

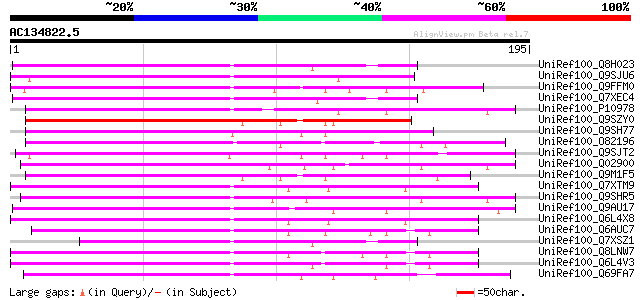
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.5 - phase: 0
(195 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Or... 110 2e-23
UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidop... 110 3e-23
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 110 3e-23
UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa] 105 6e-22
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 102 4e-21
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 101 9e-21
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 99 6e-20
UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabid... 97 2e-19
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 97 2e-19
UniRef100_Q02900 Orf 1 [Arabidopsis thaliana] 96 5e-19
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 94 2e-18
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 94 3e-18
UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana] 92 7e-18
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 92 7e-18
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 92 7e-18
UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa] 91 1e-17
UniRef100_Q7XSZ1 OSJNBb0056F09.5 protein [Oryza sativa] 90 4e-17
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 89 5e-17
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 89 8e-17
UniRef100_Q69FA7 Retrovirus-related pol polyprotein [Phaseolus v... 89 8e-17
>UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Oryza sativa]
Length = 556
Score = 110 bits (275), Expect = 2e-23
Identities = 62/154 (40%), Positives = 92/154 (59%), Gaps = 7/154 (4%)
Query: 2 KVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKAL 61
K+ Y +KSL + LKQQLY +M E + + + FN+++ DL+ ++V ++DEDKA+
Sbjct: 219 KLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSKLDVKLDDEDKAI 278
Query: 62 LLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENS--EGLCVSR 119
+LLCSLP SFEH T+ +GK+ T EEI S+L + L +SK A E S E L V
Sbjct: 279 ILLCSLPPSFEHVVTTLTHGKD-TVKTEEIISSLLARDLRRSKKNEAMEASQAESLLVKA 337
Query: 120 GNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRD 153
+ G S + G +C+KCH+FGH +R+
Sbjct: 338 KHDHEAGVSKSKEKG----ARCYKCHEFGHIRRN 367
>UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 838
Score = 110 bits (274), Expect = 3e-23
Identities = 62/160 (38%), Positives = 96/160 (59%), Gaps = 9/160 (5%)
Query: 1 MKVLQK-YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDK 59
++VL K + KSL +R LKQ+LY +KM +S +I E + +F K++ DL N++V++ DED+
Sbjct: 97 IRVLDKLFMAKSLPNRIYLKQRLYGYKMSDSMTIEENVNDFFKLISDLENVKVSVPDEDQ 156
Query: 60 ALLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVSR 119
A++LL SLPK F+ KDT+ YGK T L+EI A+R+K L + +NS +
Sbjct: 157 AIVLLMSLPKQFDQLKDTLKYGKT-TLALDEITGAIRSKVLELGASGKMLKNSSDALFVQ 215
Query: 120 GNG-------GGRGNRGSSKSGNKERYKCFKCHKFGHFKR 152
G N+ S+S ++E+ C+ C K GHFK+
Sbjct: 216 DRGRSEKRDKSSERNKSQSRSKSREKKVCWVCGKEGHFKK 255
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 110 bits (274), Expect = 3e-23
Identities = 81/196 (41%), Positives = 113/196 (57%), Gaps = 20/196 (10%)
Query: 1 MKVL-QKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDK 59
+KVL Q + KSL +R LKQ+LY +KM E+ ++ E + +F K++ DL N++V + DED+
Sbjct: 105 IKVLDQLFMAKSLPNRIYLKQRLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQ 164
Query: 60 ALLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTK--KLTKSKDLRANENSEGLCV 117
A++LL SLP+ F+ K+T+ Y K T LEEI SA+R+K +L S L N NS+GL V
Sbjct: 165 AIVLLMSLPRQFDQLKETLKYCKT-TLHLEEITSAIRSKILELGASGKLLKN-NSDGLFV 222
Query: 118 -SRGNGGGRG---NRGSSKSGNKERYK-CFKCHKFGHFKRDF----------SEDNENFA 162
RG RG N+ S+S +K K C+ C K GHFK+ S A
Sbjct: 223 QDRGRSETRGKGPNKNKSRSKSKGAGKTCWICGKEGHFKKQCYVWKERNKQGSTSERGEA 282
Query: 163 QVVSEEYEDAGALVVS 178
V+ DA ALVVS
Sbjct: 283 STVTARVTDAAALVVS 298
>UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa]
Length = 382
Score = 105 bits (262), Expect = 6e-22
Identities = 58/154 (37%), Positives = 93/154 (59%), Gaps = 7/154 (4%)
Query: 2 KVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKAL 61
K+ + +KSL + LKQQLY ++ E + + + FN+++ DL+ ++V ++DEDKA+
Sbjct: 88 KLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRKHVDVFNQLVVDLSKLDVKLDDEDKAI 147
Query: 62 LLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEG--LCVSR 119
+LLCSLP S+EH T+ +GK+ T EEI S+L + L +SK A + S+G L V
Sbjct: 148 ILLCSLPLSYEHVVTTLTHGKD-TVKTEEIISSLLARDLRRSKKNEATKASQGKSLLVKD 206
Query: 120 GNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRD 153
+ G S + G +C+KCH+FGH +R+
Sbjct: 207 KHDHEAGVSKSKEKG----ARCYKCHEFGHIRRN 236
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 102 bits (255), Expect = 4e-21
Identities = 68/201 (33%), Positives = 106/201 (51%), Gaps = 22/201 (10%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L ++ LK+QLY+ M E + L FN ++ LAN+ V +E+EDKA+LLL S
Sbjct: 92 YMSKTLTNKLYLKKQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNS 151
Query: 67 LPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVSRGNG---- 122
LP S+++ TIL+GK T L+++ SAL L K + EN ++ G G
Sbjct: 152 LPSSYDNLATTILHGKT-TIELKDVTSAL----LLNEKMRKKPENQGQALITEGRGRSYQ 206
Query: 123 ------GGRGNRGSSKSGNKERYK-CFKCHKFGHFKRDFSEDNENFAQVVSEEYEDAGAL 175
G G RG SK+ +K R + C+ C++ GHFKRD + + ++ +D A
Sbjct: 207 RSSNNYGRSGARGKSKNRSKSRVRNCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAA 266
Query: 176 VVS------CWEDDEGEVSHL 190
+V + ++E E HL
Sbjct: 267 MVQNNDNVVLFINEEEECMHL 287
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 101 bits (252), Expect = 9e-21
Identities = 63/156 (40%), Positives = 97/156 (61%), Gaps = 13/156 (8%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R LKQ+LYS+KM E+ S+ + EF +++ DL N V + DED+A+LLL S
Sbjct: 105 YMSKALPNRIYLKQKLYSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMS 164
Query: 67 LPKSFEHFKDTILYGKEGTT-TLEEIQSALRTKKL---TKSKDLRANENSEGLCV----- 117
LPK F+ KDT+ YG TT +++E+ +A+ +K+L + K +R +EGL V
Sbjct: 165 LPKQFDQLKDTLKYGSGRTTLSVDEVVAAIYSKELELGSNKKSIRG--QAEGLYVKDKPE 222
Query: 118 SRG--NGGGRGNRGSSKSGNKERYKCFKCHKFGHFK 151
+RG +GN+G S+S +K C+ C + GHFK
Sbjct: 223 TRGMSEQKEKGNKGRSRSRSKGWKGCWICGEEGHFK 258
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 99.0 bits (245), Expect = 6e-20
Identities = 57/160 (35%), Positives = 94/160 (58%), Gaps = 7/160 (4%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R KQ+LYSFKM E+ S+ + EF +I+ DL N+ V + DED+A+LLL +
Sbjct: 107 YMSKALPNRIYPKQKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTA 166
Query: 67 LPKSFEHFKDTILYGK-EGTTTLEEIQSALRTKKLTKSKDLRA-NENSEGLCV-----SR 119
LPK+F+ KDT+ Y + TL+E+ +A+ +K+L ++ +EGL V ++
Sbjct: 167 LPKAFDQLKDTLKYSSGKSILTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENK 226
Query: 120 GNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRDFSEDNE 159
G G +G K +K++ C+ C + GHF+ N+
Sbjct: 227 GKGEQKGKGKGKKGKSKKKPGCWTCGEEGHFRSSCPNQNK 266
>UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1137
Score = 97.4 bits (241), Expect = 2e-19
Identities = 63/188 (33%), Positives = 108/188 (56%), Gaps = 12/188 (6%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y KSL +R L+ ++Y+++M +SK++ E + EF K++ DL N+++ + DE +A+L+L +
Sbjct: 77 YLVKSLPNRVYLQLKVYNYRMQDSKTLEENVDEFQKMISDLNNLQIQVPDEVQAILILSA 136
Query: 67 LPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKL-TKSKDLRANENSEGLCVSRGNGGGR 125
LP S++ K+T+ YG+EG L+++ SA ++K+L + + EGL V RG R
Sbjct: 137 LPDSYDMLKETLKYGREG-IKLDDVISAAKSKELELRDSSGGSRPVGEGLYV-RGKSQAR 194
Query: 126 GNRGSSKSGNKERYKCFKCHKFGHFKRD----FSEDNENFA---QVVSEEYEDAGALVVS 178
G+ G + K+ C+ C K GHFKR ++ N A +V ++ +D LV S
Sbjct: 195 GSDGPKSTEGKK--VCWICGKEGHFKRQCYKWLEKNKANGAGETALVKDDAQDLVGLVAS 252
Query: 179 CWEDDEGE 186
EG+
Sbjct: 253 EVNMSEGK 260
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 97.1 bits (240), Expect = 2e-19
Identities = 73/199 (36%), Positives = 110/199 (54%), Gaps = 14/199 (7%)
Query: 3 VLQK-YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKAL 61
VL K Y +K+L +R KQ+LYSFKM E+ SI + EF +I+ DL N V + DED+A+
Sbjct: 102 VLDKLYMSKALPNRIYQKQKLYSFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAI 161
Query: 62 LLLCSLPKSFEHFKDTILYG-KEGTTTLEEIQSALRTKKLTKSKDLRA-NENSEGLCV-- 117
LLL SLPK F+ +DT+ YG T +L+E+ +A+ +K+L + ++ +EGL V
Sbjct: 162 LLLMSLPKPFDQLRDTLKYGLGRVTLSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKE 221
Query: 118 ---SRGNGGGRGNRGSS-KSGNKERYK--CFKCHKFGHFKRDFSEDNENFAQVVSEEYED 171
+RG RGN ++ KS +K R K C+ C + + ++SE N + VSE
Sbjct: 222 KTETRGRTEQRGNNNNNKKSRSKSRSKKGCWICGESSNGSSNYSEANGLY---VSEALSS 278
Query: 172 AGALVVSCWEDDEGEVSHL 190
+ W D G H+
Sbjct: 279 TDIHLEDEWVMDTGCSYHM 297
>UniRef100_Q02900 Orf 1 [Arabidopsis thaliana]
Length = 560
Score = 95.9 bits (237), Expect = 5e-19
Identities = 68/209 (32%), Positives = 110/209 (52%), Gaps = 24/209 (11%)
Query: 5 QKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLL 64
++Y SL +R ++ + YSFKM +SKSI+E + EF KI+ +L+++E+N+ +E +A+L L
Sbjct: 109 KQYMETSLPNRIYVQLKFYSFKMNDSKSINENVNEFLKIVAELSSLEINVVEEVRAILFL 168
Query: 65 CSLPKSFEHFKDTILYGKEGTTTLEEIQSALR-TKKLTKSKDLRAN------ENSEGLCV 117
L + K T+ YG + + + I SA ++L + K+ N N G +
Sbjct: 169 NGLSSRYSQLKHTLKYGNKALSLQDVISSARSLERELDEQKETDKNTSTVLYTNERGRPL 228
Query: 118 SRG---NGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRD-------FSEDNENFAQVVSE 167
+R N GG+G RG SKS + + C+ C K GH K+D +N A V++E
Sbjct: 229 TRNQNQNKGGQG-RGRSKSNSNAKLTCWYCKKEGHVKKDCFARKRKLESENPGEAGVITE 287
Query: 168 EYEDAGALVVS------CWEDDEGEVSHL 190
+ + AL V+ WE D G SH+
Sbjct: 288 KLVFSEALSVNDLAVRDIWELDSGCPSHM 316
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 94.0 bits (232), Expect = 2e-18
Identities = 63/177 (35%), Positives = 98/177 (54%), Gaps = 12/177 (6%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R LKQ+LYSFKM E+ SI + EF I+ DL N+ V + DED+A+LLL S
Sbjct: 107 YMSKALPNRIYLKQKLYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMS 166
Query: 67 LPKSFEHFKDTILYGKEGTT-TLEEIQSALRTKKL---TKSKDLRANENSEGLCV----- 117
LPK F+ KDT+ Y T +L+E+ +A+ +++L + K ++ +EGL V
Sbjct: 167 LPKPFDQLKDTLKYSSGKTVLSLDEVAAAIYSRELEFGSVKKSIKG--QAEGLYVKDKAE 224
Query: 118 SRGNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRDFSEDNE-NFAQVVSEEYEDAG 173
+RG + +S +K + C+ C + GH K N+ F S + E +G
Sbjct: 225 NRGRSEQKDKGKGKRSKSKSKRGCWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSG 281
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 93.6 bits (231), Expect = 3e-18
Identities = 66/205 (32%), Positives = 102/205 (49%), Gaps = 30/205 (14%)
Query: 1 MKVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKA 60
+K+ Q TK L + LKQ+L+ K+ + S+ + L+ F +I+ DL ++EV +++D A
Sbjct: 85 LKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKEIVADLESMEVKYDEKDLA 144
Query: 61 LLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKS--KDLRANENSEGLCVS 118
L+LLCSLP S+ +F+DTILY ++ T TL+E+ AL K+ K +N +EGL V
Sbjct: 145 LILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHAKEKMKKMVPSEGSNSQAEGLVVR 203
Query: 119 --------------------RGNGGGRGNRGSSKSGNKERYKCFKCHKF-------GHFK 151
RG RG S K ++ + KC K G +
Sbjct: 204 GSQQEKNTNNKSRDKSSSSYRGRSKSRGRYKSCKYCKRDGHDISKCWKLQDKDKRTGKYI 263
Query: 152 RDFSEDNENFAQVVSEEYEDAGALV 176
++ E A VV++E DA LV
Sbjct: 264 PKGKKEEEGKAAVVTDEKSDAELLV 288
>UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana]
Length = 1356
Score = 92.0 bits (227), Expect = 7e-18
Identities = 61/207 (29%), Positives = 111/207 (53%), Gaps = 22/207 (10%)
Query: 5 QKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLL 64
+KY SL +R + +LYSFKM+ + +I + + EF +I+ +L ++E+ +++E +A+L+L
Sbjct: 113 KKYMETSLPNRIYTQLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILIL 172
Query: 65 CSLPKSFEHFKDTILYGKEGTTTLEEIQSALRT--KKLTKSKDLRANE------NSEGLC 116
SLP S K T+ YG + T T++++ S+ ++ ++L ++ DL + G
Sbjct: 173 NSLPASHIQLKHTLKYGNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRP 231
Query: 117 VSRGNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRD-------FSEDNENFAQVVSEEY 169
+ R N G +G S+S +K + C+ C K GH K+D + + A V++E+
Sbjct: 232 LVRNNQKGGQGKGRSRSNSKTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKL 291
Query: 170 EDAGALVVS------CWEDDEGEVSHL 190
+ AL V+ W D G SH+
Sbjct: 292 VFSEALSVNEQMVKDLWILDSGCTSHM 318
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 92.0 bits (227), Expect = 7e-18
Identities = 65/204 (31%), Positives = 103/204 (49%), Gaps = 18/204 (8%)
Query: 2 KVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKAL 61
K+ Y +K+L ++ LK+QLY+ M E + L N ++ LAN+ V +E+EDK +
Sbjct: 88 KLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSHLNVLNGLITQLANLGVKIEEEDKRI 147
Query: 62 LLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVSRG- 120
+LL SLP S++ TIL+GK+ + L+++ SAL + + K N + SRG
Sbjct: 148 VLLNSLPSSYDTLSTTILHGKD-SIQLKDVTSALLLNEKMRKKP--ENHGQVFITESRGR 204
Query: 121 -------NGGGRGNRGSSKSGNKERYK-CFKCHKFGHFKRDFSEDNENFAQVVSEEYEDA 172
N G G RG SK +K + + C+ C + GHFKRD + ++ +D
Sbjct: 205 SYQRSSSNYGRSGARGKSKVRSKSKARNCYNCDQPGHFKRDCPNPKRGKGESSGQKNDDN 264
Query: 173 GALVVSCWED------DEGEVSHL 190
A +V +D +E E HL
Sbjct: 265 TAAMVQNNDDVVLLINEEEECMHL 288
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 92.0 bits (227), Expect = 7e-18
Identities = 66/205 (32%), Positives = 100/205 (48%), Gaps = 30/205 (14%)
Query: 1 MKVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKA 60
+K+ Q TK L + LKQ+L+ K+ + S+ + L+ F KI+ DL ++EV ++ED
Sbjct: 85 LKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKKIVADLESMEVKYDEEDLC 144
Query: 61 LLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKS--KDLRANENSEGLCVS 118
L+LLCSLP S+ +F+DTILY + T TL+E+ AL K+ K +N +EGL V
Sbjct: 145 LILLCSLPSSYANFRDTILYSCD-TLTLKEVYDALHAKEKIKKMVPSEGSNSQAEGLVVR 203
Query: 119 --------------------RGNGGGRGNRGSSKSGNKERYKCFKCHKF-------GHFK 151
RG RG S K ++ + +C K G +
Sbjct: 204 GRQQEKNTNSKSRDKSSSSYRGRSKSRGRYKSCKYYKRDGHDISECWKLQDKDKRTGKYV 263
Query: 152 RDFSEDNENFAQVVSEEYEDAGALV 176
++ E A VV++E DA LV
Sbjct: 264 PKGKKEEEGKAAVVTDEKSDAELLV 288
>UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa]
Length = 1241
Score = 91.3 bits (225), Expect = 1e-17
Identities = 68/200 (34%), Positives = 100/200 (50%), Gaps = 36/200 (18%)
Query: 9 TKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCSLP 68
TK L + LKQ+L+ K+ + S+ + L+ F +I+ DL ++EV ++ED L+LLCSLP
Sbjct: 2 TKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKEIVADLESMEVKYDEEDLGLILLCSLP 61
Query: 69 KSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKS--KDLRANENSEGLCV-SRGNGGGR 125
S+ +F+DTILY ++ T TL+E+ AL K+ K +N +EGL V R
Sbjct: 62 SSYANFRDTILYSRD-TLTLKEVYDALHAKEKMKKMVPSEGSNSQAEGLVVRGRQQEKNT 120
Query: 126 GNRGSSKSG--------NKERYK-CFKCHKFGHFKRDFSE-------------------- 156
N+ KS ++ RYK C C + GH D SE
Sbjct: 121 NNKSRDKSSSIYRGRSKSRGRYKSCKYCKRDGH---DISECWKLQDKDKRTRKYIPKGKK 177
Query: 157 DNENFAQVVSEEYEDAGALV 176
+ E A VV++E DA LV
Sbjct: 178 EEEGKAAVVTDEKSDAELLV 197
>UniRef100_Q7XSZ1 OSJNBb0056F09.5 protein [Oryza sativa]
Length = 371
Score = 89.7 bits (221), Expect = 4e-17
Identities = 50/129 (38%), Positives = 78/129 (59%), Gaps = 7/129 (5%)
Query: 27 MLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYGKEGTT 86
M E + + + FN+++ DL+ ++V ++DEDKA++LLCSLP S+EH T+++GK+ T
Sbjct: 133 MQEESDLRKHVDVFNQLVVDLSKLDVKLDDEDKAIILLCSLPPSYEHVVTTLMHGKD-TI 191
Query: 87 TLEEIQSALRTKKLTKSKDLRANENS--EGLCVSRGNGGGRGNRGSSKSGNKERYKCFKC 144
EEI S+L + L +SK A E S E L V + G S + G +C+KC
Sbjct: 192 KTEEIISSLLARDLRRSKKNEAMEASQAESLLVKAKHDHEAGVLKSKEKG----ARCYKC 247
Query: 145 HKFGHFKRD 153
H+FGH +R+
Sbjct: 248 HEFGHIRRN 256
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 89.4 bits (220), Expect = 5e-17
Identities = 69/209 (33%), Positives = 103/209 (49%), Gaps = 38/209 (18%)
Query: 1 MKVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKA 60
+K+ Q TK L + LKQ+L+ K+ + S+ + L+ F +I+ DL +IEV ++ED
Sbjct: 120 LKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSTFKEIVADLESIEVKYDEEDLG 179
Query: 61 LLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKS--KDLRANENSEGLCVS 118
L+LLCSLP S+ +F+DTILY + T L+E+ AL K+ K +N +EGL V
Sbjct: 180 LILLCSLPSSYANFRDTILYSHD-TLILKEVYDALHAKEKMKKMVPSEGSNSQAEGLVV- 237
Query: 119 RGNGGGRGNRGSS----------KSGNKERYK-CFKCHKFGHFKRDFSE----------- 156
RG + + S +S ++ RYK C C + GH D SE
Sbjct: 238 RGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCKYCKRDGH---DISECWKLQDKDKRT 294
Query: 157 ---------DNENFAQVVSEEYEDAGALV 176
+ E A VV++E D LV
Sbjct: 295 GKYIPKGKKEEEGKAAVVTDEKSDTELLV 323
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 88.6 bits (218), Expect = 8e-17
Identities = 65/209 (31%), Positives = 101/209 (48%), Gaps = 38/209 (18%)
Query: 1 MKVLQKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKA 60
+K+ Q TK L + LKQ+L+ K+ + +S+ + L+ F +I+ DL ++EV +++D
Sbjct: 85 LKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSAFKEIVADLESMEVKYDEDDLG 144
Query: 61 LLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENS--EGLCVS 118
L+LLCSLP S+ +F+ TILY ++ T TL+E+ A K+ K NS EGL V
Sbjct: 145 LILLCSLPSSYANFRGTILYSRD-TLTLKEVYDAFHAKEKMKKMVTSEGSNSQAEGLVV- 202
Query: 119 RGNGGGRGNRGSSKSGNKERYK-----------CFKCHKFGHFKRDFSE----------- 156
RG + + S+ + Y+ C C + GH D SE
Sbjct: 203 RGRQQKKNTKNQSRDKSSSSYRGRTKSRGRYKSCKYCKRDGH---DISECWKLQDKDKRT 259
Query: 157 ---------DNENFAQVVSEEYEDAGALV 176
+ E A VV++E DA LV
Sbjct: 260 GKYIPKGKKEEEGKAAVVTDEKSDAELLV 288
>UniRef100_Q69FA7 Retrovirus-related pol polyprotein [Phaseolus vulgaris]
Length = 324
Score = 88.6 bits (218), Expect = 8e-17
Identities = 58/188 (30%), Positives = 102/188 (53%), Gaps = 13/188 (6%)
Query: 6 KYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLC 65
K+ KS ++ L+K++L+ F + +++ + FN+++ L N++V EDED AL+L+
Sbjct: 71 KFLKKSGQNKLLMKKRLFRFDYQQGTTMNAHITMFNQLVAYLLNLDVKFEDEDLALMLVS 130
Query: 66 SLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRA-NENSEGLCVSRG--NG 122
SLP FEH + T+L+GK+ +L+ + SAL + +L K ++ + SE V+RG
Sbjct: 131 SLPDEFEHLETTLLHGKD-NVSLDVVCSALYSHELRKQDKMKTKSTTSEEALVTRGGQQS 189
Query: 123 GGRGNRGSSKSGNK--ERYKCFKCHKFGHFKRDFSEDNENFAQVVSEEYEDAGALVVSCW 180
+ RG KS + + +C CH+ GH+K+D ++ +E A V C
Sbjct: 190 QTKERRGMCKSKGRVVAKDECAFCHEKGHWKKD-------CPKLQKKEKVPQDANVAECK 242
Query: 181 EDDEGEVS 188
D E ++S
Sbjct: 243 SDVESDIS 250
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.132 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 327,637,773
Number of Sequences: 2790947
Number of extensions: 14030491
Number of successful extensions: 55585
Number of sequences better than 10.0: 513
Number of HSP's better than 10.0 without gapping: 185
Number of HSP's successfully gapped in prelim test: 328
Number of HSP's that attempted gapping in prelim test: 55035
Number of HSP's gapped (non-prelim): 657
length of query: 195
length of database: 848,049,833
effective HSP length: 121
effective length of query: 74
effective length of database: 510,345,246
effective search space: 37765548204
effective search space used: 37765548204
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC134822.5


