

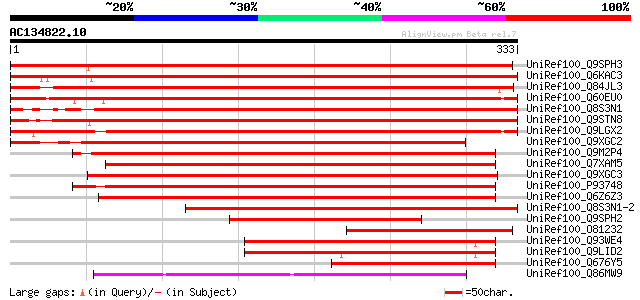
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SPH3 SINAH1 protein [Gossypium hirsutum] 588 e-167
UniRef100_Q6KAC3 Putative Ubiquitin ligase SINAT5 [Oryza sativa] 581 e-164
UniRef100_Q84JL3 Ubiquitin ligase SINAT3 [Arabidopsis thaliana] 569 e-161
UniRef100_Q60EU0 Putative ubiquitin ligase SINAT5 [Oryza sativa] 568 e-161
UniRef100_Q8S3N1 Ubiquitin ligase SINAT5 [Arabidopsis thaliana] 562 e-159
UniRef100_Q9STN8 Ubiquitin ligase SINAT4 [Arabidopsis thaliana] 554 e-156
UniRef100_Q9LGX2 ESTs AU082689 [Oryza sativa] 549 e-155
UniRef100_Q9XGC2 SINA1p [Vitis vinifera] 531 e-149
UniRef100_Q9M2P4 Ubiquitin ligase SINAT2 [Arabidopsis thaliana] 469 e-131
UniRef100_Q7XAM5 Putative developmental protein sina [Oryza sativa] 468 e-130
UniRef100_Q9XGC3 SINA2p [Vitis vinifera] 464 e-129
UniRef100_P93748 Putative ubiquitin ligase SINAT1 [Arabidopsis t... 457 e-127
UniRef100_Q6Z6Z3 Putative SINA2 protein,seven in absentia [Oryza... 449 e-125
UniRef100_Q8S3N1-2 Splice isoform 2 of Q8S3N1 [Arabidopsis thali... 426 e-118
UniRef100_Q9SPH2 SINAH2 protein [Gossypium hirsutum] 234 3e-60
UniRef100_O81232 Seven in absentia homolog [Zea mays] 221 2e-56
UniRef100_Q93WE4 Putative seven in absentia protein [Arabidopsis... 219 1e-55
UniRef100_Q9LID2 Similarity to unknown protein [Arabidopsis thal... 206 9e-52
UniRef100_Q676Y5 SINA [Hyacinthus orientalis] 190 4e-47
UniRef100_Q86MW9 Ubiquitin ligase sina [Schistosoma mansoni] 164 2e-39
>UniRef100_Q9SPH3 SINAH1 protein [Gossypium hirsutum]
Length = 336
Score = 588 bits (1517), Expect = e-167
Identities = 279/336 (83%), Positives = 294/336 (87%), Gaps = 6/336 (1%)
Query: 1 MDLDSIECVSSSDGMDED-EI-QHRILHPHHQQHHHHSEFSSLKPRSGGNNN----HGVI 54
M+ D+IECVSS D + ED EI H +LH H +H H +FSS KP G NN ++
Sbjct: 1 MESDTIECVSSIDEIVEDHEIPHHNLLHHPHPRHAPHHQFSSSKPHHNGTNNVNSISNIV 60
Query: 55 GSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIR 114
G TAIAPA SVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK RVH+RCPTCRQELGDIR
Sbjct: 61 GPTAIAPAASVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKIRVHDRCPTCRQELGDIR 120
Query: 115 CLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINF 174
CLALEKVAESLELPCKYY LGCPE FPYYSKLKHE IC +RPY CPYAGSECS VGDI F
Sbjct: 121 CLALEKVAESLELPCKYYKLGCPETFPYYSKLKHEGICIYRPYNCPYAGSECSVVGDIPF 180
Query: 175 LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMA 234
LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMA
Sbjct: 181 LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMA 240
Query: 235 PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRN 294
PVYMAF+RF+GDE EARNY+YSLE GANGRKLI + PRSIRDSHRKVRDSHDGLIIQRN
Sbjct: 241 PVYMAFIRFVGDETEARNYSYSLEFGANGRKLIRKSAPRSIRDSHRKVRDSHDGLIIQRN 300
Query: 295 MALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPN 330
MALFFSGGDRKELKLRVTGRIWKEQQNPD +CIPN
Sbjct: 301 MALFFSGGDRKELKLRVTGRIWKEQQNPDANMCIPN 336
>UniRef100_Q6KAC3 Putative Ubiquitin ligase SINAT5 [Oryza sativa]
Length = 349
Score = 581 bits (1497), Expect = e-164
Identities = 279/349 (79%), Positives = 295/349 (83%), Gaps = 16/349 (4%)
Query: 1 MDLDSIECVSSSDGMDEDE--------IQHR----ILHPHHQQHHHHSEFSSLKPRSGGN 48
MD+ SIECVS SD MD+D+ + H ++ P + S G
Sbjct: 1 MDMASIECVSYSDSMDDDDDDGVGVGGVSHLPRPILVKPSSAAAAVNVVVVSAGSGGGAG 60
Query: 49 NNHG----VIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCP 104
G V G+ A+ PATSVHELLECPVCTNSMYPPIHQC NGHTLCSTCKTRVHNRCP
Sbjct: 61 GGGGGVGVVAGAPAVPPATSVHELLECPVCTNSMYPPIHQCQNGHTLCSTCKTRVHNRCP 120
Query: 105 TCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGS 164
TCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHE+ CNFRPY CPYAGS
Sbjct: 121 TCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHESQCNFRPYNCPYAGS 180
Query: 165 ECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCL 224
ECS VGDI FLVAHLRDDHKVDMH+GCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCL
Sbjct: 181 ECSVVGDIPFLVAHLRDDHKVDMHSGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCL 240
Query: 225 HFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRD 284
HFEAFQLG+APVYMAFLRFMGDEN+ARNY+YSLEVGANGRK+IWEGTPRSIRDSHRKVRD
Sbjct: 241 HFEAFQLGVAPVYMAFLRFMGDENDARNYSYSLEVGANGRKMIWEGTPRSIRDSHRKVRD 300
Query: 285 SHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
SHDGLIIQRNMALFFSGG+RKELKLRVTGRIWKEQQNPD G CIPNL S
Sbjct: 301 SHDGLIIQRNMALFFSGGERKELKLRVTGRIWKEQQNPDSGACIPNLFS 349
>UniRef100_Q84JL3 Ubiquitin ligase SINAT3 [Arabidopsis thaliana]
Length = 326
Score = 569 bits (1467), Expect = e-161
Identities = 268/333 (80%), Positives = 282/333 (84%), Gaps = 10/333 (3%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDLDS++C S+ D D++EI HQ H ++ S + N S +
Sbjct: 1 MDLDSMDCTSTMDVTDDEEI--------HQDRHSYASVSKHHHTNNNTTNVNAAASGLLP 52
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCLALEK
Sbjct: 53 TTTSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKARVHNRCPTCRQELGDIRCLALEK 112
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCK+ SLGCPEIFPYYSKLKHET+CNFRPY+CPYAGSECS GDI FLVAHLR
Sbjct: 113 VAESLELPCKHMSLGCPEIFPYYSKLKHETVCNFRPYSCPYAGSECSVTGDIPFLVAHLR 172
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMH+GCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 173 DDHKVDMHSGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 232
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE EARNY YSLEVG GRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 233 LRFMGDETEARNYNYSLEVGGYGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFS 292
Query: 301 GGDRKELKLRVTGRIWKEQQ--NPDGGVCIPNL 331
GGDRKELKLRVTGRIWKEQQ GG CIPNL
Sbjct: 293 GGDRKELKLRVTGRIWKEQQQSGEGGGACIPNL 325
>UniRef100_Q60EU0 Putative ubiquitin ligase SINAT5 [Oryza sativa]
Length = 361
Score = 568 bits (1465), Expect = e-161
Identities = 276/363 (76%), Positives = 290/363 (79%), Gaps = 32/363 (8%)
Query: 1 MDLDSIECVSSSDG-MDEDEIQHRILHPHHQQHHHHSEFSSL----------------KP 43
MD+DS+EC+S D MD D++ HH HHHH+ L K
Sbjct: 1 MDMDSVECLSLPDSSMDVDDVDGGG-SVHHHHHHHHALPPHLPAGVAVGVGPGGRAFPKA 59
Query: 44 RSGGNNNHGVIGSTAIA-------------PATSVHELLECPVCTNSMYPPIHQCHNGHT 90
G G G+ A PATSVHELLECPVCTNSM+PPIHQC NGHT
Sbjct: 60 NVAGVGGGGAAGAPAAGGAVAGGGGPGGGPPATSVHELLECPVCTNSMFPPIHQCQNGHT 119
Query: 91 LCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHET 150
LCSTCK RVHNRCPTCRQELGDIRCLALEKVAESLELPCKY SLGCPEIFPYYSK+KHE
Sbjct: 120 LCSTCKARVHNRCPTCRQELGDIRCLALEKVAESLELPCKYCSLGCPEIFPYYSKIKHEA 179
Query: 151 ICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATW 210
C+FRPY CPYAGSEC+ GDI FLVAHLRDDHKVDMH+GCTFNHRYVKSNPREVENATW
Sbjct: 180 QCSFRPYNCPYAGSECAVAGDIPFLVAHLRDDHKVDMHSGCTFNHRYVKSNPREVENATW 239
Query: 211 MLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEG 270
MLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVG NGRK++WEG
Sbjct: 240 MLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGGNGRKMVWEG 299
Query: 271 TPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPN 330
TPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ NPD G CIPN
Sbjct: 300 TPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQTNPD-GACIPN 358
Query: 331 LCS 333
LCS
Sbjct: 359 LCS 361
>UniRef100_Q8S3N1 Ubiquitin ligase SINAT5 [Arabidopsis thaliana]
Length = 309
Score = 562 bits (1449), Expect = e-159
Identities = 268/333 (80%), Positives = 290/333 (86%), Gaps = 25/333 (7%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
M+ DSI+ V +D+DEI HQ+H +FSS K + G + I+
Sbjct: 1 METDSIDSV-----IDDDEI--------HQKH----QFSSTKSQGGA--------TVVIS 35
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
PATSV+ELLECPVCTNSMYPPIHQCHNGHTLCSTCK+RVHNRCPTCRQELGDIRCLALEK
Sbjct: 36 PATSVYELLECPVCTNSMYPPIHQCHNGHTLCSTCKSRVHNRCPTCRQELGDIRCLALEK 95
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCKYY+LGC IFPYYSKLKHE+ CNFRPY+CPYAGSEC+AVGDI FLVAHLR
Sbjct: 96 VAESLELPCKYYNLGCLGIFPYYSKLKHESQCNFRPYSCPYAGSECAAVGDITFLVAHLR 155
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF CFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 156 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFQCFGQYFCLHFEAFQLGMAPVYMAF 215
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE++ARNYTYSLEVG +GRK WEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 216 LRFMGDEDDARNYTYSLEVGGSGRKQTWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFS 275
Query: 301 GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
GGD+KELKLRVTGRIWKEQQNPD GVCI ++CS
Sbjct: 276 GGDKKELKLRVTGRIWKEQQNPDSGVCITSMCS 308
>UniRef100_Q9STN8 Ubiquitin ligase SINAT4 [Arabidopsis thaliana]
Length = 327
Score = 554 bits (1428), Expect = e-156
Identities = 266/337 (78%), Positives = 288/337 (84%), Gaps = 15/337 (4%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHS-EFSSLKPRSGGNNNH---GVIGS 56
M+ DS+ECVSS+ +EI H + H S +FSS K G ++G
Sbjct: 1 METDSMECVSSTG----NEI-------HQNGNGHQSYQFSSTKTHGGAAAAAVVTNIVGP 49
Query: 57 TAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCL 116
TA APATSV+ELLECPVCT SMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCL
Sbjct: 50 TATAPATSVYELLECPVCTYSMYPPIHQCHNGHTLCSTCKVRVHNRCPTCRQELGDIRCL 109
Query: 117 ALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLV 176
ALEKVAESLELPCK+Y+LGCPEIFPYYSKLKHE++CNFRPY+CPYAGSEC VGDI FLV
Sbjct: 110 ALEKVAESLELPCKFYNLGCPEIFPYYSKLKHESLCNFRPYSCPYAGSECGIVGDIPFLV 169
Query: 177 AHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPV 236
AHLRDDHKVDMH G TFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGM PV
Sbjct: 170 AHLRDDHKVDMHAGSTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMGPV 229
Query: 237 YMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMA 296
YMAFLRFMGDE +AR+Y+YSLEVG +GRKL WEGTPRSIRDSHRKVRDS+DGLIIQRNMA
Sbjct: 230 YMAFLRFMGDEEDARSYSYSLEVGGSGRKLTWEGTPRSIRDSHRKVRDSNDGLIIQRNMA 289
Query: 297 LFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
LFFSGGDRKELKLRVTG+IWKEQ +PD G+ IPNL S
Sbjct: 290 LFFSGGDRKELKLRVTGKIWKEQHSPDSGLSIPNLSS 326
>UniRef100_Q9LGX2 ESTs AU082689 [Oryza sativa]
Length = 381
Score = 549 bits (1414), Expect = e-155
Identities = 265/355 (74%), Positives = 280/355 (78%), Gaps = 30/355 (8%)
Query: 1 MDLDSIECVSSSDG----------------------MDEDEIQHRILHPHHQQHHHHSEF 38
MD+DS+EC+S D MD + LH H +
Sbjct: 35 MDVDSVECLSLPDSSSSSGAGAGGGGGGGGGGGVGAMDAADDVGLALHAHGALLAAAAAA 94
Query: 39 SSLKPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
+ GG GS VHELLECPVCTNSMYPPIHQC NGHTLCSTCK R
Sbjct: 95 RAAACSKGGGGGGAAAGS-------GVHELLECPVCTNSMYPPIHQCQNGHTLCSTCKAR 147
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
VHNRCPTCRQELGDIRCLALEKVAESLELPCKY SLGCPEIFPYYSK+KHE C FRPY
Sbjct: 148 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYCSLGCPEIFPYYSKIKHEAQCMFRPYN 207
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCF 218
CPYAGSEC+ VGDI +LVAHLRDDHKVDMH+GCTFNHRYVKSNPREVENATWMLTVFHCF
Sbjct: 208 CPYAGSECAVVGDIPYLVAHLRDDHKVDMHSGCTFNHRYVKSNPREVENATWMLTVFHCF 267
Query: 219 GQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDS 278
GQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNY+YSLEVGANGRK++WEGTPRS+RDS
Sbjct: 268 GQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYSYSLEVGANGRKMVWEGTPRSVRDS 327
Query: 279 HRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
HRKVRDSHDGLIIQRNMALFFSGGDRKELKLR+TGRIWKEQQ PD G CIPNLCS
Sbjct: 328 HRKVRDSHDGLIIQRNMALFFSGGDRKELKLRITGRIWKEQQTPD-GACIPNLCS 381
>UniRef100_Q9XGC2 SINA1p [Vitis vinifera]
Length = 315
Score = 531 bits (1367), Expect = e-149
Identities = 250/299 (83%), Positives = 266/299 (88%), Gaps = 17/299 (5%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
M++DSIECVSS+DG+ E+EI HHHH +FSS + H + S AI+
Sbjct: 1 MEIDSIECVSSTDGIYEEEI-----------HHHHHQFSS------SSKPHSNVVSPAIS 43
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK+RVHNRCPTCRQELGDIRCLALEK
Sbjct: 44 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKSRVHNRCPTCRQELGDIRCLALEK 103
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCKY SLGCPEIFPYYSKLKHE CNFRPY CPYAGSEC+ VGDI FLV+HLR
Sbjct: 104 VAESLELPCKYCSLGCPEIFPYYSKLKHEAQCNFRPYNCPYAGSECAVVGDIPFLVSHLR 163
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF+CFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 164 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFNCFGQYFCLHFEAFQLGMAPVYMAF 223
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFF 299
LRFMGDE EARN++YSLEVGANGRKLIWEGTPRSIRDSH+KVRDSHDGLIIQRNMALFF
Sbjct: 224 LRFMGDEIEARNFSYSLEVGANGRKLIWEGTPRSIRDSHKKVRDSHDGLIIQRNMALFF 282
>UniRef100_Q9M2P4 Ubiquitin ligase SINAT2 [Arabidopsis thaliana]
Length = 308
Score = 469 bits (1207), Expect = e-131
Identities = 219/278 (78%), Positives = 237/278 (84%), Gaps = 6/278 (2%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP S G IG I V+ELLECPVCTN MYPPIHQC NGHTLCS CK RV N
Sbjct: 37 KPGSAG------IGKYGIHSNNGVYELLECPVCTNLMYPPIHQCPNGHTLCSNCKLRVQN 90
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FRPYTCPY
Sbjct: 91 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCHDIFPYYSKLKHEQHCRFRPYTCPY 150
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 151 AGSECSVTGDIPTLVVHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 210
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+GRKL W+G PRSIRDSHRK
Sbjct: 211 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHGRKLTWQGIPRSIRDSHRK 270
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSGGDR+ELKLRVTGRIWKE+
Sbjct: 271 VRDSQDGLIIPRNLALYFSGGDRQELKLRVTGRIWKEE 308
>UniRef100_Q7XAM5 Putative developmental protein sina [Oryza sativa]
Length = 302
Score = 468 bits (1203), Expect = e-130
Identities = 211/256 (82%), Positives = 231/256 (89%)
Query: 64 SVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAE 123
+V ELLECPVC N+MYPPIHQC NGHTLCS CK RVHNRCPTCR ELG+IRCLALEKVA
Sbjct: 47 NVRELLECPVCLNAMYPPIHQCSNGHTLCSGCKPRVHNRCPTCRHELGNIRCLALEKVAA 106
Query: 124 SLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDH 183
SLELPCKY + GC I+PYY KLKHE+ C +RPYTCPYAGSEC+ GDI +LV+HL+DDH
Sbjct: 107 SLELPCKYQNFGCLGIYPYYCKLKHESQCQYRPYTCPYAGSECTVAGDIQYLVSHLKDDH 166
Query: 184 KVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRF 243
KVDMH G TFNHRYVKSNP EVENATWMLTVF CFGQYFCLHFEAFQLGMAPVY+AFLRF
Sbjct: 167 KVDMHNGSTFNHRYVKSNPHEVENATWMLTVFSCFGQYFCLHFEAFQLGMAPVYIAFLRF 226
Query: 244 MGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGD 303
MGD+ EA+NY+YSLEVG +GRK+ W+G PRSIRDSHRKVRDS+DGLIIQRNMALFFSGGD
Sbjct: 227 MGDDAEAKNYSYSLEVGGSGRKMTWQGVPRSIRDSHRKVRDSYDGLIIQRNMALFFSGGD 286
Query: 304 RKELKLRVTGRIWKEQ 319
+KELKLRVTGRIWKEQ
Sbjct: 287 KKELKLRVTGRIWKEQ 302
>UniRef100_Q9XGC3 SINA2p [Vitis vinifera]
Length = 315
Score = 464 bits (1195), Expect = e-129
Identities = 212/269 (78%), Positives = 233/269 (85%)
Query: 52 GVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELG 111
G+ G I+ VHELLECPVCT+ MYPPI+QC +GHTLCS CK+RVHN CPTCR ELG
Sbjct: 40 GLSGKKGISSPNGVHELLECPVCTSLMYPPIYQCPSGHTLCSNCKSRVHNCCPTCRHELG 99
Query: 112 DIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGD 171
DIRCLALEKVAESLELPC+Y SLGC +IFPYYSKLKHE C F PY CPYAG ECS GD
Sbjct: 100 DIRCLALEKVAESLELPCRYQSLGCHDIFPYYSKLKHEQQCRFHPYNCPYAGFECSVTGD 159
Query: 172 INFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQL 231
I LV HL+ DHKVDMH GCTFNHRYVKSNP+EVENATWMLTVF+CFG+ FCLHFEAFQL
Sbjct: 160 IPTLVEHLKGDHKVDMHDGCTFNHRYVKSNPQEVENATWMLTVFNCFGKQFCLHFEAFQL 219
Query: 232 GMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLII 291
G APVYMAFLRFMGD+NEA+ ++YSLEVG N RKLIW+G PRSIRDSHRKVRDS DGLII
Sbjct: 220 GTAPVYMAFLRFMGDDNEAKKFSYSLEVGGNSRKLIWQGVPRSIRDSHRKVRDSQDGLII 279
Query: 292 QRNMALFFSGGDRKELKLRVTGRIWKEQQ 320
QRN+AL+FSGGDR+ELKLRVTGRIWKE +
Sbjct: 280 QRNLALYFSGGDRQELKLRVTGRIWKESE 308
>UniRef100_P93748 Putative ubiquitin ligase SINAT1 [Arabidopsis thaliana]
Length = 305
Score = 457 bits (1177), Expect = e-127
Identities = 212/278 (76%), Positives = 236/278 (84%), Gaps = 5/278 (1%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP G+ + G S+ V+ELLECPVCTN MYPPIHQC NGHTLCS+CK RV N
Sbjct: 33 KPTKSGSGSIGKFHSS-----NGVYELLECPVCTNLMYPPIHQCPNGHTLCSSCKLRVQN 87
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FR Y+CPY
Sbjct: 88 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCQDIFPYYSKLKHEQHCRFRSYSCPY 147
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 148 AGSECSVTGDIPTLVDHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 207
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+ RKL W+G PRSIRDSHRK
Sbjct: 208 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHSRKLTWQGIPRSIRDSHRK 267
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSG D++ELKLRVTGRIWKE+
Sbjct: 268 VRDSQDGLIIPRNLALYFSGSDKEELKLRVTGRIWKEE 305
>UniRef100_Q6Z6Z3 Putative SINA2 protein,seven in absentia [Oryza sativa]
Length = 308
Score = 449 bits (1154), Expect = e-125
Identities = 205/261 (78%), Positives = 229/261 (87%)
Query: 59 IAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLAL 118
++ T +++LLECPVCTNSM PPI QC NGHT+CS CK RV N CPTCRQELG+IRCLAL
Sbjct: 48 LSSLTGLNDLLECPVCTNSMRPPILQCPNGHTICSNCKHRVENHCPTCRQELGNIRCLAL 107
Query: 119 EKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAH 178
EKVAESL+LPCKY SLGC EI PY +KLKHE +C FRPY+CPYAGSEC GD+ LV+H
Sbjct: 108 EKVAESLQLPCKYQSLGCAEIHPYQNKLKHEELCRFRPYSCPYAGSECLIAGDVPMLVSH 167
Query: 179 LRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYM 238
L +DHKVD+H GCTFNHRYVKSNP EVENATWMLTVF CFGQ+FCLHFEAF LGMAPVYM
Sbjct: 168 LINDHKVDLHEGCTFNHRYVKSNPYEVENATWMLTVFKCFGQHFCLHFEAFLLGMAPVYM 227
Query: 239 AFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALF 298
AFLRFMG+++EARN+ YSLEVG NGRKL W+G PRSIRDSH+KVRDS DGLII RNMALF
Sbjct: 228 AFLRFMGEDSEARNFCYSLEVGGNGRKLTWQGIPRSIRDSHKKVRDSFDGLIIHRNMALF 287
Query: 299 FSGGDRKELKLRVTGRIWKEQ 319
FSGG+R+ELKLRVTGRIWKEQ
Sbjct: 288 FSGGNRQELKLRVTGRIWKEQ 308
>UniRef100_Q8S3N1-2 Splice isoform 2 of Q8S3N1 [Arabidopsis thaliana]
Length = 233
Score = 426 bits (1096), Expect = e-118
Identities = 196/218 (89%), Positives = 208/218 (94%)
Query: 116 LALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFL 175
+ALEKVAESLELPCKYY+LGC IFPYYSKLKHE+ CNFRPY+CPYAGSEC+AVGDI FL
Sbjct: 15 IALEKVAESLELPCKYYNLGCLGIFPYYSKLKHESQCNFRPYSCPYAGSECAAVGDITFL 74
Query: 176 VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAP 235
VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF CFGQYFCLHFEAFQLGMAP
Sbjct: 75 VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFQCFGQYFCLHFEAFQLGMAP 134
Query: 236 VYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNM 295
VYMAFLRFMGDE++ARNYTYSLEVG +GRK WEGTPRS+RDSHRKVRDSHDGLIIQRNM
Sbjct: 135 VYMAFLRFMGDEDDARNYTYSLEVGGSGRKQTWEGTPRSVRDSHRKVRDSHDGLIIQRNM 194
Query: 296 ALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
ALFFSGGD+KELKLRVTGRIWKEQQNPD GVCI ++CS
Sbjct: 195 ALFFSGGDKKELKLRVTGRIWKEQQNPDSGVCITSMCS 232
>UniRef100_Q9SPH2 SINAH2 protein [Gossypium hirsutum]
Length = 143
Score = 234 bits (596), Expect = 3e-60
Identities = 100/126 (79%), Positives = 117/126 (92%)
Query: 145 KLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPRE 204
+LKHE+ C++RPY+CPYAGSEC+ +GD +LVAHL+DDHKVDMH G TFNHRYVKSNP E
Sbjct: 18 ELKHESQCSYRPYSCPYAGSECTVIGDFPYLVAHLKDDHKVDMHNGSTFNHRYVKSNPHE 77
Query: 205 VENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGR 264
VENATWMLTVF CFGQYFCLHFEAFQLG++PVY+AFLRFMGD+NEA+NY+YSLEVG NGR
Sbjct: 78 VENATWMLTVFSCFGQYFCLHFEAFQLGISPVYIAFLRFMGDDNEAKNYSYSLEVGGNGR 137
Query: 265 KLIWEG 270
K+IW+G
Sbjct: 138 KMIWQG 143
>UniRef100_O81232 Seven in absentia homolog [Zea mays]
Length = 113
Score = 221 bits (564), Expect = 2e-56
Identities = 103/109 (94%), Positives = 107/109 (97%)
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDEN+ARNY+YSLEVGANGRK+IWEGTPRSIRDSHRK
Sbjct: 5 FCLHFEAFQLGMAPVYMAFLRFMGDENDARNYSYSLEVGANGRKMIWEGTPRSIRDSHRK 64
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPN 330
VRDSHDGLIIQRNMALFFSGG+RKELKLRVTGRIWKEQQNPD G CIPN
Sbjct: 65 VRDSHDGLIIQRNMALFFSGGERKELKLRVTGRIWKEQQNPDSGACIPN 113
>UniRef100_Q93WE4 Putative seven in absentia protein [Arabidopsis thaliana]
Length = 216
Score = 219 bits (557), Expect = 1e-55
Identities = 98/168 (58%), Positives = 130/168 (77%), Gaps = 3/168 (1%)
Query: 155 RPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTV 214
+PY CP++G++C GDI L+ HLR+DH V+M G +F+HRYV +P+ + +ATWMLT+
Sbjct: 45 KPYNCPHSGAKCDVTGDIQRLLLHLRNDHNVEMSDGRSFSHRYVHHDPKHLHHATWMLTL 104
Query: 215 FHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRS 274
C G+ FCL+FEAF L P+YMAF++FMGDE EA +++YSL+VG NGRKL W+G PRS
Sbjct: 105 LDCCGRKFCLYFEAFHLRKTPMYMAFMQFMGDEEEAMSFSYSLQVGGNGRKLTWQGVPRS 164
Query: 275 IRDSHRKVRDSHDGLIIQRNMALFFSGGDR---KELKLRVTGRIWKEQ 319
IRDSH+ VRDS DGLII R +ALFFS + KELKL+V+GR+W+EQ
Sbjct: 165 IRDSHKTVRDSQDGLIITRKLALFFSTDNNTTDKELKLKVSGRVWREQ 212
>UniRef100_Q9LID2 Similarity to unknown protein [Arabidopsis thaliana]
Length = 273
Score = 206 bits (523), Expect = 9e-52
Identities = 99/195 (50%), Positives = 131/195 (66%), Gaps = 30/195 (15%)
Query: 155 RPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTV 214
+PY CP++G++C GDI L+ HLR+DH V+M G +F+HRYV +P+ + +ATWMLTV
Sbjct: 75 KPYNCPHSGAKCDVTGDIQRLLLHLRNDHNVEMSDGRSFSHRYVHHDPKHLHHATWMLTV 134
Query: 215 FH---------------------------CFGQYFCLHFEAFQLGMAPVYMAFLRFMGDE 247
+ C G+ FCL+FEAF L P+YMAF++FMGDE
Sbjct: 135 SYITDYLALFLQLCEFLSFNPLETMQLLDCCGRKFCLYFEAFHLRKTPMYMAFMQFMGDE 194
Query: 248 NEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDR--- 304
EA +++YSL+VG NGRKL W+G PRSIRDSH+ VRDS DGLII R +ALFFS +
Sbjct: 195 EEAMSFSYSLQVGGNGRKLTWQGVPRSIRDSHKTVRDSQDGLIITRKLALFFSTDNNTTD 254
Query: 305 KELKLRVTGRIWKEQ 319
KELKL+V+GR+W+EQ
Sbjct: 255 KELKLKVSGRVWREQ 269
>UniRef100_Q676Y5 SINA [Hyacinthus orientalis]
Length = 108
Score = 190 bits (483), Expect = 4e-47
Identities = 89/108 (82%), Positives = 99/108 (91%)
Query: 212 LTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGT 271
LTV GQYFCLHFEAFQLGMAPVY AFLRFMGD++EA+NY+YS EVGA GRK+IW+G
Sbjct: 1 LTVSVASGQYFCLHFEAFQLGMAPVYRAFLRFMGDDSEAKNYSYSREVGATGRKMIWQGV 60
Query: 272 PRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
PRSIRDSHRKVRDS+D ++IQRN+ALFFSGGDRKELKLRVTGRIWKEQ
Sbjct: 61 PRSIRDSHRKVRDSYDAIVIQRNIALFFSGGDRKELKLRVTGRIWKEQ 108
>UniRef100_Q86MW9 Ubiquitin ligase sina [Schistosoma mansoni]
Length = 371
Score = 164 bits (416), Expect = 2e-39
Identities = 85/245 (34%), Positives = 131/245 (52%), Gaps = 3/245 (1%)
Query: 56 STAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRC 115
+T+ + + + L ECPVC + PPI QC +GH +C++C++++ + CPTCR L +IR
Sbjct: 111 NTSDSSSIDLASLFECPVCMDYALPPIMQCQSGHIVCASCRSKLSS-CPTCRGNLDNIRN 169
Query: 116 LALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFL 175
LA+EK+A S+ PCKY + GCPE F Y SK +HE C +RPY CP G+ C +G++ +
Sbjct: 170 LAMEKLASSVLFPCKYSTSGCPETFHYTSKSEHEAACEYRPYDCPCPGASCKWLGELEQV 229
Query: 176 VAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAP 235
+ HL HK T + + A + + CFG F L E +
Sbjct: 230 MPHLVHHHK--SITTLQGEDIVFLATDISLPGAVDWVMMQSCFGHSFMLVLEKQERVPDQ 287
Query: 236 VYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNM 295
++ A ++ +G +A + Y LE+ + R+L WE PRSI D + D L+ N
Sbjct: 288 IFFALVQLIGTRKQADQFVYRLELNGHRRRLTWEACPRSIHDGVQSAIAVSDCLVFDSNT 347
Query: 296 ALFFS 300
A F+
Sbjct: 348 AHSFA 352
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 616,588,900
Number of Sequences: 2790947
Number of extensions: 26104460
Number of successful extensions: 77177
Number of sequences better than 10.0: 554
Number of HSP's better than 10.0 without gapping: 148
Number of HSP's successfully gapped in prelim test: 411
Number of HSP's that attempted gapping in prelim test: 76355
Number of HSP's gapped (non-prelim): 769
length of query: 333
length of database: 848,049,833
effective HSP length: 128
effective length of query: 205
effective length of database: 490,808,617
effective search space: 100615766485
effective search space used: 100615766485
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC134822.10


