

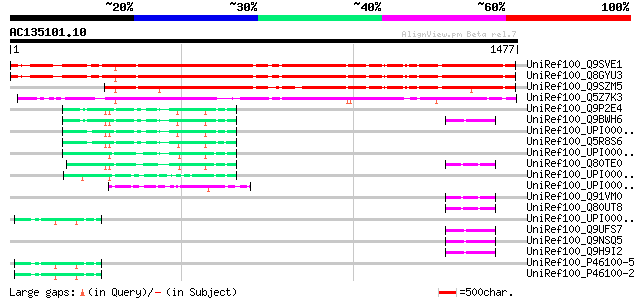
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135101.10 + phase: 0 /pseudo
(1477 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SVE1 Hypothetical protein F22I13.210 [Arabidopsis th... 1171 0.0
UniRef100_Q8GYU3 Hypothetical protein At4g38440/F22I13_210 [Arab... 1165 0.0
UniRef100_Q9SZM5 Hypothetical protein F20M13.10 [Arabidopsis tha... 1045 0.0
UniRef100_Q5Z7K3 Hypothetical protein OSJNBa0006A22.41 [Oryza sa... 905 0.0
UniRef100_Q9P2E4 KIAA1403 protein [Homo sapiens] 97 5e-18
UniRef100_Q9BWH6 DKFZP727M111 protein [Homo sapiens] 97 5e-18
UniRef100_UPI000036A014 UPI000036A014 UniRef100 entry 96 1e-17
UniRef100_Q5R8S6 Hypothetical protein DKFZp469H2113 [Pongo pygma... 94 2e-17
UniRef100_UPI00001CF220 UPI00001CF220 UniRef100 entry 89 1e-15
UniRef100_Q80TE0 MKIAA1403 protein [Mus musculus] 87 3e-15
UniRef100_UPI0000364055 UPI0000364055 UniRef100 entry 76 7e-12
UniRef100_UPI0000439692 UPI0000439692 UniRef100 entry 75 2e-11
UniRef100_Q91VM0 Rpap1 protein [Mus musculus] 54 3e-05
UniRef100_Q80UT8 Rpap1 protein [Mus musculus] 54 3e-05
UniRef100_UPI00004578C3 UPI00004578C3 UniRef100 entry 53 8e-05
UniRef100_Q9UFS7 Hypothetical protein DKFZp727M111 [Homo sapiens] 53 8e-05
UniRef100_Q9NSQ5 Hypothetical protein DKFZp434E1120 [Homo sapiens] 53 8e-05
UniRef100_Q9H9I2 Hypothetical protein FLJ12732 [Homo sapiens] 53 8e-05
UniRef100_P46100-5 Splice isoform 5 of P46100 [Homo sapiens] 53 8e-05
UniRef100_P46100-2 Splice isoform 1 of P46100 [Homo sapiens] 53 8e-05
>UniRef100_Q9SVE1 Hypothetical protein F22I13.210 [Arabidopsis thaliana]
Length = 1468
Score = 1171 bits (3030), Expect = 0.0
Identities = 674/1490 (45%), Positives = 915/1490 (61%), Gaps = 137/1490 (9%)
Query: 1 MGFEKAAAFANPV*RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSS 60
M + AAFA P+ RK+ K MD G+W K M G DD + S+
Sbjct: 94 MNADSIAAFAKPLQRKEKKDMDLGRW-----KDMVSG-------DDPA----------ST 131
Query: 61 YGPTTGRKKN--ENGGKNTSKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSATSMELD 118
+ P RK E + ++ + + + + + V FI + + E
Sbjct: 132 HVPQQSRKLKIIETRPPYVASADAATTSSNTLLAARASDQREFVSDKAPFIKNLGTKERV 191
Query: 119 TSNKDDKKEVFAAERDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMS 178
N V N S+SLE++ID EN A++Q MS
Sbjct: 192 PLNASPPLAV---------------------SNGLGTRHASSSLESDIDVENHAKLQTMS 230
Query: 179 TEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQQVQITQGAKH 238
+EI EA+A++++K+ PALL +L+KRG+ KLKK V E+ AK+
Sbjct: 231 PDEIAEAQAELLDKMDPALLSILKKRGEAKLKKRKHSVQGVSITDET----------AKN 280
Query: 239 LQTEDD-ISHTIMAPPSKKQLDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDV 297
+TE ++ +MA P +K + K W+AW+ RVEA R+LRFS G+V
Sbjct: 281 SRTEGHFVTPKVMAIPKEKSVVQK------PGIAQGFVWDAWTERVEAARDLRFSFDGNV 334
Query: 298 VDTEQ-EPV--------YDNIAERDYLRTEGDPGAAGYTIKEALEITRSVRALGLHLLSS 348
V+ + P ++ AERD+LRTEGDPGAAGYTIKEA+ + RSVR L LHLL+S
Sbjct: 335 VEEDVVSPAETGGKWSGVESAAERDFLRTEGDPGAAGYTIKEAIALARSVRCLALHLLAS 394
Query: 349 VLDKALCYICKDRTENMTKKGNKVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVV 408
VLDKAL +C+ R ++ DKS DWEA+W YALGP+PEL L+LRM LDDNH SVV
Sbjct: 395 VLDKALNKLCQSRIGYAREEK---DKSTDWEAIWAYALGPEPELVLALRMALDDNHASVV 451
Query: 409 LACAKVVQSALSCDVNENYFDISENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAK 468
+AC KV+Q LSC +NEN+F+I ENM + KDI TA VFRS+P+I LGFL+G YWKYSAK
Sbjct: 452 IACVKVIQCLLSCSLNENFFNILENMGPHGKDIFTASVFRSKPEIDLGFLRGCYWKYSAK 511
Query: 469 PSNIQPFSEDSMDNESDDKHTIQDDVFVAGQDFTAGLVRMGILPRLRYLLETDPTAALEE 528
PSNI F E+ +D+ ++D TIQ DVFVAGQD AGLVRM ILPR+ +LLET+PTAALE+
Sbjct: 512 PSNIVAFREEILDDGTEDTDTIQKDVFVAGQDVAAGLVRMDILPRIYHLLETEPTAALED 571
Query: 529 CIVSILIAIVRHSPSCANAVLKCERLIQTIVQRFTVG-NFEIRSSMIKSVKLLKVLARLD 587
I+S+ IAI RHSP C A+LK + +QTIV+RF + ++ SS I SV+LLKVLAR D
Sbjct: 572 SIISVTIAIARHSPKCTTAILKYPKFVQTIVKRFQLNKRMDVLSSQINSVRLLKVLARYD 631
Query: 588 RKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGY 647
+ TC+EF+KNG FNA+TW+L+Q S+D W+KLGK+ CKL S L +EQLRFW+VCI G
Sbjct: 632 QSTCMEFVKNGTFNAVTWHLFQFTSSLDSWVKLGKQNCKLSSTLMVEQLRFWKVCIHSGC 691
Query: 648 CVSHFSKIFPALCFWLDLPSFEKLTKNNVLNESTCISREAYLVLESLAERLRNLFSQQCL 707
CVS F ++FPALC WL PSFEKL + N+++E T +S EAYLVLE+ AE L N++SQ
Sbjct: 692 CVSRFPELFPALCLWLSCPSFEKLREKNLISEFTSVSNEAYLVLEAFAETLPNMYSQNIP 751
Query: 708 TNQHPESTDDAEFWSWSYVGPMVDLAIKWIARRSDPEVYKLFEGQEEGVNHFTLGDLSST 767
N ++ W WSYV PM+D A+ WI P++ K +G E +S+T
Sbjct: 752 RN-------ESGTWDWSYVSPMIDSALSWITLA--PQLLKWEKGIES-------VSVSTT 795
Query: 768 PLLWVYAAVTHMLFRVLEKVTLGDAISLQEANGHVPWLPKFVPKIGLELINYWHLGFSVA 827
LLW+Y+ V + +VLEK IS + +PWLP+FVPKIGL +I + L FSVA
Sbjct: 796 TLLWLYSGVMRTISKVLEK------ISAEGEEEPLPWLPEFVPKIGLAIIKHKLLSFSVA 849
Query: 828 SVTKSGRDSGD-ESFMKELIHLRQKG-DIEMSLASTCCLNGIINVITKIDNLIRSAKTGI 885
V++ G+DS SFM+ L LR++ D E++LAS CL+G+ I I NLI SA++ +
Sbjct: 850 DVSRFGKDSSRCSSFMEYLCFLRERSQDDELALASVNCLHGLTRTIVSIQNLIESARSKM 909
Query: 886 CNPPVTEQSLSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAP 945
P S E VL GI++ L EL S+ F S SS W +QSIE+ RGG AP
Sbjct: 910 KAPHQVSISTGDE-SVLANGILAESLAELTSVSCSFRDSVSSEWPIVQSIELHKRGGLAP 968
Query: 946 GMGVGWGAHGGGFWSKTVLPVKTDARLLVCLLQIFENTSNDAPETEQMTFSMQQVNTALG 1005
G+G+GWGA GGGFWS VL + A LL L I + +D+ + M +VN+AL
Sbjct: 969 GVGLGWGASGGGFWSTRVLLAQAGAGLLSLFLNI---SLSDSQNDQGSVGFMDKVNSALA 1025
Query: 1006 LCLTAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMHFSRM 1065
+CL AGP D +++E+ + + L++L CI++ N++ +F W+ + DY S M
Sbjct: 1026 MCLIAGPRDYLLVERAFEYVLRPHALEHLACCIKS---NKKNISFEWECSEGDYHRMSSM 1082
Query: 1066 LSSHFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSDM--SSTTSPCCNSLM 1123
L+SHFR RWL + +S A +G SGV+ K V L+TI+ED +M SST +S
Sbjct: 1083 LASHFRHRWLQQKGRSIAEEG--VSGVR---KGTVGLETIHEDGEMSNSSTQDKKSDSST 1137
Query: 1124 IEWARQNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANLLEVAKCGLFFVLGIE 1183
IEWA Q +PLP H++LS IS + +G G P + LLEVAK G+FF+ G+E
Sbjct: 1138 IEWAHQRMPLPPHWFLSAISAV----HSGKTSTG----PPESTELLEVAKAGVFFLAGLE 1189
Query: 1184 TMSSFIGTGIPSPIQRVSLTWKLHSLSVNFLVGMEILEQDQGRETFEALQDLYGELLDKE 1243
+ S F +PSP+ V L WK H+LS LVGM+I+E R + LQ+LYG+ LD+
Sbjct: 1190 SSSGF--GSLPSPVVSVPLVWKFHALSTVLLVGMDIIEDKNTRNLYNYLQELYGQFLDEA 1247
Query: 1244 RFNQNKEAISDDKKHIEFLRFKSDIHESYSTFIEELVEQFSSISYGDLIFGRQVSVYLHC 1303
R N + E LRFKSDIHE+YSTF+E +VEQ++++SYGD+++GRQVSVYLH
Sbjct: 1248 RLNH---------RDTELLRFKSDIHENYSTFLEMVVEQYAAVSYGDVVYGRQVSVYLHQ 1298
Query: 1304 CVESSIRLATWNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALD 1363
CVE S+RL+ W LSNARVLELLP L+KC A+GYLEP E+NE +LEAY KSW ALD
Sbjct: 1299 CVEHSVRLSAWTVLSNARVLELLPSLDKCLGEADGYLEPVEENEAVLEAYLKSWTCGALD 1358
Query: 1364 RAEIRGSVSYTMAVHHLSSFIFNACPVDKLLLRNNLVRSLLRDYAGKQQHEGMLMNLISH 1423
RA RGSV+YT+ VHH SS +F DK+ LRN +V++L+RD + K+ EGM+++L+ +
Sbjct: 1359 RAATRGSVAYTLVVHHFSSLVFCNQAKDKVSLRNKIVKTLVRDLSRKRHREGMMLDLLRY 1418
Query: 1424 NRQSTSNMDEQLDGLLHEESWLESRMKVLIEACEGNSSLLIQVKKLKDAA 1473
+ S + M+E++ + E RM+VL E CEGNS+LL++++KLK AA
Sbjct: 1419 KKGSANAMEEEVIA-----AETEKRMEVLKEGCEGNSTLLLELEKLKSAA 1463
>UniRef100_Q8GYU3 Hypothetical protein At4g38440/F22I13_210 [Arabidopsis thaliana]
Length = 1465
Score = 1165 bits (3015), Expect = 0.0
Identities = 674/1494 (45%), Positives = 915/1494 (61%), Gaps = 141/1494 (9%)
Query: 1 MGFEKAAAFANPV*RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSS 60
M + AAFA P+ RK+ K MD G+W K M G DD + S+
Sbjct: 87 MNADSIAAFAKPLQRKEKKDMDLGRW-----KDMVSG-------DDPA----------ST 124
Query: 61 YGPTTGRKKN--ENGGKNTSKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSATSMELD 118
+ P RK E + ++ + + + + + V FI + + E
Sbjct: 125 HVPQQSRKLKIIETRPPYVASADAATTSSNTLLAARASDQREFVSDKAPFIKNLGTKERV 184
Query: 119 TSNKDDKKEVFAAERDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMS 178
N V N S+SLE++ID EN A++Q MS
Sbjct: 185 PLNASPPLAV---------------------SNGLGTRHASSSLESDIDVENHAKLQTMS 223
Query: 179 TEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQQVQITQGAKH 238
+EI EA+A++++K+ PALL +L+KRG+ KLKK V E+ AK+
Sbjct: 224 PDEIAEAQAELLDKMDPALLSILKKRGEAKLKKRKHSVQGVSITDET----------AKN 273
Query: 239 LQTEDD-ISHTIMAPPSKKQLDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDV 297
+TE ++ +MA P +K + K W+AW+ RVEA R+LRFS G+V
Sbjct: 274 SRTEGHFVTPKVMAIPKEKSVVQK------PGIAQGFVWDAWTERVEAARDLRFSFDGNV 327
Query: 298 VDTEQ-EPV--------YDNIAERDYLRTEGDPGAAGYTIKEALEITRSV----RALGLH 344
V+ + P ++ AERD+LRTEGDPGAAGYTIKEA+ + RSV R L LH
Sbjct: 328 VEEDVVSPAETGGKWSGVESAAERDFLRTEGDPGAAGYTIKEAIALARSVIPGQRCLALH 387
Query: 345 LLSSVLDKALCYICKDRTENMTKKGNKVDKSVDWEAVWTYALGPQPELALSLRM*LDDNH 404
LL+SVLDKAL +C+ R ++ DKS DWEA+W YALGP+PEL L+LRM LDDNH
Sbjct: 388 LLASVLDKALNKLCQSRIGYAREEK---DKSTDWEAIWAYALGPEPELVLALRMALDDNH 444
Query: 405 NSVVLACAKVVQSALSCDVNENYFDISENMATYDKDICTAPVFRSRPDISLGFLQGGYWK 464
SVV+AC KV+Q LSC +NEN+F+I ENM + KDI TA VFRS+P+I LGFL+G YWK
Sbjct: 445 ASVVIACVKVIQCLLSCSLNENFFNILENMGPHGKDIFTASVFRSKPEIDLGFLRGCYWK 504
Query: 465 YSAKPSNIQPFSEDSMDNESDDKHTIQDDVFVAGQDFTAGLVRMGILPRLRYLLETDPTA 524
YSAKPSNI F E+ +D+ ++D TIQ DVFVAGQD AGLVRM ILPR+ +LLET+PTA
Sbjct: 505 YSAKPSNIVAFREEILDDGTEDTDTIQKDVFVAGQDVAAGLVRMDILPRIYHLLETEPTA 564
Query: 525 ALEECIVSILIAIVRHSPSCANAVLKCERLIQTIVQRFTVG-NFEIRSSMIKSVKLLKVL 583
ALE+ I+S+ IAI RHSP C A+LK + +QTIV+RF + ++ SS I SV+LLKVL
Sbjct: 565 ALEDSIISVTIAIARHSPKCTTAILKYPKFVQTIVKRFQLNKRMDVLSSQINSVRLLKVL 624
Query: 584 ARLDRKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCI 643
AR D+ TC+EF+KNG FNA+TW+L+Q S+D W+KLGK+ CKL S L +EQLRFW+VCI
Sbjct: 625 ARYDQSTCMEFVKNGTFNAVTWHLFQFTSSLDSWVKLGKQNCKLSSTLMVEQLRFWKVCI 684
Query: 644 RYGYCVSHFSKIFPALCFWLDLPSFEKLTKNNVLNESTCISREAYLVLESLAERLRNLFS 703
G CVS F ++FPALC WL PSFEKL + N+++E T +S EAYLVLE+ AE L N++S
Sbjct: 685 HSGCCVSRFPELFPALCLWLSCPSFEKLREKNLISEFTSVSNEAYLVLEAFAETLPNMYS 744
Query: 704 QQCLTNQHPESTDDAEFWSWSYVGPMVDLAIKWIARRSDPEVYKLFEGQEEGVNHFTLGD 763
Q N ++ W WSYV PM+D A+ WI P++ K +G E
Sbjct: 745 QNIPRN-------ESGTWDWSYVSPMIDSALSWITLA--PQLLKWEKGIES-------VS 788
Query: 764 LSSTPLLWVYAAVTHMLFRVLEKVTLGDAISLQEANGHVPWLPKFVPKIGLELINYWHLG 823
+S+T LLW+Y+ V + +VLEK IS + +PWLP+FVPKIGL +I + L
Sbjct: 789 VSTTTLLWLYSGVMRTISKVLEK------ISAEGEEEPLPWLPEFVPKIGLAIIKHKLLS 842
Query: 824 FSVASVTKSGRDSGD-ESFMKELIHLRQKG-DIEMSLASTCCLNGIINVITKIDNLIRSA 881
FSVA V++ G+DS SFM+ L LR++ D E++LAS CL+G+ I I NLI SA
Sbjct: 843 FSVADVSRFGKDSSRCSSFMEYLCFLRERSQDDELALASVNCLHGLTRTIVSIQNLIESA 902
Query: 882 KTGICNPPVTEQSLSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRG 941
++ + P S E VL GI++ L EL S+ F S SS W +QSIE+ RG
Sbjct: 903 RSKMKAPHQVSISTGDE-SVLANGILAESLAELTSVSCSFRDSVSSEWPIVQSIELHKRG 961
Query: 942 GPAPGMGVGWGAHGGGFWSKTVLPVKTDARLLVCLLQIFENTSNDAPETEQMTFSMQQVN 1001
G APG+G+GWGA GGGFWS VL + A LL L I + +D+ + M +VN
Sbjct: 962 GLAPGVGLGWGASGGGFWSTRVLLAQAGAGLLSLFLNI---SLSDSQNDQGSVGFMDKVN 1018
Query: 1002 TALGLCLTAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMH 1061
+AL +CL AGP D +++E+ + + L++L CI++ N++ +F W+ + DY
Sbjct: 1019 SALAMCLIAGPRDYLLVERAFEYVLRPHALEHLACCIKS---NKKNISFEWECSEGDYHR 1075
Query: 1062 FSRMLSSHFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSDM--SSTTSPCC 1119
S ML+SHFR RWL + +S A +G SGV+ K V L+TI+ED +M SST
Sbjct: 1076 MSSMLASHFRHRWLQQKGRSIAEEG--VSGVR---KGTVGLETIHEDGEMSNSSTQDKKS 1130
Query: 1120 NSLMIEWARQNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANLLEVAKCGLFFV 1179
+S IEWA Q +PLP H++LS IS + +G G P + LLEVAK G+FF+
Sbjct: 1131 DSSTIEWAHQRMPLPPHWFLSAISAV----HSGKTSTG----PPESTELLEVAKAGVFFL 1182
Query: 1180 LGIETMSSFIGTGIPSPIQRVSLTWKLHSLSVNFLVGMEILEQDQGRETFEALQDLYGEL 1239
G+E+ S F +PSP+ V L WK H+LS LVGM+I+E R + LQ+LYG+
Sbjct: 1183 AGLESSSGF--GSLPSPVVSVPLVWKFHALSTVLLVGMDIIEDKNTRNLYNYLQELYGQF 1240
Query: 1240 LDKERFNQNKEAISDDKKHIEFLRFKSDIHESYSTFIEELVEQFSSISYGDLIFGRQVSV 1299
LD+ R N + E LRFKSDIHE+YSTF+E +VEQ++++SYGD+++GRQVSV
Sbjct: 1241 LDEARLNH---------RDTELLRFKSDIHENYSTFLEMVVEQYAAVSYGDVVYGRQVSV 1291
Query: 1300 YLHCCVESSIRLATWNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEAYAKSWVS 1359
YLH CVE S+RL+ W LSNARVLELLP L+KC A+GYLEP E+NE +LEAY KSW
Sbjct: 1292 YLHQCVEHSVRLSAWTVLSNARVLELLPSLDKCLGEADGYLEPVEENEAVLEAYLKSWTC 1351
Query: 1360 DALDRAEIRGSVSYTMAVHHLSSFIFNACPVDKLLLRNNLVRSLLRDYAGKQQHEGMLMN 1419
ALDRA RGSV+YT+ VHH SS +F DK+ LRN +V++L+RD + K+ EGM+++
Sbjct: 1352 GALDRAATRGSVAYTLVVHHFSSLVFCNQAKDKVSLRNKIVKTLVRDLSRKRHREGMMLD 1411
Query: 1420 LISHNRQSTSNMDEQLDGLLHEESWLESRMKVLIEACEGNSSLLIQVKKLKDAA 1473
L+ + + S + M+E++ + E RM+VL E CEGNS+LL++++KLK AA
Sbjct: 1412 LLRYKKGSANAMEEEVIA-----AETEKRMEVLKEGCEGNSTLLLELEKLKSAA 1460
>UniRef100_Q9SZM5 Hypothetical protein F20M13.10 [Arabidopsis thaliana]
Length = 1179
Score = 1045 bits (2703), Expect = 0.0
Identities = 588/1246 (47%), Positives = 783/1246 (62%), Gaps = 140/1246 (11%)
Query: 276 WNAWSNRVEAIRELRFSLAGDVVDTEQ-EPV--------YDNIAERDYLRTEGDPGAAGY 326
W+AW+ RVEA R+LRFS G+VV+ + P ++ AERD+LRTEGDPGAAGY
Sbjct: 21 WDAWTERVEAARDLRFSFDGNVVEEDVVSPAETGGKWSGVESAAERDFLRTEGDPGAAGY 80
Query: 327 TIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGNKVDKSVDWEAVW 382
TIKEA+ + RSV R L LHLL+SVLDKAL +C+ R ++ DKS DWEA+W
Sbjct: 81 TIKEAIALARSVIPGQRCLALHLLASVLDKALNKLCQSRIGYAREEK---DKSTDWEAIW 137
Query: 383 TYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDISEN--------- 433
YALGP+PEL L+LRM LDDNH SVV+AC KV+Q LSC +NEN+F+I EN
Sbjct: 138 AYALGPEPELVLALRMALDDNHASVVIACVKVIQCLLSCSLNENFFNILENVIIHVQTDK 197
Query: 434 -------MATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDD 486
M + KDI TA VFRS+P+I LGFL+G YWKYSAKPSNI F E+ +D+ ++D
Sbjct: 198 THFYFQNMGPHGKDIFTASVFRSKPEIDLGFLRGCYWKYSAKPSNIVAFREEILDDGTED 257
Query: 487 KHTIQDDVFVAGQDFTAGLVRMGILPRLRYLLETDPTAALEECIVSILIAIVRHSPSCAN 546
TIQ DVFVAGQD AGLVRM ILPR+ +LLET+PTAALE+ I+S+ IAI RHSP C
Sbjct: 258 TDTIQKDVFVAGQDVAAGLVRMDILPRIYHLLETEPTAALEDSIISVTIAIARHSPKCTT 317
Query: 547 AVLKCERLIQTIVQRFTVGN-FEIRSSMIKSVKLLK-----VLARLDRKTCLEFIKNGYF 600
A+LK + +QTIV+RF + ++ SS I SV+LLK VLAR D+ TC+EF+KNG F
Sbjct: 318 AILKYPKFVQTIVKRFQLNKRMDVLSSQINSVRLLKNLMIRVLARYDQSTCMEFVKNGTF 377
Query: 601 NAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCVSHFSKIFPALC 660
NA+TW+L+Q S+D W+KLGK+ CKL S L +EQLRFW+VCI G CVS F ++FPALC
Sbjct: 378 NAVTWHLFQFTSSLDSWVKLGKQNCKLSSTLMVEQLRFWKVCIHSGCCVSRFPELFPALC 437
Query: 661 FWLDLPSFEKLTKNNVLNESTCISREAYLVLESLAERLRNLFSQQCLTNQHPESTDDAEF 720
WL PSFEKL + N+++E T +S EAYLVLE+ AE L N++SQ N ++
Sbjct: 438 LWLSCPSFEKLREKNLISEFTSVSNEAYLVLEAFAETLPNMYSQNIPRN-------ESGT 490
Query: 721 WSWSYVGPMVDLAIKWIARRSDPEVYKLFEGQEEGVNHFTLGDLSSTPLLWVYAAVTHML 780
W WSYV PM+D A+ WI P++ K +G E +V +
Sbjct: 491 WDWSYVSPMIDSALSWITLA--PQLLKWEKGIE---------------------SVMRTI 527
Query: 781 FRVLEKVTLGDAISLQEANGHVPWLPKFVPKIGLELINYWHLGFSVASVTKSGRDSGDES 840
+VLEK IS + +PWLP+FVPKIGL +I + L FSVA V++
Sbjct: 528 SKVLEK------ISAEGEEEPLPWLPEFVPKIGLAIIKHKLLSFSVADVSR--------- 572
Query: 841 FMKELIHLRQKGDIEMSLASTCCLNGIINVITKIDNLIRSAKTGICNPPVTEQSLSKEGK 900
+ D E++LAS CL+G+ I I NLI SA++ + P S E
Sbjct: 573 --------ERSQDDELALASVNCLHGLTRTIVSIQNLIESARSKMKAPHQVSISTGDE-S 623
Query: 901 VLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAPGMGVGWGAHGGGFWS 960
VL GI++ L EL S+ F S SS W +QSIE+ RGG APG+G+GWGA GGGFWS
Sbjct: 624 VLANGILAESLAELTSVSCSFRDSVSSEWPIVQSIELHKRGGLAPGVGLGWGASGGGFWS 683
Query: 961 KTVLPVKTDARLLVCLLQIFENTSNDAPETEQMTFSMQQVNTALGLCLTAGPADMVVIEK 1020
VL + A LL L I + +D+ + M +VN+AL +CL AGP D +++E+
Sbjct: 684 TRVLLAQAGAGLLSLFLNI---SLSDSQNDQGSVGFMDKVNSALAMCLIAGPRDYLLVER 740
Query: 1021 TLDLLFHVSILKYLDLCIQNFLLNRRGKAFGWKYEDDDYMHFSRMLSSHFRSRWLSVRVK 1080
+ + L++L CI++ N++ +F W+ + DY S ML+SHFR RWL + +
Sbjct: 741 AFEYVLRPHALEHLACCIKS---NKKNISFEWECSEGDYHRMSSMLASHFRHRWLQQKGR 797
Query: 1081 SKAVDGSSSSGVKATPKADVRLDTIYEDSDM--SSTTSPCCNSLMIEWARQNLPLPVHFY 1138
S A +G SGV+ K V L+TI+ED +M SST +S IEWA Q +PLP H++
Sbjct: 798 SIAEEG--VSGVR---KGTVGLETIHEDGEMSNSSTQDKKSDSSTIEWAHQRMPLPPHWF 852
Query: 1139 LSPISTIPLTKRAGPQKVGSVHNPHDPANLLEVAKCGLFFVLGIETMSSFIGTGIPSPIQ 1198
LS IS + +G G P + LLEVAK G+FF+ G+E+ S F +PSP+
Sbjct: 853 LSAISAV----HSGKTSTG----PPESTELLEVAKAGVFFLAGLESSSGF--GSLPSPVV 902
Query: 1199 RVSLTWKLHSLSVNFLVGMEILEQDQGRETFEALQDLYGELLDKERFNQNKEAISDDKKH 1258
V L WK H+LS LVGM+I+E R + LQ+LYG+ LD+ R N +
Sbjct: 903 SVPLVWKFHALSTVLLVGMDIIEDKNTRNLYNYLQELYGQFLDEARLNH---------RD 953
Query: 1259 IEFLRFKSDIHESYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLS 1318
E LRFKSDIHE+YSTF+E +VEQ++++SYGD+++GRQVSVYLH CVE S+RL+ W LS
Sbjct: 954 TELLRFKSDIHENYSTFLEMVVEQYAAVSYGDVVYGRQVSVYLHQCVEHSVRLSAWTVLS 1013
Query: 1319 NARVLELLPPLEKCFSGAEGYLEPAE-----------DNEEILEAYAKSWVSDALDRAEI 1367
NARVLELLP L+KC A+GYLEP E +NE +LEAY KSW ALDRA
Sbjct: 1014 NARVLELLPSLDKCLGEADGYLEPVEVIYKNNKMLIVENEAVLEAYLKSWTCGALDRAAT 1073
Query: 1368 RGSVSYTMAVHHLSSFIFNACPVDKLLLRNNLVRSLLRDYAGKQQHEGMLMNLISHNRQS 1427
RGSV+YT+ VHH SS +F DK+ LRN +V++L+RD + K+ EGM+++L+ + + S
Sbjct: 1074 RGSVAYTLVVHHFSSLVFCNQAKDKVSLRNKIVKTLVRDLSRKRHREGMMLDLLRYKKGS 1133
Query: 1428 TSNMDEQLDGLLHEESWLESRMKVLIEACEGNSSLLIQVKKLKDAA 1473
+ M+E++ + E RM+VL E CEGNS+LL++++KLK AA
Sbjct: 1134 ANAMEEEVIA-----AETEKRMEVLKEGCEGNSTLLLELEKLKSAA 1174
>UniRef100_Q5Z7K3 Hypothetical protein OSJNBa0006A22.41 [Oryza sativa]
Length = 1487
Score = 905 bits (2339), Expect = 0.0
Identities = 566/1520 (37%), Positives = 829/1520 (54%), Gaps = 225/1520 (14%)
Query: 24 GKWREKKTKGMDFGKWREFTQDD---KSFLGKDLE-KDVSSYGPTTGRKKNENGGKNTSK 79
G + K+ KGMDF +WREF DD K K L+ K ++ TG GG K
Sbjct: 125 GPVKRKEKKGMDFSRWREFVADDAPPKRRQAKPLQPKKQTAQKIDTGVVAATTGGTAQEK 184
Query: 80 KISSYSDGSVFASMEV-DAKPQLVKLDGGFINSATSMELDTSNKDDKKEVFAAERDKIFS 138
+ G + +EV + K +L GG ++ D + + K+V A RD + +
Sbjct: 185 R-----SGGIGMQLEVGNGKEEL----GG-----AALMSDVAPRKPMKQVDA--RDDVRN 228
Query: 139 DRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALL 198
+ S+ SL EI++EN AR+ MS EI EA+A+I+ ++ PA +
Sbjct: 229 VELRGEGMESDNG-------EPSLTAEINAENMARLAGMSAGEIAEAQAEILNRMDPAFV 281
Query: 199 KVLQKRGKEKL-KKPNSLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQ 257
++L++RGKEK + + K + G + S ++ + L + HT
Sbjct: 282 EMLKRRGKEKSGSRKDGGKGKGGGI--SGPGKISKAMPGEWLSAGEHSGHT--------- 330
Query: 258 LDDKNVSGKTSTTTSSSSWNAWSNRVEAIRELRFSLAGDVVD----TEQEPVY------- 306
W AWS RVE IR RF+L GD++ EQ+ V+
Sbjct: 331 ------------------WKAWSERVERIRSCRFTLEGDILGFQSCQEQQHVFWYPLHVN 372
Query: 307 ------------DNIAERDYLRTEGDPGAAGYTIKEALEITRSV----RALGLHLLSSVL 350
+ + ERD+LRTEGDP A GYTI EA+ ++RS+ R L L LL+ +L
Sbjct: 373 LAFPLTGKKAHVETVGERDFLRTEGDPAAVGYTINEAVALSRSMVPGQRVLALQLLALIL 432
Query: 351 DKALCYICKDRTENMTKKGNKVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLA 410
++AL + K + K+ N DK DW+AVW YA+GP+PEL LSLRM LDDNH+SVVL
Sbjct: 433 NRALQNLHKTDLIDNFKESNDDDKFNDWQAVWAYAIGPEPELVLSLRMSLDDNHDSVVLT 492
Query: 411 CAKVVQSALSCDVNENYFDISENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPS 470
CAKV+ + LS ++NE YFD+ E + KDICTAPVFRS+PD + GFL+GG+WKY+ KPS
Sbjct: 493 CAKVINAMLSYEMNEMYFDVLEKVVDQGKDICTAPVFRSKPDQNGGFLEGGFWKYNTKPS 552
Query: 471 NIQPFSEDSMDNESDDKHTIQDDVFVAGQDFTAGLVRMGILPRLRYLLETDPTAALEECI 530
NI P ++ + E D+KHTIQDDV V+GQD AGLVRMGILPR+ +LLE DP LE+ +
Sbjct: 553 NILPHYGENDEEEGDEKHTIQDDVVVSGQDVAAGLVRMGILPRICFLLEMDPHPILEDNL 612
Query: 531 VSILIAIVRHSPSCANAVLKCERLIQTIVQRFT-VGNFEIRSSMIKSVKLLKVLARLDRK 589
VSIL+ + RHSP A+A+L C RL+Q++V+ G+ EI SS IK V LLKVL++ +R+
Sbjct: 613 VSILLGLARHSPQSADAILNCPRLVQSVVKLLVKQGSMEIHSSQIKGVNLLKVLSKYNRQ 672
Query: 590 TCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCV 649
TC F+ G F+ W +C
Sbjct: 673 TCFNFVNTGVFHQAMW-----------------------------------------HCP 691
Query: 650 SHFSKIFPALCFWLDLPSFEKLTKNNVLNESTCISREAYLVLESLAERLRNLFSQQCLTN 709
S F +KL+++NV+ E + I+ E+YLVL +LA+RL L S + L+
Sbjct: 692 SMF----------------QKLSESNVVAEFSSIATESYLVLGALAQRLPLLHSVEQLSK 735
Query: 710 QHPE-STDDAEFWSWSYVGPMVDLAIKWIARRSDPEVYKLFEGQEEGVNHFTLGDLSSTP 768
Q S E WSWS+ PMVDLA+ W+ P V L GQ + + L +
Sbjct: 736 QDMGLSGIQVETWSWSHAVPMVDLALSWLCLNDIPYVCLLISGQSKNI-------LEGSY 788
Query: 769 LLWVYAAVTHMLFRVLEKVTLGDAISLQEANGH-VPWLPKFVPKIGLELINYWHLGFSVA 827
V ++V ML +LE+++ S + + +PW+P FVPKIGL +I F
Sbjct: 789 FALVISSVLGMLDSILERIS---PDSTHDGKSYCLPWIPDFVPKIGLGVITNGFFNFLDD 845
Query: 828 SVTKSGRDSG--DESFMKELIHLRQKGDIEMSLASTCCLNGIINVITKIDNLIRSAKTGI 885
+ + + + S ++ L HLR +G+++ SL S C ++ + ID +I++A T
Sbjct: 846 NAVELEQHTSFHGSSLVQGLFHLRSQGNVDTSLCSISCFQRLLQLSCSIDRVIQNATTN- 904
Query: 886 CNPPVTEQSLSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGPAP 945
C + E G++LE+GI + L ML SS W +Q+IE+FGRGGPAP
Sbjct: 905 CTEHLKESKTGIAGRILEQGICNFWRNNLLDMLTSLLPMISSQWSILQNIEMFGRGGPAP 964
Query: 946 GMGVGWGAHGGGFWSKTVLPVKTDARLLVCLLQIFEN------TSNDAPE---------T 990
G+G GWGA+GGGFWS L + D+ ++ L++I T N + T
Sbjct: 965 GVGFGWGAYGGGFWSLNFLLAQLDSHFVLELMKILSTGPEGLVTVNKSVNPIVQEGNNVT 1024
Query: 991 EQMTFSMQQVNTALGLCLTAGPADMVVIEKTLDLLFHVSILKYLDLCIQNFLLNRRGKAF 1050
+ + + +++++ L + L AGP + +EK D+LFH S+LK+L + + + + KAF
Sbjct: 1025 DSVAITSERISSVLSVSLMAGPGQISTLEKAFDILFHPSVLKFLKSSVLDSHM-KLAKAF 1083
Query: 1051 GWKYEDDDYMHFSRMLSSHFRSRWLSVRVKSKAVDGSSSSGVKATPKADVRLDTIYEDSD 1110
W +D+Y+HFS +L+SHFRSRWL ++ K +++G PK L+TI E+++
Sbjct: 1084 EWDITEDEYLHFSSVLNSHFRSRWLVIKKKHSDEFTRNNNGTN-VPKIPETLETIQEETE 1142
Query: 1111 MSSTTSPCCNSLMIEWARQNLPLPVHFYLSPISTIPLTKRAGPQKVGSVHNPHDPANL-- 1168
++ +P C+ L +EWA Q LPLPVH+ LS + I K ANL
Sbjct: 1143 LAEAVNPPCSVLAVEWAHQRLPLPVHWILSAVCCIDDPK----------------ANLST 1186
Query: 1169 ---LEVAKCGLFFVLGIETMSSFIGTGIPSPIQRVSLTWKLHSLSVNFLVGMEILEQDQG 1225
++V+K GLFF+LG+E +S+ +P L WK+H+LS + M++L +D+
Sbjct: 1187 SYAVDVSKAGLFFLLGLEAISA-------APCLHAPLVWKMHALSASIRSSMDLLLEDRS 1239
Query: 1226 RETFEALQDLYGELLDK--------ERFNQNKEAISDDKK--HIEFLRFKSDIHESYSTF 1275
R+ F ALQ+LYG LD+ + A D++K E LRF+ IH +Y+TF
Sbjct: 1240 RDIFHALQELYGLHLDRLCQKYDSAHSVKKEGSASVDEEKVTRTEVLRFQEKIHANYTTF 1299
Query: 1276 IEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNARVLELLPPLEKCFSG 1335
+E L+EQF+++SYGD +FGRQV++YLH VE +IRLA WN LSNA VLELLPPL+KC
Sbjct: 1300 VESLIEQFAAVSYGDALFGRQVAIYLHRSVEPTIRLAAWNALSNAYVLELLPPLDKCVGD 1359
Query: 1336 AEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACPVDKLLL 1395
+GYLEP ED+E ILE+YAKSW S ALD+A R ++S+T+A HHLS F+F K +
Sbjct: 1360 VQGYLEPLEDDEGILESYAKSWTSGALDKAFQRDAMSFTVARHHLSGFVFQCSGSGK--V 1417
Query: 1396 RNNLVRSLLRDYAGKQQHEGMLMNLISHNRQSTSNMDEQLDGLLHEESWLESRMKVLIEA 1455
RN LV+SL+R Y K+ HE ML + S +++ + R +++ +A
Sbjct: 1418 RNKLVKSLIRCYGQKRHHEDMLKGFVLQGIAQDSQRNDE----------VSRRFEIMKDA 1467
Query: 1456 CEGNSSLLIQVKKLKDAAEK 1475
CE NSSLL +V++LK + ++
Sbjct: 1468 CEMNSSLLAEVRRLKTSIDR 1487
>UniRef100_Q9P2E4 KIAA1403 protein [Homo sapiens]
Length = 1337
Score = 96.7 bits (239), Expect = 5e-18
Identities = 120/552 (21%), Positives = 220/552 (39%), Gaps = 114/552 (20%)
Query: 154 MHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPN 213
+ +QE+ I EN AR+Q M+ EEI + + ++ ++ P+L+ L+ +
Sbjct: 237 LRDQEAEQEAQTIHEENIARLQAMAPEEILQEQQRLLAQLDPSLVAFLRSH--------S 288
Query: 214 SLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGKTSTTTSS 273
+ + G E+ +++ + + ++ E+ + + P K+ + T
Sbjct: 289 HTQEQTG---ETASEEQRPGGPSANVTKEEPLMSAFASEPRKRDKLEPEAPALALPVTPQ 345
Query: 274 SSWNA----------WSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYL 315
W W+ + +R + RFSL G+++ + + + L
Sbjct: 346 KEWLHMDTVELEKLHWTQDLPPVRRQQTQERMQARFSLQGELLAPDVD-----LPTHLGL 400
Query: 316 RTEGDPGA-AGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGN 370
G+ AGY+++E +TRS RAL LH+L+ V+ +A DR G+
Sbjct: 401 HHHGEEAERAGYSLQELFHLTRSQVSQQRALALHVLAQVISRAQAGEFGDRLA-----GS 455
Query: 371 KVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDI 430
+ +D ++ LR LDD + V+ + +++ L +E D
Sbjct: 456 VLSLLLDAGFLFL------------LRFSLDDRVDGVIATAIRALRALLVAPGDEELLD- 502
Query: 431 SENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDK--- 487
+T+ W + A + P ED D + D++
Sbjct: 503 ----STFS------------------------WYHGALTFPLMPSQEDKEDEDKDEECPA 534
Query: 488 ---------HTIQDDVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAI 537
+ +A D GL+ +LPRLRY+LE T P A+ I+++LI +
Sbjct: 535 GKAKRKSPEEESRPPPDLARHDVIKGLLATSLLPRLRYVLEVTYPGPAVVLDILAVLIRL 594
Query: 538 VRHSPSCANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLD 587
RHS A VL+C RLI+TIV+ F ++ + ++KLL+VLA
Sbjct: 595 ARHSLESATRVLECPRLIETIVREFLPTSWSPVGAGPTPSLYKVPCATAMKLLRVLASAG 654
Query: 588 RKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGY 647
R + + + L ++ L L E+ ++ L+ E LR W V YG
Sbjct: 655 RNIAARLLSSFDLRS---RLCRIIAEAPQELALPPEEAEM---LSTEALRLWAVAASYGQ 708
Query: 648 CVSHFSKIFPAL 659
+ +++P L
Sbjct: 709 GGYLYRELYPVL 720
>UniRef100_Q9BWH6 DKFZP727M111 protein [Homo sapiens]
Length = 1393
Score = 96.7 bits (239), Expect = 5e-18
Identities = 120/552 (21%), Positives = 220/552 (39%), Gaps = 114/552 (20%)
Query: 154 MHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPN 213
+ +QE+ I EN AR+Q M+ EEI + + ++ ++ P+L+ L+ +
Sbjct: 215 LRDQEAEQEAQTIHEENIARLQAMAPEEILQEQQRLLAQLDPSLVAFLRSH--------S 266
Query: 214 SLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGKTSTTTSS 273
+ + G E+ +++ + + ++ E+ + + P K+ + T
Sbjct: 267 HTQEQTG---ETASEEQRPGGPSANVTKEEPLMSAFASEPRKRDKLEPEAPALALPVTPQ 323
Query: 274 SSWNA----------WSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYL 315
W W+ + +R + RFSL G+++ + + + L
Sbjct: 324 KEWLHMDTVELEKLHWTQDLPPVRRQQTQERMQARFSLQGELLAPDVD-----LPTHLGL 378
Query: 316 RTEGDPGA-AGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGN 370
G+ AGY+++E +TRS RAL LH+L+ V+ +A DR G+
Sbjct: 379 HHHGEEAERAGYSLQELFHLTRSQVSQQRALALHVLAQVISRAQAGEFGDRLA-----GS 433
Query: 371 KVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDI 430
+ +D ++ LR LDD + V+ + +++ L +E D
Sbjct: 434 VLSLLLDAGFLFL------------LRFSLDDRVDGVIATAIRALRALLVAPGDEELLD- 480
Query: 431 SENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDK--- 487
+T+ W + A + P ED D + D++
Sbjct: 481 ----STFS------------------------WYHGALTFPLMPSQEDKEDEDKDEECPA 512
Query: 488 ---------HTIQDDVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAI 537
+ +A D GL+ +LPRLRY+LE T P A+ I+++LI +
Sbjct: 513 GKAKRKSPEEESRPPPDLARHDVIKGLLATSLLPRLRYVLEVTYPGPAVVLDILAVLIRL 572
Query: 538 VRHSPSCANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLD 587
RHS A VL+C RLI+TIV+ F ++ + ++KLL+VLA
Sbjct: 573 ARHSLESATRVLECPRLIETIVREFLPTSWSPVGAGPTPSLYKVPCATAMKLLRVLASAG 632
Query: 588 RKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGY 647
R + + + L ++ L L E+ ++ L+ E LR W V YG
Sbjct: 633 RNIAARLLSSFDLRS---RLCRIIAEAPQELALPPEEAEM---LSTEALRLWAVAASYGQ 686
Query: 648 CVSHFSKIFPAL 659
+ +++P L
Sbjct: 687 GGYLYRELYPVL 698
Score = 52.8 bits (125), Expect = 8e-05
Identities = 46/148 (31%), Positives = 71/148 (47%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNA-RVLELLPPL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + A R L L PL
Sbjct: 1206 SFPDLYANFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFGEHVGALRALSL--PL 1263
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y P EDN +L+ Y ++ V+ AL V Y +AV H++SFIF+ P
Sbjct: 1264 TQLPVSLECYTVPPEDNLALLQLYFRTLVTGALRPRWC--PVLYAVAVAHVNSFIFSQDP 1321
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L ++ L G +QH
Sbjct: 1322 QSSDEVKAARRSMLQKTWLLADEGLRQH 1349
>UniRef100_UPI000036A014 UPI000036A014 UniRef100 entry
Length = 1013
Score = 95.5 bits (236), Expect = 1e-17
Identities = 121/552 (21%), Positives = 213/552 (37%), Gaps = 114/552 (20%)
Query: 154 MHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPN 213
+ +QE+ I EN AR+Q M+ EEI + + ++ ++ P+L+ L+ + + +
Sbjct: 215 LRDQEAEQEAQTIHEENIARLQAMAPEEILQEQQRLLAQLDPSLVAFLRSHSRTQEQTGE 274
Query: 214 SLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGKTSTTTSS 273
+ E S N + E+ + + P K + T
Sbjct: 275 TASEEQRPGGPSAN-----------VTKEEPLMSAFASEPRKGDKLEPEAPALALPVTPQ 323
Query: 274 SSWNA----------WSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYL 315
W W+ + +R + RFSL G+++ + + + L
Sbjct: 324 KEWLHMDTVELEKLHWTQDLPPVRRQQTQERMQARFSLQGELLAPDAD-----LPTHLGL 378
Query: 316 RTEGDPGA-AGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGN 370
G+ AGY+++E +TRS RAL LH+L+ V+ +A DR G+
Sbjct: 379 HHHGEEAERAGYSLQELFHLTRSQVSQQRALALHVLAQVISRAQAGEFGDRLA-----GS 433
Query: 371 KVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDI 430
+ +D ++ LR LDD + V+ + +++ L +E D
Sbjct: 434 VLSLLLDAGFLFL------------LRFSLDDRVDGVIATAIRALRALLVAPGDEELLD- 480
Query: 431 SENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDK--- 487
+T+ W + A + P ED D + D++
Sbjct: 481 ----STFS------------------------WYHGALTFPLMPSQEDKEDEDEDEECPA 512
Query: 488 ---------HTIQDDVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAI 537
+ +A D GL+ +LPRLRY+LE T P A+ I+++LI +
Sbjct: 513 GNAKRKSPEEESRPPPDLARHDVIKGLLATSLLPRLRYVLEVTYPGPAVVLDILAVLIRL 572
Query: 538 VRHSPSCANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLD 587
RHS A VL+C RLI+TIV+ F ++ + ++KLL+VLA
Sbjct: 573 ARHSLESATRVLECPRLIETIVREFLPTSWSPVGAGPTPSLYKVPCATAMKLLRVLASAG 632
Query: 588 RKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGY 647
R + + + L + L L E+ ++ L+ E LR W V YG
Sbjct: 633 RNIAARLLSSFDLRS---RLCRFIAEAPQELALPPEEAEM---LSTEALRLWAVAASYGQ 686
Query: 648 CVSHFSKIFPAL 659
+ +++P L
Sbjct: 687 GGYLYRELYPVL 698
>UniRef100_Q5R8S6 Hypothetical protein DKFZp469H2113 [Pongo pygmaeus]
Length = 1074
Score = 94.4 bits (233), Expect = 2e-17
Identities = 122/552 (22%), Positives = 211/552 (38%), Gaps = 114/552 (20%)
Query: 154 MHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKLKKPN 213
+ +QE+ I EN AR+Q M+ EEI + + ++ ++ P+L+ L+ + + +
Sbjct: 215 LRDQEAEQEAQTIHEENIARLQAMAPEEILQEQQRLLAQLDPSLVAFLRSHSRTQEQTGE 274
Query: 214 SLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGKTSTTTSS 273
+ E S N + E+ + + P K D T
Sbjct: 275 TASEEQRPGGPSAN-----------VTKEEPLMSAFASEPRKGDKLDSEAPALALPVTPQ 323
Query: 274 SSWNA----------WSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYL 315
W W+ + ++ + RFSL G+++ + + + L
Sbjct: 324 KEWLHMDTVELEKLHWTQDLPPVQRRQTQERMQARFSLQGELLAPDVD-----LPTHLGL 378
Query: 316 RTEGDPGA-AGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGN 370
G+ AGY+ +E +TRS RAL LH+L+ V+ +A DR G+
Sbjct: 379 HHHGEEAERAGYSPQELFHLTRSQVSQQRALALHVLAQVISRAQAGEFGDRLA-----GS 433
Query: 371 KVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDI 430
+ +D ++ LR LDD + V+ + +++ L +E D
Sbjct: 434 VLSLLLDAGFLFL------------LRFSLDDRVDGVIATAIRALRALLVAPGDEELLD- 480
Query: 431 SENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDKHTI 490
+T+ W + A + P ED D + D++ T
Sbjct: 481 ----STFS------------------------WYHGALTFPLMPSQEDKEDEDEDEECTA 512
Query: 491 ------------QDDVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAI 537
+ +A D GL+ +LPRLRY+LE T P A+ I+++LI +
Sbjct: 513 GKAKRKSPEEESRPPPDLARHDVIKGLLATSLLPRLRYVLEVTYPGPAVVLDILAVLIRL 572
Query: 538 VRHSPSCANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLD 587
RHS A VL+C RLI+TIV+ F ++ + ++KLL+VLA
Sbjct: 573 ARHSLESATRVLECPRLIETIVREFLPTSWSPVGAGPTPSLYKVPCATAMKLLRVLASAG 632
Query: 588 RKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGY 647
R + + + L L L E+ + L+ E LR W V YG
Sbjct: 633 RNIAARLLSSFDLRS---RLCHFIAEAPQELALPPEEAE---TLSTEALRLWAVAASYGQ 686
Query: 648 CVSHFSKIFPAL 659
+ +++P L
Sbjct: 687 GGHLYRELYPVL 698
>UniRef100_UPI00001CF220 UPI00001CF220 UniRef100 entry
Length = 1355
Score = 88.6 bits (218), Expect = 1e-15
Identities = 124/549 (22%), Positives = 212/549 (38%), Gaps = 100/549 (18%)
Query: 154 MHEQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQK----RGKEKL 209
+ Q + I EN AR+Q M EEI + + ++ ++ P+L+ L+ R + +
Sbjct: 232 LRSQAAVQEVQTIHEENVARLQAMDPEEILKEQQQLLAQLDPSLVAFLRAHNHTREQTET 291
Query: 210 KKPNSLKSEVGAVTESVNQQVQIT---QGAKHLQTEDDISHTIMA-PPSKKQLDDKNVSG 265
K E +V S + + T + + ED + + P+ K N
Sbjct: 292 KATKEQNPERPSVPVSKEEPIMSTCTGESGTRDKLEDKLEDKLQPRTPALKLPMTPNKEW 351
Query: 266 KTSTTTSSSSWNAWSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYLRT 317
T + W+ + +R + RFSL G+++ EP D
Sbjct: 352 LHMDTVELEKLH-WTQDLPPLRRQQTQERMQARFSLQGELL----EPDVDLPTHLGLHHH 406
Query: 318 EGDPGAAGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGNKVD 373
+ AGY+++E +TRS RAL LH+LS ++ +A DR +
Sbjct: 407 GEEAERAGYSLQELFHLTRSQVSQQRALALHVLSHIVGRAQAGEFGDRLVGSVLR----- 461
Query: 374 KSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDISEN 433
L LR LDD +SV+ A + +++ L +E D
Sbjct: 462 ------------LLLDAGFLFLLRFSLDDRIDSVIAAAVRALRALLVAPGDEELLD---- 505
Query: 434 MATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDKHTIQD- 492
+T+ W + A + P +D D + D++ T +
Sbjct: 506 -STFS------------------------WYHGASVFPMMPSHDDKEDEDEDEELTKEKV 540
Query: 493 -----------DVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAIVRH 540
+A D GL+ +LPR RY+LE T P ++ I+++LI + RH
Sbjct: 541 NRKTPEEGSRPPPDLARHDVIKGLLATNLLPRFRYVLEVTCPGPSVVLDILAVLIRLARH 600
Query: 541 SPSCANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLDRKT 590
S A VL+C RL++TIV+ F ++ + ++KLL+VLA R
Sbjct: 601 SLESAMRVLECPRLMETIVREFLPTSWSPIGVGPAPSLYKVPCAAAMKLLRVLASAGRNI 660
Query: 591 CLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCVS 650
+ + F+ + L + L L E+ ++ LT E R W V YG
Sbjct: 661 AARLLSS--FDVRS-RLCRFIAEAPRDLALPFEEAEI---LTTEAFRLWAVAASYGQGGD 714
Query: 651 HFSKIFPAL 659
+ +++P L
Sbjct: 715 LYRELYPVL 723
Score = 40.8 bits (94), Expect = 0.32
Identities = 25/63 (39%), Positives = 37/63 (58%), Gaps = 2/63 (3%)
Query: 1323 LELLPPLEKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSS 1382
L +L PL + E Y EPAED+ +L+ Y ++ V+ AL V YT+AV H++S
Sbjct: 1219 LSILFPLSQLPVPLECYTEPAEDSLALLQLYFRALVTGALHARWC--PVLYTVAVAHVNS 1276
Query: 1383 FIF 1385
F+F
Sbjct: 1277 FVF 1279
>UniRef100_Q80TE0 MKIAA1403 protein [Mus musculus]
Length = 1444
Score = 87.4 bits (215), Expect = 3e-15
Identities = 123/546 (22%), Positives = 208/546 (37%), Gaps = 110/546 (20%)
Query: 166 IDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEK----LKKPNSLKSEVGA 221
I EN AR+Q M EEI + + ++ ++ P+L+ L+ + + K E +
Sbjct: 262 IHEENVARLQAMDPEEILKEQQQLLAQLDPSLVAFLRSHSQVQEQTGTKATKKQSPERPS 321
Query: 222 VTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKKQLD--DKNVSGKTSTTTSSSSWNA- 278
V + + V T+ + +T D + A K D T S W
Sbjct: 322 VLVTKEEPVTSTR-TREPRTGDKLEEKPEATVEDKMEDKLQPRTPALKLPMTPSKDWLHM 380
Query: 279 ---------WSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDYLRTEGDP 321
W+ + +R + RFSL G+++ + + + L G+
Sbjct: 381 DTVELDKLHWTQDLPPLRRQQTQERMQARFSLQGELLAPDVD-----LPTHLGLHHHGEE 435
Query: 322 GA-AGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTKKGNKVDKSV 376
AGY+++E +TRS RAL L +LS ++ +A DR +
Sbjct: 436 AERAGYSLQELFHLTRSQVSQQRALALQVLSQIVGRAQAGEFGDRLVGSVLR-------- 487
Query: 377 DWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENYFDISENMAT 436
L LR LDD +SV+ A + +++ L +E D + +
Sbjct: 488 ---------LLLDAGFLFLLRFSLDDRVDSVIAAAVRALRTLLVAPGDEELLDRTFS--- 535
Query: 437 YDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDKHTIQD---- 492
W + A + P +D D + D++ T +
Sbjct: 536 --------------------------WYHGASVFPLMPSQDDKEDEDEDEELTTEKVKRK 569
Query: 493 --------DVFVAGQDFTAGLVRMGILPRLRYLLE-TDPTAALEECIVSILIAIVRHSPS 543
+A D GL+ +LPRLRY+LE T P ++ I+++LI + RHS
Sbjct: 570 TPEEGSRPPPDLARHDVIKGLLATNLLPRLRYVLEVTCPGPSVVLDILAVLIRLARHSLE 629
Query: 544 CANAVLKCERLIQTIVQRFTVGNFE----------IRSSMIKSVKLLKVLARLDRKTCLE 593
A VL+C RL++TIVQ F ++ + ++KLL+VLA R
Sbjct: 630 SAMRVLECPRLMETIVQEFLPTSWSPIGVGPTPSLYKVPCASAMKLLRVLASAGRNIAAR 689
Query: 594 FIKNGYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCVSHFS 653
+ F+ + L + L L E+ ++ LT E R W V YG +
Sbjct: 690 LLSG--FDVRS-RLCRFIAEAPHDLALPPEEAEI---LTTEAFRLWAVAASYGQGGDLYR 743
Query: 654 KIFPAL 659
+++P L
Sbjct: 744 ELYPVL 749
Score = 54.3 bits (129), Expect = 3e-05
Identities = 48/148 (32%), Positives = 75/148 (50%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNARVLELLP-PL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + + VL L PL
Sbjct: 1257 SFPDLYASFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFG--EHVGVLRALGLPL 1314
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y EPAED+ +L+ Y ++ V+ +L RA + YT+AV H++SFIF P
Sbjct: 1315 TQLPVPLECYTEPAEDSLPLLQLYFRALVTGSL-RAR-WCPILYTVAVAHVNSFIFCQDP 1372
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L R+ L G +QH
Sbjct: 1373 KSSDEVKTARRSMLQRTWLLTDEGLRQH 1400
>UniRef100_UPI0000364055 UPI0000364055 UniRef100 entry
Length = 1381
Score = 76.3 bits (186), Expect = 7e-12
Identities = 123/542 (22%), Positives = 217/542 (39%), Gaps = 92/542 (16%)
Query: 156 EQESTSLENEIDSENRARIQQMSTEEIEEAKADIMEKISPALLKVLQKRGKEKL------ 209
+ +ST I EN+A++Q MS EI + + ++ ++ P L+K ++ + + +
Sbjct: 210 DPDSTGETQRIHRENQAKLQGMSQSEILDEQKKLLSQLDPRLVKFIRSQKAQSVPSSTSP 269
Query: 210 -KKPNSLKSEVGAVTESVNQQVQITQGAKHLQTE----DDISHTIMAPPS--KKQLDDKN 262
K P S E+V++Q A Q + + PP+ ++ L +
Sbjct: 270 SKLPRGGSSRENIPLENVSKQSDSLSAAPEPQEVTMEGEGEEEGVPRPPAVMEEVLPVRP 329
Query: 263 VSGKTSTTTSSSSWNAWSNRVEAIR--------ELRFSLAGDVVDTEQEPVYDNIAERDY 314
W + A R + RF+ AG ++ P +++
Sbjct: 330 QKDWVHMDKVEPEKLEWMRDLPAPRKKGTKKAMQARFNFAGTLI-----PPNEDLPTHLG 384
Query: 315 LRTEGD-PGAAGYTIKEALEITRSV----RALGLHLLSSVLDKALCYICKDRTENMTK-- 367
L G+ P AGY+++E ++RS R L L L+++L KA R E T
Sbjct: 385 LHHHGEEPERAGYSLQELFMLSRSQFNQQRMLALSTLANILSKA-------RAEGYTSVL 437
Query: 368 KGNKVDKSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVVQSALSCDVNENY 427
KG+ + +D L LR LDD+ V+ A +++ L C +E
Sbjct: 438 KGSVLSVLLD------------AGLLFLLRFALDDSVEGVMSAAVHALRAFLVCAEDEEC 485
Query: 428 FDISENMATYDKDICTAPVFRSRPDISLGFLQGGYWKYSAKPSNIQPFSEDSMDNESDDK 487
D ++ + + + P+ S D +G P N++ + ++
Sbjct: 486 LD---GTFSWFRGLASFPLLPSAQDDEDEEDEG-------VPENLR--------ESAKER 527
Query: 488 HTIQDDVFVAGQDFTAGLVRMGILPRLRYLLETDPTAALEECIVSILIAIVRHSPSCANA 547
+ D VA QD +V+ LR ++ P + + IL RHS S A
Sbjct: 528 EARKSDYDVARQD----VVKAKTEVPLRSIVFHPPPRVQD--VXEILTRXARHSSSSATQ 581
Query: 548 VLKCERLIQTIVQRF-----TVGNFEIRSS-----MIKSVKLLKVLARLDRKTCLEFIKN 597
VL C RL++T++ F T + R S + ++KLL+V+A R C + +
Sbjct: 582 VLDCPRLMETLLSEFFPTSWTAPSLTPRQSAYGFPLASAMKLLRVVATSGRHACARLLNS 641
Query: 598 GYFNAMTWNLYQLPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCVSHFSKIFP 657
+T L + LSI+ L +E ++ +T E R W V YG F +++P
Sbjct: 642 ---FGVTERLSHV-LSIEPSELLLEESEAVR--ITAEAYRLWAVAAGYGQACKLFIELYP 695
Query: 658 AL 659
+L
Sbjct: 696 SL 697
Score = 45.1 bits (105), Expect = 0.017
Identities = 32/116 (27%), Positives = 60/116 (51%), Gaps = 3/116 (2%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNARVLELLPPLE 1330
S+ L+ Q+ ++S+GD +FG + + L ++++LA + + L +E
Sbjct: 1207 SFQDLYLALLTQYEAVSFGDRLFGCWLLLPLQRRYSATMKLAVFGE-HVGMLRSLGVTIE 1265
Query: 1331 KCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFN 1386
+ E + PAED+ +L Y +S V+ L A R V Y +A+ H+++FIF+
Sbjct: 1266 QLSIPIERFTSPAEDSLPLLRLYFRSLVTGTL--APRRCPVLYVVALSHVNAFIFS 1319
>UniRef100_UPI0000439692 UPI0000439692 UniRef100 entry
Length = 1027
Score = 74.7 bits (182), Expect = 2e-11
Identities = 107/431 (24%), Positives = 176/431 (40%), Gaps = 79/431 (18%)
Query: 288 ELRFSLAGDVVDTEQEPVYDNIAERDYLRTEGD-PGAAGYTIKEALEITRSV----RALG 342
+ RF AG ++ P ++ L GD P AGY+++E ++RS R L
Sbjct: 2 QARFDFAGTLI-----PPTKDLPTHLGLHHHGDEPELAGYSLQELFLLSRSKLNQQRNLA 56
Query: 343 LHLLSSVLDKALCYICKDRTENMTKKGNKVDKSVDWEAVWTYALGPQPELALSLRM*LDD 402
+ L++VL KA + + K + + +D L LR LDD
Sbjct: 57 ISTLANVLTKA-----RAGEFIFSLKSSVLSSLLD------------AGLLFLLRFSLDD 99
Query: 403 NHNSVVLACAKVVQSALSCDVNENYFDISENMATYDKDICTAPVFRSRPDISLGFLQGGY 462
+V+ A +++ L +E+ +N ++ + P+ + +
Sbjct: 100 GVEAVMSAAVHALRALLVSSEDESL----DNTFSWLLGMANFPLLPAAEEDE-------- 147
Query: 463 WKYSAKPSNIQPFSEDSMDNESDDKHTIQDDVFVAGQDFTAGLVRMGILPRLRYLLE-TD 521
+ +++ NE ++K T D VA QD G +RM LPRLRY+LE
Sbjct: 148 ---DEDDEGLDEVMKETA-NEKEEKKTDHD---VARQDVVKGFLRMQGLPRLRYILEVVR 200
Query: 522 PTAALEECIVSILIAIVRHSPSCANAVLKCERLIQTIVQRFTVGNFEIRSSMIKS----- 576
P+ + I+ +LI I RHS + A +L C RL+ T++ F ++ SS S
Sbjct: 201 PSPGVVLDILEVLIRIARHSTASAVQILDCPRLMDTVISEFLPCSWAPASSQAPSSLYGL 260
Query: 577 -----VKLLKVLARLDRKTCLEFIKNGYFNAMTWNLYQLPLSIDDWLKLGKE-KCKLKSA 630
+KLL+VLA R C I N + + + + L+ G+ +C L
Sbjct: 261 PVSLAMKLLRVLASAGRHICAR-ILNSLRGKELLSRFVVVEPSEMLLEEGEALRCSL--- 316
Query: 631 LTIEQLRFWRVCIRYGYCVSHFSKIFPALCFWLDLPSFEKLTKNNVLNESTCISREAYLV 690
E LR + V YG + ++ ++P L K + C S EA L
Sbjct: 317 ---EALRLFAVAASYGQACNLYTDLYPV------------LVKRLQAGQQCCSSSEAVLR 361
Query: 691 LESLAERLRNL 701
LE +R+R L
Sbjct: 362 LE--LDRIRAL 370
Score = 47.0 bits (110), Expect = 0.004
Identities = 51/181 (28%), Positives = 84/181 (46%), Gaps = 21/181 (11%)
Query: 1266 SDIHESYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWN-----TLSNA 1320
+ H+ YS L+ Q+ ++S+GD +FG V + L +++LA + S
Sbjct: 849 ASFHDLYSA----LLAQYEAVSFGDPLFGCFVLLPLQRRYSVTMKLAVFGEHVGMLRSLG 904
Query: 1321 RVLELLP-PLEKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHH 1379
L+ LP PLEK + P ED+ +L Y ++ V+ AL R V Y +AV H
Sbjct: 905 VSLQQLPIPLEK-------FTSPMEDSLPLLRLYFRALVTGALKRNWC--PVLYVVAVAH 955
Query: 1380 LSSFIF--NACPVDKLLLRNNLVRSLLRDYAGKQQHEGMLMNLISHNRQSTSNMDEQLDG 1437
L++FIF +A + L R NL++ ++ +L L N + + EQL
Sbjct: 956 LNAFIFSQDAVTEEVDLARRNLLQKTFYLTDEVLKNHLLLFRLPQQNSELGFSTYEQLPS 1015
Query: 1438 L 1438
+
Sbjct: 1016 I 1016
>UniRef100_Q91VM0 Rpap1 protein [Mus musculus]
Length = 295
Score = 54.3 bits (129), Expect = 3e-05
Identities = 48/148 (32%), Positives = 75/148 (50%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNARVLELLP-PL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + + VL L PL
Sbjct: 108 SFPDLYASFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFG--EHVGVLRALGLPL 165
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y EPAED+ +L+ Y ++ V+ +L RA + YT+AV H++SFIF P
Sbjct: 166 TQLPVPLECYTEPAEDSLPLLQLYFRALVTGSL-RAR-WCPILYTVAVAHVNSFIFCQDP 223
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L R+ L G +QH
Sbjct: 224 KSSDEVKTARRSMLQRTWLLTDEGLRQH 251
>UniRef100_Q80UT8 Rpap1 protein [Mus musculus]
Length = 799
Score = 54.3 bits (129), Expect = 3e-05
Identities = 48/148 (32%), Positives = 75/148 (50%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNARVLELLP-PL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + + VL L PL
Sbjct: 612 SFPDLYASFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFG--EHVGVLRALGLPL 669
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y EPAED+ +L+ Y ++ V+ +L RA + YT+AV H++SFIF P
Sbjct: 670 TQLPVPLECYTEPAEDSLPLLQLYFRALVTGSL-RAR-WCPILYTVAVAHVNSFIFCQDP 727
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L R+ L G +QH
Sbjct: 728 KSSDEVKTARRSMLQRTWLLTDEGLRQH 755
>UniRef100_UPI00004578C3 UPI00004578C3 UniRef100 entry
Length = 2486
Score = 52.8 bits (125), Expect = 8e-05
Identities = 62/278 (22%), Positives = 109/278 (38%), Gaps = 42/278 (15%)
Query: 15 RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGG 74
+ T G DF + K K K + TQ + S +LEK++ S
Sbjct: 776 KSSTSGSDFDTKKGKSAKSSIISKKKRQTQSESSNYDSELEKEIKSMSKI-------GAA 828
Query: 75 KNTSKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSATSME--LDTSNKDDKKEVFAAE 132
+ T K+I + D F S E + + + G N TS E D + + ++E F++
Sbjct: 829 RTTKKRIPNTKD---FDSSEDEKHSKKGMDNQGHKNLKTSQEGSSDDAERKQERETFSSA 885
Query: 133 -------------RDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMST 179
RD++ + S+ EQ TSLE +E + + + + T
Sbjct: 886 EGTVDKDTTIMELRDRLPKKQQASASTDGVDKLSGKEQSFTSLEVRKVAETKEKSKHLKT 945
Query: 180 ---EEIEEAKADIMEKI--------SPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQ 228
+++++ +DI EK + K K+G E+ KKP+ K + V + Q
Sbjct: 946 KTCKKVQDGLSDIAEKFLKKDQSDETSEDDKKQSKKGTEEKKKPSDFKKK---VIKMEQQ 1002
Query: 229 QVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGK 266
+ G + L ++I H P KQ+ + G+
Sbjct: 1003 YESSSDGTEKLPEREEICH---FPKGIKQIKNGTTDGE 1037
>UniRef100_Q9UFS7 Hypothetical protein DKFZp727M111 [Homo sapiens]
Length = 633
Score = 52.8 bits (125), Expect = 8e-05
Identities = 46/148 (31%), Positives = 71/148 (47%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNA-RVLELLPPL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + A R L L PL
Sbjct: 446 SFPDLYANFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFGEHVGALRALSL--PL 503
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y P EDN +L+ Y ++ V+ AL V Y +AV H++SFIF+ P
Sbjct: 504 TQLPVSLECYTVPPEDNLALLQLYFRTLVTGALRPRWC--PVLYAVAVAHVNSFIFSQDP 561
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L ++ L G +QH
Sbjct: 562 QSSDEVKAARRSMLQKTWLLADEGLRQH 589
>UniRef100_Q9NSQ5 Hypothetical protein DKFZp434E1120 [Homo sapiens]
Length = 398
Score = 52.8 bits (125), Expect = 8e-05
Identities = 46/148 (31%), Positives = 71/148 (47%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNA-RVLELLPPL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + A R L L PL
Sbjct: 211 SFPDLYANFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFGEHVGALRALSL--PL 268
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y P EDN +L+ Y ++ V+ AL V Y +AV H++SFIF+ P
Sbjct: 269 TQLPVSLECYTVPPEDNLALLQLYFRTLVTGALRPRWC--PVLYAVAVAHVNSFIFSQDP 326
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L ++ L G +QH
Sbjct: 327 QSSDEVKAARRSMLQKTWLLADEGLRQH 354
>UniRef100_Q9H9I2 Hypothetical protein FLJ12732 [Homo sapiens]
Length = 772
Score = 52.8 bits (125), Expect = 8e-05
Identities = 46/148 (31%), Positives = 71/148 (47%), Gaps = 9/148 (6%)
Query: 1271 SYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLATWNTLSNA-RVLELLPPL 1329
S+ ++ F ++S+GD +FG V + L ++RLA + A R L L PL
Sbjct: 585 SFPDLYANFLDHFEAVSFGDHLFGALVLLPLQRRFSVTLRLALFGEHVGALRALSL--PL 642
Query: 1330 EKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSYTMAVHHLSSFIFNACP 1389
+ E Y P EDN +L+ Y ++ V+ AL V Y +AV H++SFIF+ P
Sbjct: 643 TQLPVSLECYTVPPEDNLALLQLYFRTLVTGALRPRWC--PVLYAVAVAHVNSFIFSQDP 700
Query: 1390 VD----KLLLRNNLVRSLLRDYAGKQQH 1413
K R+ L ++ L G +QH
Sbjct: 701 QSSDEVKAARRSMLQKTWLLADEGLRQH 728
>UniRef100_P46100-5 Splice isoform 5 of P46100 [Homo sapiens]
Length = 2337
Score = 52.8 bits (125), Expect = 8e-05
Identities = 62/278 (22%), Positives = 109/278 (38%), Gaps = 42/278 (15%)
Query: 15 RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGG 74
+ T G DF + K K K + TQ + S +LEK++ S
Sbjct: 627 KSSTSGSDFDTKKGKSAKSSIISKKKRQTQSESSNYDSELEKEIKSMSKI-------GAA 679
Query: 75 KNTSKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSATSME--LDTSNKDDKKEVFAAE 132
+ T K+I + D F S E + + + G N TS E D + + ++E F++
Sbjct: 680 RTTKKRIPNTKD---FDSSEDEKHSKKGMDNQGHKNLKTSQEGSSDDAERKQERETFSSA 736
Query: 133 -------------RDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMST 179
RD++ + S+ EQ TSLE +E + + + + T
Sbjct: 737 EGTVDKDTTIMELRDRLPKKQQASASTDGVDKLSGKEQSFTSLEVRKVAETKEKSKHLKT 796
Query: 180 ---EEIEEAKADIMEKI--------SPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQ 228
+++++ +DI EK + K K+G E+ KKP+ K + V + Q
Sbjct: 797 KTCKKVQDGLSDIAEKFLKKDQSDETSEDDKKQSKKGTEEKKKPSDFKKK---VIKMEQQ 853
Query: 229 QVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGK 266
+ G + L ++I H P KQ+ + G+
Sbjct: 854 YESSSDGTEKLPEREEICH---FPKGIKQIKNGTTDGE 888
>UniRef100_P46100-2 Splice isoform 1 of P46100 [Homo sapiens]
Length = 2288
Score = 52.8 bits (125), Expect = 8e-05
Identities = 62/278 (22%), Positives = 109/278 (38%), Gaps = 42/278 (15%)
Query: 15 RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGG 74
+ T G DF + K K K + TQ + S +LEK++ S
Sbjct: 578 KSSTSGSDFDTKKGKSAKSSIISKKKRQTQSESSNYDSELEKEIKSMSKI-------GAA 630
Query: 75 KNTSKKISSYSDGSVFASMEVDAKPQLVKLDGGFINSATSME--LDTSNKDDKKEVFAAE 132
+ T K+I + D F S E + + + G N TS E D + + ++E F++
Sbjct: 631 RTTKKRIPNTKD---FDSSEDEKHSKKGMDNQGHKNLKTSQEGSSDDAERKQERETFSSA 687
Query: 133 -------------RDKIFSDRMTDHSSTSEKNYFMHEQESTSLENEIDSENRARIQQMST 179
RD++ + S+ EQ TSLE +E + + + + T
Sbjct: 688 EGTVDKDTTIMELRDRLPKKQQASASTDGVDKLSGKEQSFTSLEVRKVAETKEKSKHLKT 747
Query: 180 ---EEIEEAKADIMEKI--------SPALLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQ 228
+++++ +DI EK + K K+G E+ KKP+ K + V + Q
Sbjct: 748 KTCKKVQDGLSDIAEKFLKKDQSDETSEDDKKQSKKGTEEKKKPSDFKKK---VIKMEQQ 804
Query: 229 QVQITQGAKHLQTEDDISHTIMAPPSKKQLDDKNVSGK 266
+ G + L ++I H P KQ+ + G+
Sbjct: 805 YESSSDGTEKLPEREEICH---FPKGIKQIKNGTTDGE 839
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,431,969,448
Number of Sequences: 2790947
Number of extensions: 102669492
Number of successful extensions: 305765
Number of sequences better than 10.0: 411
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 412
Number of HSP's that attempted gapping in prelim test: 304877
Number of HSP's gapped (non-prelim): 1052
length of query: 1477
length of database: 848,049,833
effective HSP length: 140
effective length of query: 1337
effective length of database: 457,317,253
effective search space: 611433167261
effective search space used: 611433167261
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 82 (36.2 bits)
Medicago: description of AC135101.10


