

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.6 + phase: 0
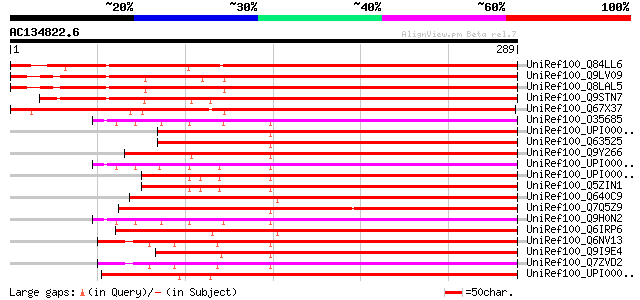
(289 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84LL6 Salt tolerance protein 5 [Beta vulgaris] 350 2e-95
UniRef100_Q9LV09 Similarity to nuclear movement protein nudC [Ar... 328 1e-88
UniRef100_Q8LAL5 Hypothetical protein [Arabidopsis thaliana] 328 1e-88
UniRef100_Q9STN7 Hypothetical protein T27E11.130 [Arabidopsis th... 311 1e-83
UniRef100_Q67X37 Putative salt tolerance protein 5 [Oryza sativa] 295 9e-79
UniRef100_O35685 MNUDC protein (Nuclear distribution gene C homo... 216 6e-55
UniRef100_UPI0000367F72 UPI0000367F72 UniRef100 entry 214 2e-54
UniRef100_Q63525 RnudC protein [Rattus norvegicus] 214 2e-54
UniRef100_Q9Y266 MNUDC protein [Homo sapiens] 213 6e-54
UniRef100_UPI00003AC55F UPI00003AC55F UniRef100 entry 211 2e-53
UniRef100_UPI00003AC560 UPI00003AC560 UniRef100 entry 210 4e-53
UniRef100_Q5ZIN1 Hypothetical protein [Gallus gallus] 210 4e-53
UniRef100_Q640C9 Hypothetical protein [Xenopus laevis] 209 7e-53
UniRef100_Q7Q5Z9 ENSANGP00000019739 [Anopheles gambiae str. PEST] 209 7e-53
UniRef100_Q9H0N2 Hypothetical protein DKFZp566P0446 [Homo sapiens] 209 9e-53
UniRef100_Q6IRP6 MGC83068 protein [Xenopus laevis] 207 3e-52
UniRef100_Q6NV13 Zgc:56099 protein [Brachydanio rerio] 206 4e-52
UniRef100_Q9I9E4 Putative nuclear movement protein PNUDC [Pleuro... 205 1e-51
UniRef100_Q7ZVD2 Similar to nuclear distribution gene C homolog ... 205 1e-51
UniRef100_UPI0000360EC2 UPI0000360EC2 UniRef100 entry 204 2e-51
>UniRef100_Q84LL6 Salt tolerance protein 5 [Beta vulgaris]
Length = 295
Score = 350 bits (899), Expect = 2e-95
Identities = 179/305 (58%), Positives = 225/305 (73%), Gaps = 27/305 (8%)
Query: 1 MAIISDYQDDNQTKTSSSSSSQPKPSKPIP---FSSTFDPSNPTTFLEKVFDFIAKESTN 57
MAI+SDY+++ QP+P K P FS+TFDPSNP FL+ +F++KES +
Sbjct: 1 MAILSDYEEEEH---------QPQPEKKQPSKKFSATFDPSNPLGFLQSTLEFVSKES-D 50
Query: 58 FFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAA-------------EKKL 104
FF K+S+ K V+S V+ + K + V +K K+ + A AAA EKK+
Sbjct: 51 FFAKESSAKDVVSLVQKVKEKYIEEVENKKKKLLDESAAAAAAAAAAAASSSSSDLEKKV 110
Query: 105 NDEKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKN 164
+D +S TEK KA PN GNG DLE YSW Q+LQE+ VNVPVP GTKS F+ C+IKKN
Sbjct: 111 DDNESAEETEKSKYKA-PNSGNGQDLENYSWIQSLQEVTVNVPVPPGTKSRFIDCQIKKN 169
Query: 165 HLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEIN 224
HLKVGLKGQPPIID EL+K +KPD+C+WS+EDQ ++S+LLTKHDQM+WW+ LVKG+PEI+
Sbjct: 170 HLKVGLKGQPPIIDGELFKPVKPDDCFWSLEDQKSISMLLTKHDQMEWWRSLVKGEPEID 229
Query: 225 TQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDF 284
TQKVEP SSKL DLD ETR TVEKMMFDQRQKSMGLPTS+++QK++M+KKFMS+HP MDF
Sbjct: 230 TQKVEPESSKLSDLDPETRSTVEKMMFDQRQKSMGLPTSDDMQKQDMLKKFMSEHPEMDF 289
Query: 285 SGAKF 289
S AKF
Sbjct: 290 SNAKF 294
>UniRef100_Q9LV09 Similarity to nuclear movement protein nudC [Arabidopsis thaliana]
Length = 304
Score = 328 bits (841), Expect = 1e-88
Identities = 177/314 (56%), Positives = 217/314 (68%), Gaps = 36/314 (11%)
Query: 1 MAIISDYQDDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFD 60
MAIIS+ +++ SSSS+P PF +T +NP FLEKVFDF+ ++S +F
Sbjct: 1 MAIISEVEEE-------SSSSRPMI---FPFRATLSSANPLGFLEKVFDFLGEQS-DFLK 49
Query: 61 KDSAEKVVLSAVRAAR--VKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEK--SEAVTEKI 116
K SAE ++ AVRAA+ +KKA+ AEK + EK KL ++K E+V I
Sbjct: 50 KPSAEDEIVVAVRAAKEKLKKAEKKKAEKESVKPVEKKAEKEIVKLVEKKVEKESVKPTI 109
Query: 117 DEKAA---------------------PNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSG 155
+A PN+GNG DLE YSW Q LQE+ VN+PVP GTK+
Sbjct: 110 AASSAEPIEVEKPKEEEEKKESGPIVPNKGNGTDLENYSWIQNLQEVTVNIPVPTGTKAR 169
Query: 156 FVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKC 215
V+CEIKKN LKVGLKGQ PI+D ELY+S+KPD+CYW+IEDQ +SILLTK DQM+WWKC
Sbjct: 170 TVVCEIKKNRLKVGLKGQDPIVDGELYRSVKPDDCYWNIEDQKVISILLTKSDQMEWWKC 229
Query: 216 LVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKF 275
VKG+PEI+TQKVEP +SKLGDLD ETR TVEKMMFDQRQK MGLPTSEELQK+E++KKF
Sbjct: 230 CVKGEPEIDTQKVEPETSKLGDLDPETRSTVEKMMFDQRQKQMGLPTSEELQKQEILKKF 289
Query: 276 MSQHPNMDFSGAKF 289
MS+HP MDFS AKF
Sbjct: 290 MSEHPEMDFSNAKF 303
>UniRef100_Q8LAL5 Hypothetical protein [Arabidopsis thaliana]
Length = 304
Score = 328 bits (841), Expect = 1e-88
Identities = 176/314 (56%), Positives = 216/314 (68%), Gaps = 36/314 (11%)
Query: 1 MAIISDYQDDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFD 60
MAIIS+ +++ SSSS+P PF +T +NP FLEKVFDF+ ++S +F
Sbjct: 1 MAIISEVEEE-------SSSSRPMI---FPFRATLSSANPLGFLEKVFDFLGEQS-DFLK 49
Query: 61 KDSAEKVVLSAVRAAR--VKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDE 118
K SAE ++ AVRAA+ +KKA+ AEK + EK KL ++K E + K
Sbjct: 50 KPSAEDEIVVAVRAAKEKLKKAEKKKAEKESVKPVEKKAEKEIVKLVEKKVEKESVKPTM 109
Query: 119 KAA-----------------------PNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSG 155
A+ PN+GNG DLE YSW Q LQE+ VN+PVP GTK+
Sbjct: 110 AASSAEPIEVEKPKDEEEKKESGPIVPNKGNGTDLENYSWIQNLQEVTVNIPVPTGTKAR 169
Query: 156 FVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKC 215
V+CEIKKN LKVGLKGQ PI+D ELY+S+KPD+CYW+IEDQ +SILLTK DQM+WWKC
Sbjct: 170 TVVCEIKKNRLKVGLKGQDPIVDGELYRSVKPDDCYWNIEDQKVISILLTKSDQMEWWKC 229
Query: 216 LVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKF 275
VKG+PEI+TQKVEP +SKLGDLD ETR TVEKMMFDQRQK MGLPTSEELQK+E++KKF
Sbjct: 230 CVKGEPEIDTQKVEPETSKLGDLDPETRSTVEKMMFDQRQKQMGLPTSEELQKQEILKKF 289
Query: 276 MSQHPNMDFSGAKF 289
MS+HP MDFS AKF
Sbjct: 290 MSEHPEMDFSNAKF 303
>UniRef100_Q9STN7 Hypothetical protein T27E11.130 [Arabidopsis thaliana]
Length = 293
Score = 311 bits (797), Expect = 1e-83
Identities = 158/290 (54%), Positives = 205/290 (70%), Gaps = 20/290 (6%)
Query: 18 SSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAA-- 75
S + +PS +PF+++FDPSNP FLEKV D I KES NF KD+AEK +++AV AA
Sbjct: 5 SEMEEARPSM-VPFTASFDPSNPIAFLEKVLDVIGKES-NFLKKDTAEKEIVAAVMAAKQ 62
Query: 76 RVKKAKSVAAEKAKIAAKEKAKAAAEK----KLNDEKSEAVT------------EKIDEK 119
R+++A+ EK + + E K + +L K E++ EK
Sbjct: 63 RLREAEKKKLEKESVKSMEVEKPKKDSLKPTELEKPKEESLMATDPMEIEKPKEEKESGP 122
Query: 120 AAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDR 179
PN+GNG+D EKYSW Q LQE+ +N+P+P GTKS V CEIKKN LKVGLKGQ I+D
Sbjct: 123 IVPNKGNGLDFEKYSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVDG 182
Query: 180 ELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLD 239
E + S+KPD+C+W+IEDQ +S+LLTK DQM+WWK VKG+PEI+TQKVEP +SKLGDLD
Sbjct: 183 EFFNSVKPDDCFWNIEDQKMISVLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLD 242
Query: 240 SETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
ETR +VEKMMFDQRQK MGLP S+E++K++M+KKFM+Q+P MDFS AKF
Sbjct: 243 PETRASVEKMMFDQRQKQMGLPRSDEIEKKDMLKKFMAQNPGMDFSNAKF 292
>UniRef100_Q67X37 Putative salt tolerance protein 5 [Oryza sativa]
Length = 308
Score = 295 bits (755), Expect = 9e-79
Identities = 159/307 (51%), Positives = 202/307 (65%), Gaps = 20/307 (6%)
Query: 1 MAIISDYQDDN----QTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKEST 56
MAIISD+Q++ Q + +S ++ + + FL+ D + S
Sbjct: 1 MAIISDFQEEEAPPRQQQQPASVAAAAGSGDEVLAAELERRGGAIPFLQAAIDVARRRSD 60
Query: 57 NFFDKDSAEKV--VLSAVRA--------ARVKKAKSVAAEKAKIAAKEKAKAAAEKKLND 106
F D + +V + SA RA AR K K+ AE+ A+ KAKA AE K
Sbjct: 61 LFRDPSAVSRVTSMASAARAVVEAEERKAREAKRKAEEAERKAAEAERKAKAPAEPKPES 120
Query: 107 EKSEAVTEKIDEKAA-----PNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEI 161
+ E +D+K PN GNG+DLEKYSW Q L E+ + VPVP GTKS FV+C+I
Sbjct: 121 SAGKDSME-VDKKEEGNVRKPNAGNGLDLEKYSWIQQLPEVTITVPVPQGTKSRFVVCDI 179
Query: 162 KKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDP 221
KKNHLKVGLKGQPPIID EL+K +K D+C+WSIED ++SILLTK +QM+WWK +VKGDP
Sbjct: 180 KKNHLKVGLKGQPPIIDGELFKPVKVDDCFWSIEDGKSLSILLTKQNQMEWWKSVVKGDP 239
Query: 222 EINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPN 281
E++TQKVEP +SKL DLD ETR TVEKMMFDQRQK MGLPTS+E+QK++M+KKFM+QHP
Sbjct: 240 EVDTQKVEPENSKLADLDPETRQTVEKMMFDQRQKQMGLPTSDEMQKQDMLKKFMAQHPE 299
Query: 282 MDFSGAK 288
MDFS AK
Sbjct: 300 MDFSNAK 306
>UniRef100_O35685 MNUDC protein (Nuclear distribution gene C homolog) (Sig 92
protein) (Mus musculus 10 days neonate cerebellum cDNA,
RIKEN full-length enriched library, clone:6530413H12
product:nuclear distribution gene C homolog
(Aspergillus), full insert sequence) [Mus musculus]
Length = 332
Score = 216 bits (550), Expect = 6e-55
Identities = 130/300 (43%), Positives = 175/300 (58%), Gaps = 59/300 (19%)
Query: 48 FDFIAKESTNFF---DKDSAEKVVLS--------AVRAARVKKAKSVAA--EKAKIAAKE 94
F F+ ++ T+FF ++ AEK++ A +A R K+A+ EKA+ AA+
Sbjct: 33 FSFLRRK-TDFFIGGEEGMAEKLITQTFNHHNQLAQKARREKRARQETERREKAERAARL 91
Query: 95 KAKAAAE------KKLNDEKSEAVTEKIDEKA---------------------------- 120
+A AE K+L DE++E + +ID+K
Sbjct: 92 AKEAKAETPGPQIKELTDEEAERLQLEIDQKKDAEDQEAQLKNGSLDSPGKQDAEDEEDE 151
Query: 121 ---------APNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVG 169
PN GNG DL Y WTQTL EL++ VP V K V+ +I++ HL+VG
Sbjct: 152 EDEKDKGKLKPNLGNGADLPNYRWTQTLAELDLAVPFRVSFRLKGKDVVVDIQRRHLRVG 211
Query: 170 LKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVE 229
LKGQPP++D ELY +K +E W IED V++ L K ++M+WW LV DPEINT+K+
Sbjct: 212 LKGQPPVVDGELYNEVKVEESSWLIEDGKVVTVHLEKINKMEWWNRLVTSDPEINTKKIN 271
Query: 230 PASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
P +SKL DLDSETR VEKMM+DQRQKSMGLPTS+E +K+E++KKFM QHP MDFS AKF
Sbjct: 272 PENSKLSDLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQEILKKFMDQHPEMDFSKAKF 331
>UniRef100_UPI0000367F72 UPI0000367F72 UniRef100 entry
Length = 305
Score = 214 bits (546), Expect = 2e-54
Identities = 111/207 (53%), Positives = 139/207 (66%), Gaps = 2/207 (0%)
Query: 85 AEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNV 144
AE + K + + K+ +E E EK K PN GNG DL Y WTQTL EL++
Sbjct: 98 AENHEAQLKNGSLDSPGKQETEEDEEEEDEKDKGKLKPNLGNGADLPNYRWTQTLSELDL 157
Query: 145 NVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSI 202
VP V K V+ +I++ HL+VGLKGQP IID ELY +K +E W IED V++
Sbjct: 158 AVPFCVNFRLKGKDVVVDIQRRHLRVGLKGQPAIIDGELYNEVKVEESSWLIEDGKVVTV 217
Query: 203 LLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPT 262
L K ++M+WW LV DPEINT+K+ P +SKL DLDSETR VEKMM+DQRQKSMGLPT
Sbjct: 218 HLEKINKMEWWSRLVSSDPEINTKKINPENSKLSDLDSETRSMVEKMMYDQRQKSMGLPT 277
Query: 263 SEELQKEEMMKKFMSQHPNMDFSGAKF 289
S+E +K+E++KKFM QHP MDFS AKF
Sbjct: 278 SDEQKKQEILKKFMDQHPEMDFSKAKF 304
>UniRef100_Q63525 RnudC protein [Rattus norvegicus]
Length = 332
Score = 214 bits (545), Expect = 2e-54
Identities = 109/207 (52%), Positives = 141/207 (67%), Gaps = 2/207 (0%)
Query: 85 AEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQELNV 144
AE ++ K + + K+ +E+ + EK K PN GNG DL Y WTQTL EL++
Sbjct: 125 AENHEVQLKNGSLDSPGKQDAEEEEDEEDEKDKGKLKPNLGNGADLPNYRWTQTLSELDL 184
Query: 145 NVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSI 202
VP V K V+ +I++ HL+VGLKGQ P+ID ELY +K +E W IED V++
Sbjct: 185 AVPFRVSFRLKGKDVVVDIQRRHLRVGLKGQAPVIDGELYNEVKVEESSWLIEDGKVVTV 244
Query: 203 LLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPT 262
L K ++M+WW LV DPEINT+K+ P +SKL DLDSETR VEKMM+DQRQKSMGLPT
Sbjct: 245 HLEKINKMEWWNRLVTSDPEINTKKINPENSKLSDLDSETRSMVEKMMYDQRQKSMGLPT 304
Query: 263 SEELQKEEMMKKFMSQHPNMDFSGAKF 289
S+E +K+E++KKFM QHP MDFS AKF
Sbjct: 305 SDEQKKQEILKKFMDQHPEMDFSKAKF 331
>UniRef100_Q9Y266 MNUDC protein [Homo sapiens]
Length = 331
Score = 213 bits (541), Expect = 6e-54
Identities = 114/238 (47%), Positives = 148/238 (61%), Gaps = 14/238 (5%)
Query: 66 KVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEK------------KLNDEKSEAVT 113
K S ++K+ AE+ ++ +K A + K + E+ E
Sbjct: 93 KEAKSETSGPQIKELTDEEAERLQLEIDQKKDAENHEAQLKNGSLDSPGKQDTEEDEEED 152
Query: 114 EKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGLK 171
EK K PN GNG DL Y WTQTL EL++ VP V K ++ +I++ HL+VGLK
Sbjct: 153 EKDKGKLKPNLGNGADLPNYRWTQTLSELDLAVPFCVNFRLKGKDMVVDIQRRHLRVGLK 212
Query: 172 GQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPA 231
GQP IID ELY +K +E W IED V++ L K ++M+WW LV DPEINT+K+ P
Sbjct: 213 GQPAIIDGELYNEVKVEESSWLIEDGKVVTVHLEKINKMEWWSRLVSSDPEINTKKINPE 272
Query: 232 SSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
+SKL DLDSETR VEKMM+DQRQKSMGLPTS+E +K+E++KKFM QHP MDFS AKF
Sbjct: 273 NSKLSDLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQEILKKFMDQHPEMDFSKAKF 330
>UniRef100_UPI00003AC55F UPI00003AC55F UniRef100 entry
Length = 325
Score = 211 bits (536), Expect = 2e-53
Identities = 127/296 (42%), Positives = 172/296 (57%), Gaps = 55/296 (18%)
Query: 48 FDFIAKESTNFF---DKDSAEKVVLS--------AVRAARVKKAKSVA-----AEKAKIA 91
F F+ ++ T+FF + AEK++ A + + KKA+ A AE+A
Sbjct: 30 FSFLRRK-TDFFTGGEDGVAEKLITDSFNHHNKLAQKERKEKKARQEAERREKAERAAKL 88
Query: 92 AKEKAKAAAE---KKLNDEKSEAVTEKIDE------------------------------ 118
AKE + A E K+L DE++E + +ID+
Sbjct: 89 AKEAKQEANEPRIKELTDEEAERLQLEIDQVRASLGTYGYQLCPACLLHAFANDEEKDEN 148
Query: 119 ---KAAPNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQ 173
K PN GNG DL Y WTQTL EL++ +P V K V+ +I++ L+VGLKG
Sbjct: 149 DKGKLKPNAGNGADLPNYRWTQTLSELDLAIPFKVTFRLKGKDVVVDIQRRRLRVGLKGH 208
Query: 174 PPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASS 233
PP+ID EL+ +K +E W IED TV++ L K ++M+WW LV DPEINT+K+ P +S
Sbjct: 209 PPVIDGELFNEVKVEESSWLIEDGKTVTVHLEKVNKMEWWNKLVSTDPEINTKKINPENS 268
Query: 234 KLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
KL DLDSETR VEKMM+DQRQKSMGLPTS+E +K++++KKFM QHP MDFS AKF
Sbjct: 269 KLSDLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQDILKKFMEQHPEMDFSKAKF 324
>UniRef100_UPI00003AC560 UPI00003AC560 UniRef100 entry
Length = 332
Score = 210 bits (534), Expect = 4e-53
Identities = 113/233 (48%), Positives = 149/233 (63%), Gaps = 19/233 (8%)
Query: 76 RVKKAKSVAAEKAKIAAKEKAKAAAE------KKLNDE-------KSEAVTEKIDE---- 118
R+K+ AE+ ++ +K +A E K D+ K E E+ DE
Sbjct: 99 RIKELTDEEAERLQLEIDQKKEAQKEVNNVPVKSSEDDGDSSDSNKQETDDEEKDENDKG 158
Query: 119 KAAPNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPI 176
K PN GNG DL Y WTQTL EL++ +P V K V+ +I++ L+VGLKG PP+
Sbjct: 159 KLKPNAGNGADLPNYRWTQTLSELDLAIPFKVTFRLKGKDVVVDIQRRRLRVGLKGHPPV 218
Query: 177 IDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLG 236
ID EL+ +K +E W IED TV++ L K ++M+WW LV DPEINT+K+ P +SKL
Sbjct: 219 IDGELFNEVKVEESSWLIEDGKTVTVHLEKVNKMEWWNKLVSTDPEINTKKINPENSKLS 278
Query: 237 DLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
DLDSETR VEKMM+DQRQKSMGLPTS+E +K++++KKFM QHP MDFS AKF
Sbjct: 279 DLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQDILKKFMEQHPEMDFSKAKF 331
>UniRef100_Q5ZIN1 Hypothetical protein [Gallus gallus]
Length = 341
Score = 210 bits (534), Expect = 4e-53
Identities = 113/233 (48%), Positives = 149/233 (63%), Gaps = 19/233 (8%)
Query: 76 RVKKAKSVAAEKAKIAAKEKAKAAAE------KKLNDE-------KSEAVTEKIDE---- 118
R+K+ AE+ ++ +K +A E K D+ K E E+ DE
Sbjct: 108 RIKELTDEEAERLQLEIDQKKEAQKEVNNVPVKSSEDDGDSSDSNKQETDDEEKDENDKG 167
Query: 119 KAAPNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPI 176
K PN GNG DL Y WTQTL EL++ +P V K V+ +I++ L+VGLKG PP+
Sbjct: 168 KLKPNAGNGADLPNYRWTQTLSELDLAIPFKVTFRLKGKDVVVDIQRRRLRVGLKGHPPV 227
Query: 177 IDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLG 236
ID EL+ +K +E W IED TV++ L K ++M+WW LV DPEINT+K+ P +SKL
Sbjct: 228 IDGELFNEVKVEESSWLIEDGKTVTVHLEKINKMEWWNKLVSTDPEINTKKINPENSKLS 287
Query: 237 DLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
DLDSETR VEKMM+DQRQKSMGLPTS+E +K++++KKFM QHP MDFS AKF
Sbjct: 288 DLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQDILKKFMEQHPEMDFSKAKF 340
>UniRef100_Q640C9 Hypothetical protein [Xenopus laevis]
Length = 327
Score = 209 bits (532), Expect = 7e-53
Identities = 109/223 (48%), Positives = 141/223 (62%), Gaps = 2/223 (0%)
Query: 69 LSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGM 128
L+ A R++K E+ + + K EK +E+ EK K PN GNG
Sbjct: 104 LTDEEAERLQKEIDKNKERENATNETEEKPEPEKNGEEEEEGEEDEKDKGKLKPNSGNGA 163
Query: 129 DLEKYSWTQTLQELNVNVPVPNG--TKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIK 186
DL Y WTQTL E+++ VP P K V +IK+ L VGL+GQ P++D EL+ IK
Sbjct: 164 DLPHYRWTQTLSEVDMIVPFPVSFRLKGKDVQVDIKRRRLTVGLRGQKPVLDGELFNDIK 223
Query: 187 PDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTV 246
+EC W IED V++ L K + M+WW +V DPEINT+K+ P +SKL DLD ETR V
Sbjct: 224 VEECSWLIEDGKVVTVHLEKINTMEWWSRIVLTDPEINTKKINPENSKLSDLDGETRSMV 283
Query: 247 EKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
EKMM+DQRQKSMGLPTS+E +K+++MKKFM QHP MDFS AKF
Sbjct: 284 EKMMYDQRQKSMGLPTSDEQKKQDIMKKFMEQHPEMDFSKAKF 326
>UniRef100_Q7Q5Z9 ENSANGP00000019739 [Anopheles gambiae str. PEST]
Length = 328
Score = 209 bits (532), Expect = 7e-53
Identities = 118/231 (51%), Positives = 147/231 (63%), Gaps = 5/231 (2%)
Query: 63 SAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAV-TEKIDE-KA 120
SA L+ A +++K +KA A EK A E K N S+ E DE K
Sbjct: 98 SATITELTEEEAEQLQKELDAKKQKAAEPAVEKVVPAEESKENKAGSDTEDVEPGDEGKL 157
Query: 121 APNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIID 178
PN+GNG DL+KYSWTQTLQEL + VP V K+ V+ I++ HLKVGLKG P IID
Sbjct: 158 KPNRGNGCDLDKYSWTQTLQELELRVPFDVKFTLKAKDVVVSIQRKHLKVGLKGHPAIID 217
Query: 179 RELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDL 238
EL IK ++ W +E +N V + + K +QM+WW LV DP INT+K+ P SSKL DL
Sbjct: 218 GELCSEIKIEDSLWHLE-KNAVVVTVEKINQMNWWDRLVTTDPPINTRKINPESSKLSDL 276
Query: 239 DSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
D TR VEKMM+DQRQK MGLPTS+E +K++M+KKFM QHP MDFS KF
Sbjct: 277 DGSTRSMVEKMMYDQRQKEMGLPTSDEQKKQDMLKKFMEQHPEMDFSKCKF 327
>UniRef100_Q9H0N2 Hypothetical protein DKFZp566P0446 [Homo sapiens]
Length = 331
Score = 209 bits (531), Expect = 9e-53
Identities = 128/299 (42%), Positives = 173/299 (57%), Gaps = 58/299 (19%)
Query: 48 FDFIAKESTNFF---DKDSAEKVVLS--------AVRAARVKKAKSVAA--EKAKIAAKE 94
F F+ ++ T+FF ++ AEK++ A + R K+A+ A EKA+ AA+
Sbjct: 33 FSFLRRK-TDFFIGGEEGMAEKLITQTFSHHNQLAQKNRREKRARQEAERREKAERAARL 91
Query: 95 KAKAAAE------KKLNDEKSEAVTEKIDEKA---------------------------- 120
+A +E K+L DE++E + +ID+K
Sbjct: 92 AKEAKSETSGPQIKELTDEEAERLQLEIDQKKDAENHEAQLKNGSLDSPGKQDTEEDVEE 151
Query: 121 --------APNQGNGMDLEKYSWTQTLQELNVNVP--VPNGTKSGFVICEIKKNHLKVGL 170
PN GNG D Y WTQTL EL++ VP V K ++ +I++ HL+VGL
Sbjct: 152 DEKDKGKLKPNLGNGADPPNYRWTQTLSELDLAVPFCVNFRLKGKDMVVDIQRRHLRVGL 211
Query: 171 KGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEP 230
KGQP IID ELY +K +E W IED V++ L K ++M+WW LV DPEINT+K+ P
Sbjct: 212 KGQPAIIDGELYNEVKVEESSWLIEDGKVVTVHLEKINKMEWWSRLVSSDPEINTKKINP 271
Query: 231 ASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
+SKL DLDSETR VEKMM+DQRQKSMGLPTS+E +K+E++KKFM QHP MDFS AKF
Sbjct: 272 ENSKLSDLDSETRSMVEKMMYDQRQKSMGLPTSDEQKKQEILKKFMDQHPEMDFSKAKF 330
>UniRef100_Q6IRP6 MGC83068 protein [Xenopus laevis]
Length = 329
Score = 207 bits (527), Expect = 3e-52
Identities = 112/239 (46%), Positives = 149/239 (61%), Gaps = 10/239 (4%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTE------ 114
K A + S + ++A+ + E K A+EKA + E+KL EK+ E
Sbjct: 90 KKKAAEAGESRITELTDEEAERLQKEIDKNKAREKATSETEEKLEPEKNGEEKEEEGGEE 149
Query: 115 --KIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNG--TKSGFVICEIKKNHLKVGL 170
K K PN GNG DL Y WTQTL E+++ VP P K V +I++ L VGL
Sbjct: 150 DEKDKGKLKPNSGNGADLPHYRWTQTLSEVDLIVPFPVSFRLKGKDVQVDIRRRRLTVGL 209
Query: 171 KGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEP 230
+GQ P++D EL+ IK +EC W IED V++ L K + M+WW +V DPEI+T+K+ P
Sbjct: 210 RGQKPVLDGELFNDIKVEECSWLIEDGKVVTVHLEKINTMEWWSRIVLTDPEISTRKINP 269
Query: 231 ASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
+SKL DLD ETR VEKMM+DQRQKSMGLPTS+E +K++++KKFM QHP MDFS AKF
Sbjct: 270 ENSKLSDLDGETRSMVEKMMYDQRQKSMGLPTSDEQKKQDILKKFMEQHPEMDFSKAKF 328
>UniRef100_Q6NV13 Zgc:56099 protein [Brachydanio rerio]
Length = 333
Score = 206 bits (525), Expect = 4e-52
Identities = 117/270 (43%), Positives = 163/270 (60%), Gaps = 35/270 (12%)
Query: 51 IAKESTNFFDKDSAEKVVLSAVRAARVK----KAKSVAAEKAKIAA-----KEKAKAAAE 101
+ KE + +K+ EK A RAA++K K + + AE+ +I E+ ++ +
Sbjct: 67 VQKEKQSRQEKERKEK----AERAAKLKEEEMKKRKIDAEEPRIKELTDEEAERLQSQLQ 122
Query: 102 KKLNDEKSEAVTEKID--------------------EKAAPNQGNGMDLEKYSWTQTLQE 141
K ++K+ V EK D +K PN GNG DL Y WTQ+L E
Sbjct: 123 TKQEEQKNSEVAEKTDADSKDKKGSDSEGEEDEKDKDKVKPNAGNGADLPNYRWTQSLSE 182
Query: 142 LNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNT 199
+++ VP V K V+ ++++ LKVGLKG PP+ID +L+ +K +E W IED
Sbjct: 183 VDLVVPFDVSFRLKGKDVVVDVQRRTLKVGLKGHPPLIDGQLFNEVKVEESSWLIEDGKV 242
Query: 200 VSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMG 259
V+I K ++M+WW LV DPEINT+K+ P +SKL DLD ETR VEKMM+DQRQKSMG
Sbjct: 243 VTIHFEKINKMEWWNKLVTTDPEINTKKICPENSKLSDLDGETRSMVEKMMYDQRQKSMG 302
Query: 260 LPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
LPTSEE +K++++KKFM+QHP MDFS AKF
Sbjct: 303 LPTSEEQKKQDILKKFMAQHPEMDFSKAKF 332
>UniRef100_Q9I9E4 Putative nuclear movement protein PNUDC [Pleurodeles waltlii]
Length = 346
Score = 205 bits (522), Expect = 1e-51
Identities = 108/212 (50%), Positives = 141/212 (65%), Gaps = 6/212 (2%)
Query: 84 AAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEK----AAPNQGNGMDLEKYSWTQTL 139
A+E + + + E K + KS A +E+ DEK PN GNG DL Y WTQTL
Sbjct: 134 ASESQQASGNGITQTPTELKDPETKSGADSEEEDEKDKGKLRPNSGNGADLPLYRWTQTL 193
Query: 140 QELNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQ 197
E+++ VP V K V+ +I++ LKVGLKGQ P+ID ELY +K +E W IED
Sbjct: 194 SEVDLVVPLNVSFRLKGKDVVVDIQRRILKVGLKGQAPVIDGELYNEVKVEESSWLIEDG 253
Query: 198 NTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKS 257
+++ L K ++M+WW +V DPEINT+K+ P +SKL DLDSETR VEKMM+DQRQKS
Sbjct: 254 KVITVHLEKINKMEWWSRIVSTDPEINTKKINPENSKLSDLDSETRSMVEKMMYDQRQKS 313
Query: 258 MGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
MGLPTSEE +K++++KKFM QHP MDF AKF
Sbjct: 314 MGLPTSEEQKKQDILKKFMEQHPEMDFFKAKF 345
>UniRef100_Q7ZVD2 Similar to nuclear distribution gene C homolog [Brachydanio rerio]
Length = 333
Score = 205 bits (522), Expect = 1e-51
Identities = 117/270 (43%), Positives = 162/270 (59%), Gaps = 35/270 (12%)
Query: 51 IAKESTNFFDKDSAEKVVLSAVRAARVK----KAKSVAAEKAKIAA-----KEKAKAAAE 101
+ KE + +K+ EK A RAA++K K + + AE+ +I E+ ++ +
Sbjct: 67 VQKEKQSRQEKERKEK----AERAAKLKEEEMKKRKIDAEEPRIKELTDEEAERLQSQLQ 122
Query: 102 KKLNDEKSEAVTEKID--------------------EKAAPNQGNGMDLEKYSWTQTLQE 141
K + K+ V EK D +K PN GNG DL Y WTQ+L E
Sbjct: 123 TKQEEPKNSEVAEKTDADSKDKKGSDSEGEEDEKDKDKVKPNAGNGADLPNYRWTQSLSE 182
Query: 142 LNVNVP--VPNGTKSGFVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNT 199
+++ VP V K V+ ++++ LKVGLKG PP+ID +L+ +K +E W IED
Sbjct: 183 VDLVVPFDVSFRLKGKDVVVDVQRRTLKVGLKGHPPLIDGQLFNEVKVEESSWLIEDGKV 242
Query: 200 VSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMG 259
V+I K ++M+WW LV DPEINT+K+ P +SKL DLD ETR VEKMM+DQRQKSMG
Sbjct: 243 VTIHFEKINKMEWWNKLVTTDPEINTKKICPENSKLSDLDGETRSMVEKMMYDQRQKSMG 302
Query: 260 LPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
LPTSEE +K++++KKFM+QHP MDFS AKF
Sbjct: 303 LPTSEEQKKQDILKKFMAQHPEMDFSKAKF 332
>UniRef100_UPI0000360EC2 UPI0000360EC2 UniRef100 entry
Length = 333
Score = 204 bits (520), Expect = 2e-51
Identities = 113/253 (44%), Positives = 158/253 (61%), Gaps = 16/253 (6%)
Query: 53 KESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEK---------AKAAAEKK 103
KE T K + E+ + R+++ AEK ++ +K A AEK
Sbjct: 80 KEKTERAAKLAEEERKKESDDGPRIQELTDEEAEKLQLELDKKKNEEDKTADAPTNAEKP 139
Query: 104 LND-EKSEAVT------EKIDEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGF 156
L+D EK++ EK +K PN GNG DL + WTQTL E++++VP K
Sbjct: 140 LDDSEKAKGSDSEGEDDEKDKDKLKPNAGNGADLPTHKWTQTLSEVDLSVPFNIRIKGRD 199
Query: 157 VICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCL 216
V+ +I++ +KVGLKG PP+++ +LY IK +E W I+D V++ L K ++M+WW +
Sbjct: 200 VVVDIQRRSIKVGLKGHPPVVEGQLYGEIKVEESSWLIDDGKVVTVHLEKINKMEWWNKV 259
Query: 217 VKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFM 276
V DPEINT+K+ P +SKL DLD ETR VEKMM+DQRQKSMGLPTSEE +K++++KKFM
Sbjct: 260 VTTDPEINTKKICPENSKLSDLDGETRGMVEKMMYDQRQKSMGLPTSEEQKKQDILKKFM 319
Query: 277 SQHPNMDFSGAKF 289
+QHP MDFS AKF
Sbjct: 320 AQHPEMDFSKAKF 332
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.126 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 473,012,516
Number of Sequences: 2790947
Number of extensions: 19727588
Number of successful extensions: 89002
Number of sequences better than 10.0: 715
Number of HSP's better than 10.0 without gapping: 214
Number of HSP's successfully gapped in prelim test: 521
Number of HSP's that attempted gapping in prelim test: 85661
Number of HSP's gapped (non-prelim): 2631
length of query: 289
length of database: 848,049,833
effective HSP length: 126
effective length of query: 163
effective length of database: 496,390,511
effective search space: 80911653293
effective search space used: 80911653293
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC134822.6


