPESIRRLTKLKKLRITNCPVLS 1196
Score = 35.8 bits (81), Expect = 5.1
Identities = 20/61 (32%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 621 FPNLEELNIDYCKDLVVLQTGICDIIS-LKKLNVTNCHKLSSLPQDIGKLENLELLSLSS 679
F +L L+I C +L L + D + L+++++ C LSSLP+ I +L L+ L +++
Sbjct: 1132 FTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCPMLSSLPESIRRLTKLKKLRITN 1191
Query: 680 C 680
C
Sbjct: 1192 C 1192
>UniRef100_Q8H6V2 Putative rp3 protein [Zea mays]
Length = 944
Score = 132 bits (333), Expect = 3e-29
Identities = 190/796 (23%), Positives = 339/796 (41%), Gaps = 119/796 (14%)
Query: 17 DVLNKISEIVETAR-KNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQPREEI------NS 69
D++ +I+ ++T K +SS+ L +L K++ +D+ H H++ +++I N+
Sbjct: 40 DLVEEINNWLQTVGDKGRSSKWLKKL--KEVVYDAEDLVH-EFHIEAEKQDIEITGGKNT 96
Query: 70 LIEESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNH--------------- 114
L++ K + + I N +E + + + N
Sbjct: 97 LVKYFITKPKATVTEFKIAHKIKKIKNRFDEIVKGRSDYSTIANSMPVDYPVQHTRKTIG 156
Query: 115 ENPKFTV-------GLDIPFSKLKMELLRGGSSTLVLT--GLGGLGKTTLATKLCWDEEV 165
E P +T+ G D +++ EL+ S +++ GLGG GKTTLA KL +++
Sbjct: 157 EVPLYTIVDETTIFGRDQAKNQIISELIETDSQQKIVSVIGLGGSGKTTLA-KLVFNDGN 215
Query: 166 NGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK-KVEGSPL 224
K E +++V S+ ++ +VE++ + ++ SD + + + K+ G
Sbjct: 216 IIKHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSDHPPLQHVSRTISDKLVGKRF 271
Query: 225 LLVLDDVWPSSESLVEKLQFQM------TGFKILVTSR---VAFPRFSTTCILKP-LAHE 274
L VLDDVW +E VE QF + G IL+T+R VA S+ P L+ E
Sbjct: 272 LAVLDDVW--TEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKE 329
Query: 275 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKE 334
D+ +F + + D +++V C G+PL IKVIA L +K
Sbjct: 330 DSWKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHG---------IKG 380
Query: 335 LSQGHSILDSNT--------ELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAAL 386
+ + SI DSN + L F L D+ K CF+ ++FP + I L
Sbjct: 381 IEEWRSICDSNLLDVQDDEHRVFACLSLSFVHLPDHLK--PCFLHCSIFPRGYVINRRHL 438
Query: 387 VDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 438
+ W + + +D GI + + K+G + I + ++
Sbjct: 439 ISQWIAHGFVPTNQARQAEDVGIGYFDSLLKVGFLQDHVQIWSTRGEVTCKMHD------ 492
Query: 439 ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNP 498
++HD+ R++ R + ++ NK + L + +LC K
Sbjct: 493 LVHDLARQI-----------LRDEFVSEIETNKQIKRCRYLSLTSCTGKLDNKLCGKVRA 541
Query: 499 QQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNY 558
+ R L D + + V +IL T SLP +++K L L I++
Sbjct: 542 LYVCGRELE--------FDKTMNKQCCVRTIILKYITDD-SLPLFVSKFEYLGYLEISDV 592
Query: 559 GFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTLKNLKKLSLYMCNTILAFEKGS 614
P L+ +LQ L + R++V S G LK L+ L L ++I + +
Sbjct: 593 NCEALPEALSRC---WNLQALHVLNCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS- 648
Query: 615 ILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLENL 672
I D NL L ++ C+ + + + + +L+ L++ +C L LP GKL NL
Sbjct: 649 --IGDC-DNLRRLYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLNL 705
Query: 673 ELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE 732
+ ++ + C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C +
Sbjct: 706 QTITFNLCYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKLR 765
Query: 733 -LPFSVVNLQNLKTIT 747
LP L L+ ++
Sbjct: 766 GLPAGCGKLTRLQQLS 781
>UniRef100_Q8H6U6 Putative rp3 protein [Zea mays]
Length = 1222
Score = 132 bits (333), Expect = 3e-29
Identities = 193/796 (24%), Positives = 340/796 (42%), Gaps = 119/796 (14%)
Query: 17 DVLNKISEIVETAR-KNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQPREEI------NS 69
D++ +I+ ++T K +SS+ L +L K++ +D+ H H++ +++I N+
Sbjct: 40 DLVEEINNWLQTVGDKGRSSKWLKKL--KEVVYDAEDLVH-EFHIEAEKQDIEITGGKNT 96
Query: 70 LIEESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNH--------------- 114
L++ K + + I N +E + + + N
Sbjct: 97 LVKYFITKPKATVTEFKIAHKIKKIKNRFDEIVKGRSDYSTIANSMPVDYPVQHTRKTIG 156
Query: 115 ENPKFTV-------GLDIPFSKLKMELLRGGSSTLVLT--GLGGLGKTTLATKLCWDEEV 165
E P +T+ G D +++ EL+ S +++ GLGG GKTTLA KL +++
Sbjct: 157 EVPLYTIVDETTIFGRDQAKNQIISELIETDSQQKIVSVIGLGGSGKTTLA-KLVFNDGN 215
Query: 166 NGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK-KVEGSPL 224
K E +++V S+ ++ +VE++ + ++ SD + + + K+ G
Sbjct: 216 IIKHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSDHPPLQHVSRTISDKLVGKRF 271
Query: 225 LLVLDDVWPSSESLVEKLQFQM------TGFKILVTSR---VAFPRFSTTCILKP-LAHE 274
L VLDDVW +E VE QF + G IL+T+R VA S+ P L+ E
Sbjct: 272 LAVLDDVW--TEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKE 329
Query: 275 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKE 334
D+ +F + + D +++V C G+PL IKVIA L +K
Sbjct: 330 DSWKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHG---------IKG 380
Query: 335 LSQGHSILDSNT--------ELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAAL 386
+ + SI DSN + L F L D+ K CF+ ++FP + I L
Sbjct: 381 IEEWRSICDSNLLDVQDDEHRVFACLSLSFVHLPDHLK--PCFLHCSIFPRGYVINRRHL 438
Query: 387 VDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 438
+ W + + +D GI + + K+G + I + ++
Sbjct: 439 ISQWIAHGFVPTNQARQAEDVGIGYFDSLLKVGFLQDHVQIWSTRGEVTCKMHD------ 492
Query: 439 ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNP 498
++HD+ R++ R + ++ NK + L + +LC K
Sbjct: 493 LVHDLARQI-----------LRDEFVSEIETNKQIKRCRYLSLTSCTGKLDNKLCGK--- 538
Query: 499 QQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNY 558
R L V E D + + V +IL T SLP +++K L L I++
Sbjct: 539 ----VRALYVCGPEL-EFDKTMNKQCCVRTIILKYITAD-SLPLFVSKFEYLGYLEISSV 592
Query: 559 GFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTLKNLKKLSLYMCNTILAFEKGS 614
P L+ +LQ L ++ R++V S G LK L+ L L ++I + +
Sbjct: 593 NCEALPEALSRC---WNLQALHVLKCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS- 648
Query: 615 ILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLENL 672
I D NL L ++ C + + + + +L+ LN+ +C L LP GKL NL
Sbjct: 649 --IGDC-DNLRRLYLEGCHGIEDIPNSLGKLENLRILNIVHCISLQKLPPSDSFGKLLNL 705
Query: 673 ELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE 732
+ ++ + C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C +
Sbjct: 706 QTITFNLCYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKLR 765
Query: 733 -LPFSVVNLQNLKTIT 747
LP L L+ ++
Sbjct: 766 GLPAGCGKLTRLQQLS 781
Score = 60.1 bits (144), Expect = 3e-07
Identities = 87/361 (24%), Positives = 149/361 (41%), Gaps = 51/361 (14%)
Query: 387 VDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRE 446
++M EL+ LD ++ I KL I +PR A +D+ + I
Sbjct: 856 LNMEKELHLLDS--LEPPSKIEKLRIRGYRGSQLPRWMAKQSDSCGPADDTHI------- 906
Query: 447 LGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARIL 506
+ Q F L++D N L E + + +I L +K+ P+ +
Sbjct: 907 --VMQRNPSEFSHLTELVLDNLPNLEHLGELVELPLVKI------LKLKRLPKLVELLTT 958
Query: 507 SVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRV------LIITNYGF 560
+ + L H V L++ K P + + LR+ L+ + F
Sbjct: 959 TTGEEGVEVLYRFH----HVSTLVIIDCPKLVVKPYFPPSLQSLRLEGNNGQLVSSGCFF 1014
Query: 561 HPSKLN--NIELLGSLQNLERIRLERISVPSFGT-----LKNLKKLSLYMCNTILAFEKG 613
HP + + +++G+ +LER+ L R++ S G L L L ++ C T L
Sbjct: 1015 HPRHHHAAHADVIGT--HLERLELRRLTGSSSGWEVLQHLTGLHTLEIFKC-TDLTHLPE 1071
Query: 614 SILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCH-----------KLSSL 662
SI F L I C +L VL + ++ SL+ LN+ +C L+ L
Sbjct: 1072 SIHCPTTFCRLL---ITGCHNLRVLPDWLVELKSLQSLNIDSCDALQHLTISSLTSLTCL 1128
Query: 663 PQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKN 722
P+ + L +L L+L C +L +P +G+L L+ L + +C L+SLP+ L L+
Sbjct: 1129 PESMQHLTSLRTLNLCRCNELTHLPEWLGELSVLQKLWLQDCRGLTSLPQSIQRLTALEE 1188
Query: 723 L 723
L
Sbjct: 1189 L 1189
Score = 43.1 bits (100), Expect = 0.032
Identities = 68/302 (22%), Positives = 130/302 (42%), Gaps = 60/302 (19%)
Query: 506 LSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSK-------LRVLIITNY 558
LS++ ++ L S P+++E L + + + LP W+AK S +++ N
Sbjct: 854 LSLNMEKELHLLDSLEPPSKIEKLRIRGY-RGSQLPRWMAKQSDSCGPADDTHIVMQRN- 911
Query: 559 GFHPSKLNNIE--LLGSLQNLERI-RLERISVPSFGTLKNLKKL---------------- 599
PS+ +++ +L +L NLE + L + + LK L KL
Sbjct: 912 ---PSEFSHLTELVLDNLPNLEHLGELVELPLVKILKLKRLPKLVELLTTTTGEEGVEVL 968
Query: 600 -SLYMCNTILAFEKGSILISDAFP-NLEELNIDYCKDLVVLQ----------TGICDIIS 647
+ +T++ + +++ FP +L+ L ++ +V D+I
Sbjct: 969 YRFHHVSTLVIIDCPKLVVKPYFPPSLQSLRLEGNNGQLVSSGCFFHPRHHHAAHADVIG 1028
Query: 648 --LKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCI 705
L++L + SS + + L L L + CTDL +P SI L I+ C
Sbjct: 1029 THLERLELRRLTGSSSGWEVLQHLTGLHTLEIFKCTDLTHLPESIHCPTTFCRLLITGCH 1088
Query: 706 SLSSLPEEFGNLCNLKNLDMATCASIE------------LPFSVVNLQNLKTIT---CDE 750
+L LP+ L +L++L++ +C +++ LP S+ +L +L+T+ C+E
Sbjct: 1089 NLRVLPDWLVELKSLQSLNIDSCDALQHLTISSLTSLTCLPESMQHLTSLRTLNLCRCNE 1148
Query: 751 ET 752
T
Sbjct: 1149 LT 1150
Score = 41.6 bits (96), Expect = 0.092
Identities = 29/98 (29%), Positives = 49/98 (49%), Gaps = 7/98 (7%)
Query: 593 LKNLKKLSLYMCNTILAFEKGSILISDAFP-------NLEELNIDYCKDLVVLQTGICDI 645
LK+L+ L++ C+ + S+ P +L LN+ C +L L + ++
Sbjct: 1100 LKSLQSLNIDSCDALQHLTISSLTSLTCLPESMQHLTSLRTLNLCRCNELTHLPEWLGEL 1159
Query: 646 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 683
L+KL + +C L+SLPQ I +L LE L +S +L
Sbjct: 1160 SVLQKLWLQDCRGLTSLPQSIQRLTALEELYISGNPNL 1197
>UniRef100_Q9C784 Disease resistance protein, putative [Arabidopsis thaliana]
Length = 1398
Score = 132 bits (331), Expect = 5e-29
Identities = 85/222 (38%), Positives = 124/222 (55%), Gaps = 23/222 (10%)
Query: 540 LPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERIS--------VPSFG 591
LP I + L++L + + +E+ S+ NL ++L +S S G
Sbjct: 826 LPSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIG 879
Query: 592 TLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLK 649
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 880 NLINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLK 933
Query: 650 KLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSS 709
LN++ C L LP IG L NL+ L LS C+ L +P+SIG L+NLK LD+S C SL
Sbjct: 934 TLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVE 993
Query: 710 LPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTITCDE 750
LP GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 994 LPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 1035
Score = 132 bits (331), Expect = 5e-29
Identities = 189/781 (24%), Positives = 325/781 (41%), Gaps = 152/781 (19%)
Query: 72 EESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNHENP----KFTVGLDIPF 127
EE A + +++ ++ EN++ ++ ++ A EI + L + +P + +G+
Sbjct: 209 EEVATIAGYDSRNW---ENEAAMIEEI------AIEISKRLINSSPFSGFEGLIGMKAHI 259
Query: 128 SKLKMELLRGGSS---TLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPML 184
K+K L + T+ ++G G+GK+T+A L +++ F+ + +F+ F K
Sbjct: 260 EKMKQLLCLDSTDERRTVGISGPSGIGKSTIARVL--HNQISDGFQMS-VFMKF-KPSYT 315
Query: 185 KTIVERIHQ-----HCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLV 239
+ I H + ++ +++LG V G +L+VLD V + LV
Sbjct: 316 RPICSDDHDVKLQLEQQFLAQLINQEDIKIHQLGTAQNFVMGKKVLIVLDGV----DQLV 371
Query: 240 EKLQFQMT-----GFKILVTSR-----VAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKN 289
+ L G +I++T++ AF + P HE A+ +F +A +
Sbjct: 372 QLLAMPKAVCLGPGSRIIITTQDQQLLKAFQIKHIYNVDFPPDHE-ALQIFCIHAFGHDS 430
Query: 290 SSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELL 349
D +K L KV R LPL ++V+ + + + W EL + LD E+
Sbjct: 431 PDDGFEK-LATKVTRLAGNLPLGLRVMGSHFRGMSKEDWK---GELPRLRIRLDG--EIG 484
Query: 350 TRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINK 409
+ L+ +DVL+D K + F+ +A F D I D + G+Q
Sbjct: 485 SILKFSYDVLDDEDK--DLFLHIACFFNDEGID-HTFEDTLRHKFSNVQRGLQ------- 534
Query: 410 LGIMNLANVIIPRKNASDTDNNNYNNHFIIL-HDILRELGIYQSTKEPFEQRKRLIIDMN 468
V++ R S+ +N + L +I+R +Y+ K F + I ++
Sbjct: 535 --------VLVQRSLISEDLTQPMHNLLVQLGREIVRNQSVYEPGKRQFLVDGKEICEVL 586
Query: 469 NNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETC-------------- 514
+ +G +E G+ ++ L + + + + DE
Sbjct: 587 TSHTG-SESVIGINFEVYWSMDELNISDRVFEGMSNLQFFRFDENSYGRLHLPQGLNYLP 645
Query: 515 ----ALDWSHMQPAQVE--------VLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHP 562
L W + + V I+ H++ L E I + L+V+ + Y H
Sbjct: 646 PKLRILHWDYYPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNLKVMDL-RYSSHL 704
Query: 563 SKLNN------------------IELLGSLQNLERIR----------------------L 582
+L N IEL S+ N I+ L
Sbjct: 705 KELPNLSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITL 764
Query: 583 ERISV----------PSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNID 630
R+ + S G L NL +L L C++++ G+++ NLE
Sbjct: 765 PRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLI------NLEAFYFH 818
Query: 631 YCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSI 690
C L+ L + I ++ISLK L + L +P IG L NL+LL+LS C+ L +P+SI
Sbjct: 819 GCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLVELPSSI 878
Query: 691 GKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTITCD 749
G L+NLK LD+S C SL LP GNL NL+ L ++ C+S +ELP S+ NL NLKT+
Sbjct: 879 GNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLS 938
Query: 750 E 750
E
Sbjct: 939 E 939
Score = 122 bits (306), Expect = 4e-26
Identities = 76/184 (41%), Positives = 111/184 (60%), Gaps = 15/184 (8%)
Query: 571 LGSLQNLERIRLERISV-----PSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 623
+G+L NL+ + L S S G L NL++L L C++++ G+++ N
Sbjct: 926 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 979
Query: 624 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 683
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L LS C+ L
Sbjct: 980 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 1039
Query: 684 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQN 742
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 1040 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 1098
Query: 743 LKTI 746
LK +
Sbjct: 1099 LKKL 1102
Score = 116 bits (290), Expect = 3e-24
Identities = 76/203 (37%), Positives = 113/203 (55%), Gaps = 19/203 (9%)
Query: 571 LGSLQNLERIRLERIS----VP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 625
+G+L NL+++ L S +P S G L NLK L+L C++++ S NL+
Sbjct: 1046 IGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLK 1100
Query: 626 ELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEA 685
+L++ C LV L + I ++I+LKKL+++ C L LP IG L NL+ L LS C+ L
Sbjct: 1101 KLDLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVE 1160
Query: 686 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI----ELPFSVVNL- 740
+P+SIG L+NL+ L +S C SL LP GNL NLK LD+ C + +LP S+ L
Sbjct: 1161 LPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLV 1220
Query: 741 ----QNLKTITCDEETAATWEDF 759
++L+T+ C W F
Sbjct: 1221 AESCESLETLACSFPNPQVWLKF 1243
>UniRef100_Q9LFN1 RPP1 disease resistance protein-like [Arabidopsis thaliana]
Length = 1189
Score = 131 bits (329), Expect = 9e-29
Identities = 163/657 (24%), Positives = 290/657 (43%), Gaps = 111/657 (16%)
Query: 146 GLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSD 205
G G+GKTT+A +++ S + L +E I + Y+ P D
Sbjct: 264 GPPGIGKTTIA---------------RVVYNQLSHSFQLSVFMENIKAN--YTRPTGSDD 306
Query: 206 EDA-------------------VNKLGLLLKKVEGSPLLLVLDDVWPSSE--SLVEKLQF 244
A + LG+ +++ +L+VLD V S + ++ ++ +
Sbjct: 307 YSAKLQLQQMFMSQITKQKDIEIPHLGVAQDRLKDKKVLVVLDGVNQSVQLDAMAKEAWW 366
Query: 245 QMTGFKILVTSR--VAFPRFSTTCILKP--LAHEDAVTLFHHYAQMEKNSSDIIDKNLVE 300
G +I++T++ F I K E+A+ +F YA + + D +NL
Sbjct: 367 FGPGSRIIITTQDQKLFRAHGINHIYKVDFPPTEEALQIFCMYAFGQNSPKDGF-QNLAW 425
Query: 301 KVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLE 360
KV+ LPL ++++ + + + W K + L S LD++ + + + +D L+
Sbjct: 426 KVINLAGNLPLGLRIMGSYFRGMSREEWKKSLPRLE---SSLDADIQSILKFS--YDALD 480
Query: 361 DNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVII 420
D K + F+ +A F I + L + A+ + +E+ +L NV+
Sbjct: 481 DEDKNL--FLHIACFFNGKEIKI--LEEHLAKKF---------VEVRQRL------NVLA 521
Query: 421 PRKNASDTDNNNYNNHFIILH---DILRELGIYQSTKEPFEQRKRLIIDMNNNKSG---- 473
+ S ++ H ++ +I+R I++ + F I D+ N +
Sbjct: 522 EKSLISFSNWGTIEMHKLLAKLGGEIVRNQSIHEPGQRQFLFDGEEICDVLNGDAAGSKS 581
Query: 474 --------LAEKQQGLMTRIFS-----KFMRLCVKQNPQQLAARILSVSTDETCALDWSH 520
+ E++ + R+F +F+R + QL+ R LS + + LDW +
Sbjct: 582 VIGIDFHYIIEEEFDMNERVFEGMSNLQFLRFDCDHDTLQLS-RGLSYLSRKLQLLDWIY 640
Query: 521 M------QPAQVEVLI-LNI-HTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLG 572
VE LI LN+ H+K L E + + LR + ++ + L + L
Sbjct: 641 FPMTCLPSTVNVEFLIELNLTHSKLDMLWEGVKPLHNLRQMDLS----YSVNLKELPDLS 696
Query: 573 SLQNLERIRLER----ISVPS-FGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEEL 627
+ NL ++ L I +PS G NL+ L L C++++ DA NL++L
Sbjct: 697 TAINLRKLILSNCSSLIKLPSCIGNAINLEDLDLNGCSSLVELPS----FGDAI-NLQKL 751
Query: 628 NIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIP 687
+ YC +LV L + I + I+L++L++ C L LP IG NL +L L+ C++L +P
Sbjct: 752 LLRYCSNLVELPSSIGNAINLRELDLYYCSSLIRLPSSIGNAINLLILDLNGCSNLLELP 811
Query: 688 TSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNL 743
+SIG +NL+ LD+ C L LP GN NL+NL + C+S +ELP S+ N NL
Sbjct: 812 SSIGNAINLQKLDLRRCAKLLELPSSIGNAINLQNLLLDDCSSLLELPSSIGNATNL 868
>UniRef100_Q8H6V1 Putative rp3 protein [Zea mays]
Length = 1222
Score = 130 bits (327), Expect = 2e-28
Identities = 192/796 (24%), Positives = 339/796 (42%), Gaps = 119/796 (14%)
Query: 17 DVLNKISEIVETAR-KNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQPREEI------NS 69
D++ +I+ ++T K +SS+ L +L K++ +D+ H H++ +++I N+
Sbjct: 40 DLVEEINNWLQTVGDKGRSSKWLKKL--KEVVYDAEDLVH-EFHIEAEKQDIEITGGKNT 96
Query: 70 LIEESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNH--------------- 114
L++ K + + I N +E + + + N
Sbjct: 97 LVKYFITKPKATVTEFKIAHKIKKIKNRFDEIVKGRSDYSTIANSMPVDYPVQHTRKTIG 156
Query: 115 ENPKFTV-------GLDIPFSKLKMELLRGGSSTLVLT--GLGGLGKTTLATKLCWDEEV 165
E P +T+ G D +++ EL+ S +++ GLGG GKTTLA KL +++
Sbjct: 157 EVPLYTIVDETTIFGRDQAKNQIISELIETDSQQKIVSVIGLGGSGKTTLA-KLVFNDGN 215
Query: 166 NGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK-KVEGSPL 224
K E +++V S+ ++ +VE++ + ++ SD + + + K+ G
Sbjct: 216 IIKHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSDHPPLQHVSRTISDKLVGKRF 271
Query: 225 LLVLDDVWPSSESLVEKLQFQM------TGFKILVTSR---VAFPRFSTTCILKP-LAHE 274
L VLDDVW E VE QF + G IL+T+R VA S+ P L+ E
Sbjct: 272 LAVLDDVW--IEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKE 329
Query: 275 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKE 334
D+ +F + + D +++V C G+PL IKVIA L +K
Sbjct: 330 DSWKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHG---------IKG 380
Query: 335 LSQGHSILDSNT--------ELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAAL 386
+ + SI DSN + L F L D+ K CF+ ++FP + I L
Sbjct: 381 IEEWRSICDSNLLDVQDDEHRVFACLSLSFVHLPDHLK--PCFLHCSIFPRGYVINRRHL 438
Query: 387 VDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 438
+ W + + +D GI + + K+G + I + ++
Sbjct: 439 ISQWIAHGFVPTNQARQAEDVGIGYFDSLLKVGFLQDHVQIWSTRGEVTCKMHD------ 492
Query: 439 ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNP 498
++HD+ R++ R + ++ NK + L + +LC K
Sbjct: 493 LVHDLARQI-----------LRDEFVSEIETNKQIKRCRYLSLTSCTGKLDNKLCGK--- 538
Query: 499 QQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNY 558
R L V E D + + V +IL T SLP +++K L L I++
Sbjct: 539 ----VRALYVCGPEL-EFDKTMNKQCCVRTIILKYITAD-SLPLFVSKFEYLGYLEISDV 592
Query: 559 GFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTLKNLKKLSLYMCNTILAFEKGS 614
P L+ +LQ L + R++V S G LK L+ L L ++I + +
Sbjct: 593 NCEALPEALSRC---WNLQALHVLNCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS- 648
Query: 615 ILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLENL 672
I D NL L ++ C+ + + + + +L+ L++ +C L LP GKL NL
Sbjct: 649 --IGDC-DNLRRLYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLNL 705
Query: 673 ELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE 732
+ ++ + C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C +
Sbjct: 706 QTITFNLCYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKLR 765
Query: 733 -LPFSVVNLQNLKTIT 747
LP L L+ ++
Sbjct: 766 GLPAGCGKLTRLQQLS 781
Score = 60.1 bits (144), Expect = 3e-07
Identities = 87/361 (24%), Positives = 149/361 (41%), Gaps = 51/361 (14%)
Query: 387 VDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRE 446
++M EL+ LD ++ I KL I +PR A +D+ + I
Sbjct: 856 LNMEKELHLLDS--LEPPSKIEKLRIRGYRGSQLPRWMAKQSDSCGPADDTHI------- 906
Query: 447 LGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARIL 506
+ Q F L++D N L E + + +I L +K+ P+ +
Sbjct: 907 --VMQRNPSEFSHLTELVLDNLPNLEHLGELVELPLVKI------LKLKRLPKLVELLTT 958
Query: 507 SVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRV------LIITNYGF 560
+ + L H V L++ K P + + LR+ L+ + F
Sbjct: 959 TTGEEGVEVLYRFH----HVSTLVIIDCPKLVVKPYFPPSLQSLRLEGNNGQLVSSGCFF 1014
Query: 561 HPSKLN--NIELLGSLQNLERIRLERISVPSFGT-----LKNLKKLSLYMCNTILAFEKG 613
HP + + +++G+ +LER+ L R++ S G L L L ++ C T L
Sbjct: 1015 HPRHHHAAHADVIGT--HLERLELRRLTGSSSGWEVLQHLTGLHTLEIFKC-TDLTHLPE 1071
Query: 614 SILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCH-----------KLSSL 662
SI F L I C +L VL + ++ SL+ LN+ +C L+ L
Sbjct: 1072 SIHCPTTFCRLL---ITGCHNLRVLPDWLVELKSLQSLNIDSCDALQHLTISSLTSLTCL 1128
Query: 663 PQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKN 722
P+ + L +L L+L C +L +P +G+L L+ L + +C L+SLP+ L L+
Sbjct: 1129 PESMQHLTSLRTLNLCRCNELTHLPEWLGELSVLQKLWLQDCRGLTSLPQSIQRLTALEE 1188
Query: 723 L 723
L
Sbjct: 1189 L 1189
Score = 43.1 bits (100), Expect = 0.032
Identities = 68/302 (22%), Positives = 130/302 (42%), Gaps = 60/302 (19%)
Query: 506 LSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSK-------LRVLIITNY 558
LS++ ++ L S P+++E L + + + LP W+AK S +++ N
Sbjct: 854 LSLNMEKELHLLDSLEPPSKIEKLRIRGY-RGSQLPRWMAKQSDSCGPADDTHIVMQRN- 911
Query: 559 GFHPSKLNNIE--LLGSLQNLERI-RLERISVPSFGTLKNLKKL---------------- 599
PS+ +++ +L +L NLE + L + + LK L KL
Sbjct: 912 ---PSEFSHLTELVLDNLPNLEHLGELVELPLVKILKLKRLPKLVELLTTTTGEEGVEVL 968
Query: 600 -SLYMCNTILAFEKGSILISDAFP-NLEELNIDYCKDLVVLQ----------TGICDIIS 647
+ +T++ + +++ FP +L+ L ++ +V D+I
Sbjct: 969 YRFHHVSTLVIIDCPKLVVKPYFPPSLQSLRLEGNNGQLVSSGCFFHPRHHHAAHADVIG 1028
Query: 648 --LKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCI 705
L++L + SS + + L L L + CTDL +P SI L I+ C
Sbjct: 1029 THLERLELRRLTGSSSGWEVLQHLTGLHTLEIFKCTDLTHLPESIHCPTTFCRLLITGCH 1088
Query: 706 SLSSLPEEFGNLCNLKNLDMATCASIE------------LPFSVVNLQNLKTIT---CDE 750
+L LP+ L +L++L++ +C +++ LP S+ +L +L+T+ C+E
Sbjct: 1089 NLRVLPDWLVELKSLQSLNIDSCDALQHLTISSLTSLTCLPESMQHLTSLRTLNLCRCNE 1148
Query: 751 ET 752
T
Sbjct: 1149 LT 1150
Score = 41.6 bits (96), Expect = 0.092
Identities = 29/98 (29%), Positives = 49/98 (49%), Gaps = 7/98 (7%)
Query: 593 LKNLKKLSLYMCNTILAFEKGSILISDAFP-------NLEELNIDYCKDLVVLQTGICDI 645
LK+L+ L++ C+ + S+ P +L LN+ C +L L + ++
Sbjct: 1100 LKSLQSLNIDSCDALQHLTISSLTSLTCLPESMQHLTSLRTLNLCRCNELTHLPEWLGEL 1159
Query: 646 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 683
L+KL + +C L+SLPQ I +L LE L +S +L
Sbjct: 1160 SVLQKLWLQDCRGLTSLPQSIQRLTALEELYISGNPNL 1197
>UniRef100_Q8H6U9 Putative rp3 protein [Zea mays]
Length = 1247
Score = 130 bits (326), Expect = 2e-28
Identities = 187/797 (23%), Positives = 340/797 (42%), Gaps = 121/797 (15%)
Query: 17 DVLNKISEIVETAR-KNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQPREEI------NS 69
D++ +I+ ++T K +SS+ L +L K++ +D+ H H++ +++I N+
Sbjct: 40 DLVEEINNWLQTVGDKGRSSKWLKKL--KEVVYDAEDLVH-EFHIEAEKQDIEITGGKNT 96
Query: 70 LIEESACKSSFENDSYVVDENQSLIVNDVEETLYKAREILELLNH--------------- 114
L++ K + + I N +E + + + N
Sbjct: 97 LVKYFITKPKATVTEFKIAHKIKKIKNRFDEIVKGRSDYSTIANSMPVDYPVQHTRKTIG 156
Query: 115 ENPKFTV-------GLDIPFSKLKMELLRGGSSTLVLT--GLGGLGKTTLATKLCWDEEV 165
E P +T+ G D +++ EL+ S +++ GLGG GKTTLA KL +++
Sbjct: 157 EVPLYTIVDETTIFGRDQAKNQIISELIETDSQQKIVSVIGLGGSGKTTLA-KLVFNDGN 215
Query: 166 NGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK-KVEGSPL 224
K E +++V S+ ++ +VE++ + ++ SD + + + K+ G
Sbjct: 216 IIKHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSDHPPLQHVSRTISDKLVGKRF 271
Query: 225 LLVLDDVWPSSESLVEKLQFQM------TGFKILVTSR---VAFPRFSTTCILKP-LAHE 274
L VLDDVW +E VE QF + G IL+T+R VA S+ P L+ E
Sbjct: 272 LAVLDDVW--TEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKE 329
Query: 275 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKE 334
D+ +F + + D +++V C G+PL IKVIA L +K
Sbjct: 330 DSWKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHG---------IKG 380
Query: 335 LSQGHSILDSNT--------ELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAAL 386
+ + SI DSN + L F L D+ K CF+ ++FP + I L
Sbjct: 381 IEEWRSICDSNLLDVQDDEHRVFACLSLSFVHLPDHLK--PCFLHCSIFPRGYVINRRHL 438
Query: 387 VDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 438
+ W + + +D GI + + K+G + I + ++
Sbjct: 439 ISQWIAHGFVPTNQARQAEDVGIGYFDSLLKVGFLQDHVQIWSTRGEVTCKMHD------ 492
Query: 439 ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNP 498
++HD+ R++ R + ++ NK + R + C +
Sbjct: 493 LVHDLARQI-----------LRDEFVSEIETNKQ---------IKRCRYLSLTSCTGKLD 532
Query: 499 QQLAARILSV-STDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITN 557
+L ++ ++ D + + V +IL T SLP +++K L L I++
Sbjct: 533 NKLCGKVRALYGCGPELEFDKTMNKQCCVRTIILKYITAD-SLPLFVSKFEYLGYLEISD 591
Query: 558 YGFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTLKNLKKLSLYMCNTILAFEKG 613
P L+ +LQ L + R++V S G LK L+ L L ++I + +
Sbjct: 592 VNCEALPEALSRC---WNLQALHVLNCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS 648
Query: 614 SILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLEN 671
I D NL L ++ C+ + + + + +L+ L++ +C L LP GKL N
Sbjct: 649 ---IGDC-DNLRRLYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLN 704
Query: 672 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI 731
L+ ++ + C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C +
Sbjct: 705 LQTITFNLCYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKL 764
Query: 732 E-LPFSVVNLQNLKTIT 747
LP L L+ ++
Sbjct: 765 RGLPAGCGKLTRLQQLS 781
Score = 77.0 bits (188), Expect = 2e-12
Identities = 59/203 (29%), Positives = 95/203 (46%), Gaps = 20/203 (9%)
Query: 576 NLERIRLERISVPSFGT-----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNID 630
+LER+ L R++ S G L L L +YMC + + + L +L I
Sbjct: 1042 HLERLELRRLTGSSSGWEVLQHLTGLHTLEIYMCTDLTHLPESIHCPT----TLCKLMII 1097
Query: 631 YCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSI 690
C +L VL + ++ SL+ LN+ +C L LP+ IG+L +L+ L + S L +P S+
Sbjct: 1098 RCDNLRVLPDWLVELKSLQSLNIDSCDALQQLPEQIGELSSLQHLHIISMPFLTCLPESM 1157
Query: 691 GKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLKTI--- 746
L +L+ L++ C +L+ LPE G L L+ L + C + LP S+ L L+ +
Sbjct: 1158 QHLTSLRTLNLCRCNALTQLPEWLGELSVLQQLWLQGCRDLTSLPQSIQRLTALEDLLIS 1217
Query: 747 -------TCDEETAATWEDFQHM 762
C E W H+
Sbjct: 1218 YNPDLVRRCREGVGEDWHLVSHI 1240
>UniRef100_Q8H6V3 Putative rp3 protein [Zea mays]
Length = 1251
Score = 128 bits (321), Expect = 7e-28
Identities = 157/636 (24%), Positives = 271/636 (41%), Gaps = 101/636 (15%)
Query: 142 LVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPE 201
+ + GLGG GKTTLA ++ D + F E +++V S+ ++ +VE++ + ++
Sbjct: 195 VAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAVEKLVEKLFE----AIAG 249
Query: 202 FQSDEDAVNKLGLLLK-KVEGSPLLLVLDDVWPSS----ESLVEKLQFQMTGFKILVTSR 256
SD + + + K+ G L VLDDVW E + L+ G IL+T+R
Sbjct: 250 HMSDHLPLQHVSRTISDKLVGKRFLAVLDDVWTEDRVEWERFMVHLKSGAPGSSILLTTR 309
Query: 257 ---VAFPRFSTTCILKP-LAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLT 312
VA S+ P L+ ED+ +F ++ + D ++V C G+PL
Sbjct: 310 SRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFRIAIQALDTEFLQAGIEIVDKCGGVPLA 369
Query: 313 IKVIATSLKN-RPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMD 371
IKVIA L + + W I S + D + L F L D+ K CF+
Sbjct: 370 IKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACLWLSFVHLPDHLK--PCFLH 425
Query: 372 LALFPEDHRIPVAALVDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRK 423
++FP + + L+ W + + +D GI + + K+G +
Sbjct: 426 CSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYFDSLLKVGFLQ--------- 476
Query: 424 NASDTDNNNYNNHFI------ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEK 477
D D N Y + ++HD+ R++ R + ++ NK +
Sbjct: 477 -DQDRDQNLYTRGEVTCKMHDLVHDLARKI-----------LRDEFVSEIETNKQIKRCR 524
Query: 478 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 537
L + +LC K + + R L D + + V +IL T +
Sbjct: 525 YLSLSSCTGKLDNKLCGKVHALYVCGRELE--------FDRTMNKQCYVRTIILKYITAE 576
Query: 538 YSLPEWIAKMSKLRVLIITNYGFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTL 593
SLP +++K L L I++ P L+ +LQ L + +++V S G L
Sbjct: 577 -SLPLFVSKFEYLGYLEISDVNCEALPEALSRC---WNLQALHVLACSKLAVVPESIGKL 632
Query: 594 KNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV 653
K L+ L L ++I + + G CD +L++L +
Sbjct: 633 KKLRTLELNGVSSIKSLPES--------------------------IGDCD--NLRRLYL 664
Query: 654 TNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIP--TSIGKLLNLKHLDISNCISLSSLP 711
C + +P +GKLENL +LS+ +C L+ + S GKLLNL+ + +C +L +LP
Sbjct: 665 EGCRGIEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLP 724
Query: 712 EEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTI 746
+ +L +L+ +D+ C +ELP + NL+NLK +
Sbjct: 725 QCMTSLSHLEMVDLGYCFELVELPEGIGNLRNLKVL 760
Score = 75.9 bits (185), Expect = 4e-12
Identities = 100/419 (23%), Positives = 171/419 (39%), Gaps = 64/419 (15%)
Query: 387 VDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRE 446
++M EL+ LD ++ I KLGI +PR A +D+ + I
Sbjct: 860 LNMEKELHLLDS--LEPPSKIEKLGIRGYRGSQLPRWMAKQSDSCGPADDTHI------- 910
Query: 447 LGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARIL 506
+ Q F L++D N L E + + +I L +K+ P+ +
Sbjct: 911 --VMQRNPSEFSHLTELVLDNLPNLEHLGELVELPLIKI------LKLKRLPKLVELLTT 962
Query: 507 SVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRV------LIITNYGF 560
+ + L H V L++ K P + A + +L + L+ + F
Sbjct: 963 TTGEEGVEVLCRFH----HVSTLVIIDCPKLVVKPYFPASLQRLTLEGNNGQLVSSGCFF 1018
Query: 561 HP--------------SKLNNIELLGSLQNLERIRLERISVPSFGT-----LKNLKKLSL 601
HP S +++G+ +LER+ L ++ S G L L L +
Sbjct: 1019 HPRHHHAAHAHGDESSSSSYFADVIGT--HLERLELRWLTGSSSGWEVLQHLTGLHTLEI 1076
Query: 602 YMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSS 661
+ C + + + L L I C +L VL + ++ SL+ L V CH L
Sbjct: 1077 FKCTGLTHLPESIHCPT----TLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFCHALQQ 1132
Query: 662 LPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLK 721
LP+ IG+L +L+ L + T L +P S+ +L +L+ LD+ C +L+ LPE G L L+
Sbjct: 1133 LPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGALTQLPEWLGELSALQ 1192
Query: 722 NLDMATCASI-ELPFSVVNLQNLKTI----------TCDEETAATWEDFQHMLHNMKIE 769
L++ C + LP S+ L L+ + C E W H + N+++E
Sbjct: 1193 KLNLGGCRGLTSLPRSIQCLTALEELFIGGNPDLLRRCREGVGEDWPLVSH-IQNLRLE 1250
Score = 67.4 bits (163), Expect = 2e-09
Identities = 37/104 (35%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
+L+ L+V C KL+ +P+ IGKL+ L L L+ + ++++P SIG NL+ L + C
Sbjct: 610 NLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNLRRLYLEGCRG 669
Query: 707 LSSLPEEFGNLCNLKNLDMATCASIEL---PFSVVNLQNLKTIT 747
+ +P G L NL+ L + C S++ S L NL+TIT
Sbjct: 670 IEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTIT 713
>UniRef100_Q8H6V4 Putative rp3 protein [Zea mays]
Length = 948
Score = 127 bits (318), Expect = 2e-27
Identities = 157/636 (24%), Positives = 270/636 (41%), Gaps = 101/636 (15%)
Query: 142 LVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPE 201
+ + GLGG GKTTLA ++ D + F E +++V S+ ++ +VE++ + ++
Sbjct: 195 VAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAVEKLVEKLFE----AIAG 249
Query: 202 FQSDEDAVNKLGLLLK-KVEGSPLLLVLDDVWPSS----ESLVEKLQFQMTGFKILVTSR 256
SD + + + K+ G L VLDDVW E + L+ G IL+T+R
Sbjct: 250 HMSDHLPLQHVSRTISDKLVGKRFLAVLDDVWTEDRVEWERFMVHLKSGAPGSSILLTTR 309
Query: 257 ---VAFPRFSTTCILKP-LAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLT 312
VA S+ P L+ ED+ +F + + D ++V C G+PL
Sbjct: 310 SRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFGIAIQALDTEFLQAGIEIVDKCGGVPLA 369
Query: 313 IKVIATSLKN-RPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMD 371
IKVIA L + + W I S + D + L F L D+ K CF+
Sbjct: 370 IKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACLWLSFVHLPDHLK--PCFLH 425
Query: 372 LALFPEDHRIPVAALVDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRK 423
++FP + + L+ W + + +D GI + + K+G +
Sbjct: 426 CSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYFDSLLKVGFLQ--------- 476
Query: 424 NASDTDNNNYNNHFI------ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEK 477
D D N Y + ++HD+ R++ R + ++ NK +
Sbjct: 477 -DQDRDQNLYTRGEVTCKMHDLVHDLARKI-----------LRDEFVSEIETNKQIKRCR 524
Query: 478 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 537
L + +LC K + + R L D + + V +IL T +
Sbjct: 525 YLSLSSCTGKLDNKLCGKVHALYVCGRELE--------FDRTMNKQCYVRTIILKYITAE 576
Query: 538 YSLPEWIAKMSKLRVLIITNYGFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTL 593
SLP +++K L L I++ P L+ +LQ L + +++V S G L
Sbjct: 577 -SLPLFVSKFEYLGYLEISDVNCEALPEALSRC---WNLQALHVLACSKLAVVPESIGKL 632
Query: 594 KNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV 653
K L+ L L ++I + + G CD +L++L +
Sbjct: 633 KKLRTLELNGVSSIKSLPES--------------------------IGDCD--NLRRLYL 664
Query: 654 TNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIP--TSIGKLLNLKHLDISNCISLSSLP 711
C + +P +GKLENL +LS+ +C L+ + S GKLLNL+ + +C +L +LP
Sbjct: 665 EGCRGIEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLP 724
Query: 712 EEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTI 746
+ +L +L+ +D+ C +ELP + NL+NLK +
Sbjct: 725 QCMTSLSHLEMVDLGYCFELVELPEGIGNLRNLKVL 760
Score = 67.4 bits (163), Expect = 2e-09
Identities = 37/104 (35%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
+L+ L+V C KL+ +P+ IGKL+ L L L+ + ++++P SIG NL+ L + C
Sbjct: 610 NLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNLRRLYLEGCRG 669
Query: 707 LSSLPEEFGNLCNLKNLDMATCASIEL---PFSVVNLQNLKTIT 747
+ +P G L NL+ L + C S++ S L NL+TIT
Sbjct: 670 IEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTIT 713
>UniRef100_Q8GS26 Putative rp3 protein [Zea mays]
Length = 1251
Score = 127 bits (318), Expect = 2e-27
Identities = 157/636 (24%), Positives = 270/636 (41%), Gaps = 101/636 (15%)
Query: 142 LVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQHCGYSVPE 201
+ + GLGG GKTTLA ++ D + F E +++V S+ ++ +VE++ + ++
Sbjct: 195 VAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAVEKLVEKLFE----AIAG 249
Query: 202 FQSDEDAVNKLGLLLK-KVEGSPLLLVLDDVWPSS----ESLVEKLQFQMTGFKILVTSR 256
SD + + + K+ G L VLDDVW E + L+ G IL+T+R
Sbjct: 250 HMSDHLPLQHVSRTISDKLVGKRFLAVLDDVWTEDRVEWERFMVHLKSGAPGSSILLTTR 309
Query: 257 ---VAFPRFSTTCILKP-LAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLT 312
VA S+ P L+ ED+ +F + + D ++V C G+PL
Sbjct: 310 SRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFGIAIQALDTEFLQAGIEIVDKCGGVPLA 369
Query: 313 IKVIATSLKN-RPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMD 371
IKVIA L + + W I S + D + L F L D+ K CF+
Sbjct: 370 IKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACLWLSFVHLPDHLK--PCFLH 425
Query: 372 LALFPEDHRIPVAALVDMWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRK 423
++FP + + L+ W + + +D GI + + K+G +
Sbjct: 426 CSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYFDSLLKVGFLQ--------- 476
Query: 424 NASDTDNNNYNNHFI------ILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEK 477
D D N Y + ++HD+ R++ R + ++ NK +
Sbjct: 477 -DQDRDQNLYTRGEVTCKMHDLVHDLARKI-----------LRDEFVSEIETNKQIKRCR 524
Query: 478 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 537
L + +LC K + + R L D + + V +IL T +
Sbjct: 525 YLSLSSCTGKLDNKLCGKVHALYVCGRELE--------FDRTMNKQCYVRTIILKYITAE 576
Query: 538 YSLPEWIAKMSKLRVLIITNYGFH--PSKLNNIELLGSLQNLERIRLERISV--PSFGTL 593
SLP +++K L L I++ P L+ +LQ L + +++V S G L
Sbjct: 577 -SLPLFVSKFEYLGYLEISDVNCEALPEALSRC---WNLQALHVLACSKLAVVPESIGKL 632
Query: 594 KNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNV 653
K L+ L L ++I + + G CD +L++L +
Sbjct: 633 KKLRTLELNGVSSIKSLPES--------------------------IGDCD--NLRRLYL 664
Query: 654 TNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIP--TSIGKLLNLKHLDISNCISLSSLP 711
C + +P +GKLENL +LS+ +C L+ + S GKLLNL+ + +C +L +LP
Sbjct: 665 EGCRGIEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLP 724
Query: 712 EEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTI 746
+ +L +L+ +D+ C +ELP + NL+NLK +
Sbjct: 725 QCMTSLSHLEMVDLGYCFELVELPEGIGNLRNLKVL 760
Score = 75.9 bits (185), Expect = 4e-12
Identities = 100/419 (23%), Positives = 171/419 (39%), Gaps = 64/419 (15%)
Query: 387 VDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRE 446
++M EL+ LD ++ I KLGI +PR A +D+ + I
Sbjct: 860 LNMEKELHLLDS--LEPPSKIEKLGIRGYRGSQLPRWMAKQSDSCGPADDTHI------- 910
Query: 447 LGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARIL 506
+ Q F L++D N L E + + +I L +K+ P+ +
Sbjct: 911 --VMQRNPSEFSHLTELVLDNLPNLEHLGELVELPLIKI------LKLKRLPKLVELLTT 962
Query: 507 SVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRV------LIITNYGF 560
+ + L H V L++ K P + A + +L + L+ + F
Sbjct: 963 TTGEEGVEVLCRFH----HVSTLVIIDCPKLVVKPYFPASLQRLTLEGNNGQLVSSGCFF 1018
Query: 561 HP--------------SKLNNIELLGSLQNLERIRLERISVPSFGT-----LKNLKKLSL 601
HP S +++G+ +LER+ L ++ S G L L L +
Sbjct: 1019 HPRHHHAAHAHGDESSSSSYFADVIGT--HLERLELRWLTGSSSGWEVLQHLTGLHTLEI 1076
Query: 602 YMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSS 661
+ C + + + L L I C +L VL + ++ SL+ L V CH L
Sbjct: 1077 FKCTGLTHLPESIHCPT----TLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFCHALQQ 1132
Query: 662 LPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLK 721
LP+ IG+L +L+ L + T L +P S+ +L +L+ LD+ C +L+ LPE G L L+
Sbjct: 1133 LPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGALTQLPEWLGELSALQ 1192
Query: 722 NLDMATCASI-ELPFSVVNLQNLKTI----------TCDEETAATWEDFQHMLHNMKIE 769
L++ C + LP S+ L L+ + C E W H + N+++E
Sbjct: 1193 KLNLGGCRGLTSLPRSIQCLTALEELFIGGNPDLLRRCREGVGEDWPLVSH-IQNLRLE 1250
Score = 67.4 bits (163), Expect = 2e-09
Identities = 37/104 (35%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
+L+ L+V C KL+ +P+ IGKL+ L L L+ + ++++P SIG NL+ L + C
Sbjct: 610 NLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNLRRLYLEGCRG 669
Query: 707 LSSLPEEFGNLCNLKNLDMATCASIEL---PFSVVNLQNLKTIT 747
+ +P G L NL+ L + C S++ S L NL+TIT
Sbjct: 670 IEDIPNSLGKLENLRILSIVACFSLKKLSPSASFGKLLNLQTIT 713
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,256,464,980
Number of Sequences: 2790947
Number of extensions: 53438069
Number of successful extensions: 209923
Number of sequences better than 10.0: 3752
Number of HSP's better than 10.0 without gapping: 1303
Number of HSP's successfully gapped in prelim test: 2479
Number of HSP's that attempted gapping in prelim test: 186488
Number of HSP's gapped (non-prelim): 13778
length of query: 781
length of database: 848,049,833
effective HSP length: 135
effective length of query: 646
effective length of database: 471,271,988
effective search space: 304441704248
effective search space used: 304441704248
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC135229.3


