

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.18 - phase: 0
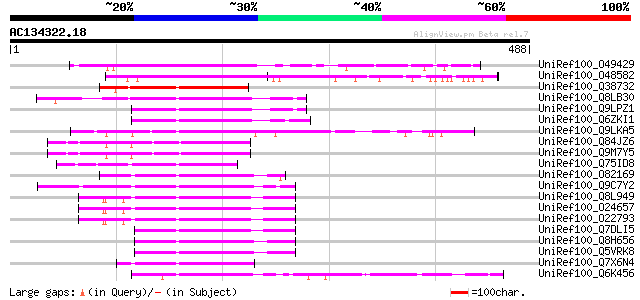
(488 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O49429 Putative DAG protein [Arabidopsis thaliana] 231 5e-59
UniRef100_O48582 Similar to pMS10 protein [Arabidopsis thaliana] 172 1e-41
UniRef100_Q38732 DAG protein, chloroplast precursor [Antirrhinum... 145 3e-33
UniRef100_Q8LB30 DAG protein, putative [Arabidopsis thaliana] 145 3e-33
UniRef100_Q9LPZ1 T23J18.10 [Arabidopsis thaliana] 144 4e-33
UniRef100_Q6ZKI1 Putative DAG protein [Oryza sativa] 144 7e-33
UniRef100_Q9LKA5 Unknown mitochondrial protein At3g15000 [Arabid... 141 5e-32
UniRef100_Q84JZ6 Putative DAG protein [Arabidopsis thaliana] 135 3e-30
UniRef100_Q9M7Y5 DAG protein, putative [Arabidopsis thaliana] 134 5e-30
UniRef100_Q75ID8 Putative chloroplast differentiation and palisa... 122 2e-26
UniRef100_O82169 Hypothetical protein At2g35240 [Arabidopsis tha... 121 4e-26
UniRef100_Q9C7Y2 Plastid protein, putative; 23108-24430 [Arabido... 120 7e-26
UniRef100_Q8L949 Plastid protein [Arabidopsis thaliana] 119 2e-25
UniRef100_O24657 DAL1 protein [Arabidopsis thaliana] 119 2e-25
UniRef100_O22793 Plastid protein [Arabidopsis thaliana] 119 2e-25
UniRef100_Q7DLI5 Plastid protein [Arabidopsis thaliana] 119 3e-25
UniRef100_Q8H656 Putative plastid protein [Oryza sativa] 116 2e-24
UniRef100_Q5VRK8 Putative DAL1 protein [Oryza sativa] 116 2e-24
UniRef100_Q7X6N4 OSJNBa0041A02.8 protein [Oryza sativa] 115 4e-24
UniRef100_Q6K456 Putative plastid protein [Oryza sativa] 115 4e-24
>UniRef100_O49429 Putative DAG protein [Arabidopsis thaliana]
Length = 419
Score = 231 bits (588), Expect = 5e-59
Identities = 165/406 (40%), Positives = 205/406 (49%), Gaps = 69/406 (16%)
Query: 57 TATSLSVPITGSISSHFS-LSKSPNVVDGIQSRSF--RSTSIS------LLSSRYGETSE 107
TATS + SISS + S P+V + RS RST ++ L S+R + +
Sbjct: 14 TATSY---VNRSISSSITPASDFPSVSAAVLKRSVIGRSTEVATRAPARLFSTRQYKLYK 70
Query: 108 LSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPP-PEEMIRIYEETCAKGLNISVEEAKKK 166
EI DT+LFEGCDYNHWL DF ++ P PEEM+ YEETCA+GL ISVEEAK++
Sbjct: 71 EGDEITEDTVLFEGCDYNHWLITMDFSKEETPKSPEEMVAAYEETCAQGLGISVEEAKQR 130
Query: 167 IYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRP 226
+YACSTTTY GFQA+MTE+ES+KF+ +PGV+F+LPDSYIDP NK+YGGD+Y G I RP
Sbjct: 131 MYACSTTTYQGFQAIMTEQESEKFKDLPGVVFILPDSYIDPQNKEYGGDKYENGVITHRP 190
Query: 227 PPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASH 286
PP+Q GR R P ++ R G G PQNF +GQ
Sbjct: 191 PPIQSGR----------------ARPRPRFD------RSGGGSGGPQNFQRNTQYGQQ-- 226
Query: 287 IPPQQNIGQQQPNIRIDSTSQRFPPQQSY-----DQASQRYPP-QQSYDQASQGYLPKQN 340
PP Q G + PQQ Y Q +Q PP Q Y+Q + P
Sbjct: 227 -PPMQGGGGS------------YGPQQGYATPGQGQGTQAPPPFQGGYNQGPRSPPPPYQ 273
Query: 341 YGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYA--PQQNFGPPGE 398
G Q +P P Q G P Y QS G+G NY+ PQ + G
Sbjct: 274 AGY-NQGQGSPVPPYQAGYNQVQGSPVPPYQGTQS--SYGQGGSGNYSQGPQGGYNQGGP 330
Query: 399 VDRRNYAPQ--QNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSE 442
RNY PQ NFGP N P G PG G+ G S P +
Sbjct: 331 ---RNYNPQGAGNFGP--ASGAGNLGPAPGAGNPGYGQ-GYSGPGQ 370
Score = 42.4 bits (98), Expect = 0.031
Identities = 34/123 (27%), Positives = 52/123 (41%), Gaps = 15/123 (12%)
Query: 345 PQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNY 404
P +Y PQ + + G + + PP QS R P+ + G +N+
Sbjct: 165 PDSYIDPQNKEYGGDKYENGVITHRPPPIQS-------GRARPRPRFDRSGGGSGGPQNF 217
Query: 405 APQQNFG--PPRQGERRNYVPQQNFGPPGQGERGNSVPS-EGGWDF-----KPSYMEEFE 456
+G PP QG +Y PQQ + PGQG+ + P +GG++ P Y +
Sbjct: 218 QRNTQYGQQPPMQGGGGSYGPQQGYATPGQGQGTQAPPPFQGGYNQGPRSPPPPYQAGYN 277
Query: 457 QGQ 459
QGQ
Sbjct: 278 QGQ 280
>UniRef100_O48582 Similar to pMS10 protein [Arabidopsis thaliana]
Length = 723
Score = 172 bits (437), Expect = 1e-41
Identities = 150/449 (33%), Positives = 196/449 (43%), Gaps = 91/449 (20%)
Query: 91 RSTSISLLSSRYGETSELS--PEIGPDTIL--FEGCDYNHWLFVCDFPRDNKPPPEEMIR 146
+ST+I+ +R T++ P G D+ + EGCD+NHWL +FP+DN P EEMI
Sbjct: 45 QSTAINRSPARLFSTTQYQYDPYTGEDSFMPDNEGCDFNHWLITMNFPKDNLPSREEMIS 104
Query: 147 IYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYID 206
I+E+TCAKGL IS+EEAKKKIYA TT+Y GFQA MT E +KF +PGV +++PDSYID
Sbjct: 105 IFEQTCAKGLAISLEEAKKKIYAICTTSYQGFQATMTIGEVEKFRDLPGVQYIIPDSYID 164
Query: 207 PVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQ-------------------- 246
NK YGGD+Y G I P P PV + + Q
Sbjct: 165 VENKVYGGDKYENGVITPGPVPVPTKEGFDSLKKESKPEQEEAEIILTPPDEGKTSGQVQ 224
Query: 247 ------LPNNRG------------NPSYNNRDSMPRDGRNYGPPQNFPPQ-QNHGQASHI 287
LP+ R + S+ G+ G + P Q Q+ GQ +
Sbjct: 225 GQGSLTLPDQRSVKERQGTLALVQGQGQRSGMSILGQGQGEGRRMSIPGQWQSRGQGNSF 284
Query: 288 PPQQNIGQQQPNIRIDST--SQRFPPQQSYDQASQRYP------PQQSYDQASQGYLPKQ 339
Q +R T S P Q + Q P Q+S +SQG L +Q
Sbjct: 285 QGSFKQSQGTLPVRKGQTQISDEIPSFQGNVKQRQEMPIHGQGQAQRSQMPSSQGTL-RQ 343
Query: 340 NYGQAPQ------NYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNF 393
Q Q Y+ Q Q Q G +P Y Q+S G+G Y N
Sbjct: 344 GQAQGSQRPSNQVGYNQGQGAQTPPYHQGQGAQTPPY--QESPNNYGQGAFVQY----NQ 397
Query: 394 GPP-GEV-----DRRNYAPQQNFGPPRQGERRNYVPQQNFGPP----GQGE--------R 435
GPP G V ++ N Q N+ P G NY P Q G P GQG+ R
Sbjct: 398 GPPQGNVVQTTQEKYNQMGQGNYAPQSGG---NYSPAQGAGSPRFGYGQGQGGQLLSPYR 454
Query: 436 GNSVPSEG------GWDFKPSYMEEFEQG 458
GN +G G + +PSY F QG
Sbjct: 455 GNYNQGQGTPLPGQGQEGQPSYQMGFSQG 483
Score = 52.4 bits (124), Expect = 3e-05
Identities = 70/261 (26%), Positives = 96/261 (35%), Gaps = 64/261 (24%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPP-QQNIGQQQPNIR 301
Q +Q+P+++G R G N GQ + PP Q G Q P +
Sbjct: 330 QRSQMPSSQGTLRQGQAQGSQRPSNQVG--------YNQGQGAQTPPYHQGQGAQTPPYQ 381
Query: 302 IDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ 361
+ Y+Q PPQ + Q +Q + Y Q Q APQ+ NYS Q
Sbjct: 382 ESPNNYGQGAFVQYNQG----PPQGNVVQTTQ-----EKYNQMGQGNYAPQSGGNYSPAQ 432
Query: 362 TYGPASPQYPPQQSFG-----------------PL-GEGERRNYAPQQNFG-------PP 396
G SP++ Q G PL G+G+ + Q F PP
Sbjct: 433 --GAGSPRFGYGQGQGGQLLSPYRGNYNQGQGTPLPGQGQEGQPSYQMGFSQGLGAPVPP 490
Query: 397 GEVDRRNYA-----------PQQNF--GPPR---QGERRNYVPQQ--NFGPPGQGERGNS 438
+V NY PQ NF GP + QG + NY PQ ++GP G+
Sbjct: 491 NQVIPGNYGQWAFVNYNQGPPQGNFLQGPQQNHNQGGQWNYSPQNGGHYGPAQFGQWYPG 550
Query: 439 VPSEGGWDFKPSYMEEFEQGQ 459
P G + P Y + QGQ
Sbjct: 551 PPQGQGIQW-PQYQLSYNQGQ 570
Score = 45.8 bits (107), Expect = 0.003
Identities = 77/258 (29%), Positives = 97/258 (36%), Gaps = 63/258 (24%)
Query: 217 YIEGQIIPRP-------PPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNY 269
Y +GQ P P P Q G + G N +P N G ++ N N
Sbjct: 457 YNQGQGTPLPGQGQEGQPSYQMGFSQGLGAPVPPNQVIPGNYGQWAFVN--------YNQ 508
Query: 270 GPPQ-NF--PPQQNHGQ----------ASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYD 316
GPPQ NF PQQNH Q H P Q GQ P Q Q SY+
Sbjct: 509 GPPQGNFLQGPQQNHNQGGQWNYSPQNGGHYGPAQ-FGQWYPGPPQGQGIQWPQYQLSYN 567
Query: 317 QASQ-------RYP-----PQQSYDQASQG-YLPKQNYGQ--APQNYSAPQAPQNYSQQQ 361
Q R P Q Y+ QG ++P+Q GQ A +Y A S Q
Sbjct: 568 QGQGTPFSGQCRCPNCGMTSYQGYNNQGQGTHIPEQWEGQDYAVLSYQA-------SYNQ 620
Query: 362 TYGPASPQYP---PQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQ--NFGPPRQG 416
+G +P Y Q + G G+G N+ Q F V+ NY Q N+GPP G
Sbjct: 621 AHGAQAPPYHGNYNQATPGGYGQGTSANF--NQRF----PVNPANYNMQNGGNYGPP-HG 673
Query: 417 ERRNYVPQQNFGPPGQGE 434
N +Q F GQ +
Sbjct: 674 LAGNPGFRQGFSGQGQNQ 691
>UniRef100_Q38732 DAG protein, chloroplast precursor [Antirrhinum majus]
Length = 230
Score = 145 bits (366), Expect = 3e-33
Identities = 78/145 (53%), Positives = 98/145 (66%), Gaps = 7/145 (4%)
Query: 85 IQSRSFRSTSISLL-----SSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKP 139
I+SRS ++ L SSR + S E +TI+ GCDYNHWL V +FP+D P
Sbjct: 43 IKSRSAAYPTVRALTDGEYSSRRNNNNNNSGE-ERETIMLPGCDYNHWLIVMEFPKDPAP 101
Query: 140 PPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFV 199
E+MI Y T A L S+EEAKK +YA STTTYTGFQ +TEE S+KF+G+PGV++V
Sbjct: 102 TREQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTTYTGFQCTVTEETSEKFKGLPGVLWV 160
Query: 200 LPDSYIDPVNKQYGGDQYIEGQIIP 224
LPDSYID NK YGGD+Y+ G+IIP
Sbjct: 161 LPDSYIDVKNKDYGGDKYVNGEIIP 185
>UniRef100_Q8LB30 DAG protein, putative [Arabidopsis thaliana]
Length = 232
Score = 145 bits (365), Expect = 3e-33
Identities = 101/262 (38%), Positives = 127/262 (47%), Gaps = 46/262 (17%)
Query: 26 LLSTTTAMASSLILLR-------RSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKS 78
+ S TT +SSL+L +LSGI S T S+ GS
Sbjct: 1 MASFTTTSSSSLLLKTLLPVSHLNRFSTLSGIRVGDSWTPLLRSISTAGS---------- 50
Query: 79 PNVVDGIQSRSFRSTSISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNK 138
R + + S Y S E +TI+ GCDYNHWL V +FP+D
Sbjct: 51 --------RRRVAIVKAATVDSDYSSKRSNSNE-QRETIMLPGCDYNHWLIVMEFPKDPA 101
Query: 139 PPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIF 198
P ++MI Y T A L S+EEAKK +YA STTTYTGFQ + EE S+KF+G+PGV++
Sbjct: 102 PSRDQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTTYTGFQCTIDEETSEKFKGLPGVLW 160
Query: 199 VLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSY-N 257
VLPDSYID NK YGGD+YI G+IIP P P R N Y +
Sbjct: 161 VLPDSYIDVKNKDYGGDKYINGEIIPCTYP----------------TYQPKQRNNTKYQS 204
Query: 258 NRDSMPRDGRNYGPPQNFPPQQ 279
R RDG PP+ P+Q
Sbjct: 205 KRYERKRDGP--PPPEQRKPRQ 224
>UniRef100_Q9LPZ1 T23J18.10 [Arabidopsis thaliana]
Length = 232
Score = 144 bits (364), Expect = 4e-33
Identities = 80/166 (48%), Positives = 99/166 (59%), Gaps = 20/166 (12%)
Query: 115 DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTT 174
+TI+ GCDYNHWL V +FP+D P ++MI Y T A L S+EEAKK +YA STTT
Sbjct: 78 ETIMLPGCDYNHWLIVMEFPKDPAPSRDQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTT 136
Query: 175 YTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRN 234
YTGFQ + EE S+KF+G+PGV++VLPDSYID NK YGGD+YI G+IIP P
Sbjct: 137 YTGFQCTIDEETSEKFKGLPGVLWVLPDSYIDVKNKDYGGDKYINGEIIPCTYP------ 190
Query: 235 LGGRRDYRQNNQLPNNRGNPSY-NNRDSMPRDGRNYGPPQNFPPQQ 279
P R N Y + R RDG PP+ P+Q
Sbjct: 191 ----------TYQPKQRNNTKYQSKRYERKRDGP--PPPEQRKPRQ 224
>UniRef100_Q6ZKI1 Putative DAG protein [Oryza sativa]
Length = 229
Score = 144 bits (362), Expect = 7e-33
Identities = 80/169 (47%), Positives = 98/169 (57%), Gaps = 20/169 (11%)
Query: 115 DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTT 174
+TIL GCDYNHWL V +FP+D P E+MI Y T A L S+EEAKK +YA STTT
Sbjct: 74 ETILLPGCDYNHWLIVMEFPKDPAPTREQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTT 132
Query: 175 YTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRN 234
YTGFQ + EE S+KF+G+PGV++VLPDSYID NK YGGD+YI G+IIP P
Sbjct: 133 YTGFQCTVDEETSEKFKGLPGVLWVLPDSYIDVKNKDYGGDKYINGEIIPCTYP------ 186
Query: 235 LGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQ 283
P R Y +R + R GPP + P+ Q
Sbjct: 187 ----------TYQPKERRTSKYESR---RYERRRDGPPASRRPRPQTAQ 222
>UniRef100_Q9LKA5 Unknown mitochondrial protein At3g15000 [Arabidopsis thaliana]
Length = 395
Score = 141 bits (355), Expect = 5e-32
Identities = 128/413 (30%), Positives = 176/413 (41%), Gaps = 69/413 (16%)
Query: 58 ATSLSVPITGSISSHFSLSKSPNVVDGIQSRS----------FRSTSISLLSSRYGETSE 107
A SLS T S +S L+KSP + SRS FR +S+ TS
Sbjct: 15 AKSLSFLFTRSFASSAPLAKSP--ASSLLSRSRPLVAAFSSVFRGGLVSVKGLSTQATSS 72
Query: 108 LSPEIGP--------DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNIS 159
+ P +TIL +GCD+ HWL V + P +P +E+I Y +T A+ + S
Sbjct: 73 SLNDPNPNWSNRPPKETILLDGCDFEHWLVVVE-PPQGEPTRDEIIDSYIKTLAQIVG-S 130
Query: 160 VEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIE 219
+EA+ KIY+ ST Y F A+++E+ S K + + V +VLPDSY+D NK YGG+ +I+
Sbjct: 131 EDEARMKIYSVSTRCYYAFGALVSEDLSHKLKELSNVRWVLPDSYLDVRNKDYGGEPFID 190
Query: 220 GQIIPRPPPV--QFGRNLGGRRDYRQNNQLP--NNRGNPSYNNRDSMPRDGRNYGPPQNF 275
G+ +P P ++ RN + + N P N+R R++M PP
Sbjct: 191 GKAVPYDPKYHEEWIRNNARANERNRRNDRPRNNDRSRNFERRRENMAGGPPPQRPPMGG 250
Query: 276 PPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGY 335
PP H S PP G P + Q + P + R+PP
Sbjct: 251 PPPPPHIGGSAPPPPHMGGSAPPPPHM---GQNYGPPPPNNMGGPRHPPP---------- 297
Query: 336 LPKQNYGQAPQ-NYSAPQAPQNYSQQQTYGPASPQY---PPQQSFGPLGEGERRNYAPQQ 391
YG PQ N P+ PQNY G P Y PP + G AP
Sbjct: 298 -----YGAPPQNNMGGPRPPQNYG-----GTPPPNYGGAPPANNMGG---------APPP 338
Query: 392 NF--GPP---GEVDRRNY--APQQNFGPPRQGERRNYVPQQNFGPPGQGERGN 437
N+ GPP G V Y AP QN +QG QN PP + GN
Sbjct: 339 NYGGGPPPQYGAVPPPQYGGAPPQNNNYQQQGSGMQQPQYQNNYPPNRDGSGN 391
>UniRef100_Q84JZ6 Putative DAG protein [Arabidopsis thaliana]
Length = 244
Score = 135 bits (340), Expect = 3e-30
Identities = 88/197 (44%), Positives = 115/197 (57%), Gaps = 10/197 (5%)
Query: 36 SLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQSRS--FRST 93
+LI RR+L +L S S+T+ S S P T S S F++ V S + ST
Sbjct: 2 ALISTRRTLSTLLNKTLS-SSTSYSSSFP-TLSSRSRFAMPLIEKVSSSRTSLGPCYIST 59
Query: 94 SISLLSSRYGETSELSPEIG----PDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYE 149
S Y ++ SP +TIL +GCDY HWL V +F D KP EEMI Y
Sbjct: 60 RPKTSGSGYSPLNDPSPNWSNRPPKETILLDGCDYEHWLIVMEFT-DPKPTEEEMINSYV 118
Query: 150 ETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVN 209
+T L EEAKKKIY+ T+TYTGF A+++EE S K + +PGV++VLPDSY+D N
Sbjct: 119 KTLTSVLGCE-EEAKKKIYSVCTSTYTGFGALISEELSCKVKALPGVLWVLPDSYLDVPN 177
Query: 210 KQYGGDQYIEGQIIPRP 226
K YGGD Y+EG++IPRP
Sbjct: 178 KDYGGDLYVEGKVIPRP 194
>UniRef100_Q9M7Y5 DAG protein, putative [Arabidopsis thaliana]
Length = 244
Score = 134 bits (338), Expect = 5e-30
Identities = 88/197 (44%), Positives = 115/197 (57%), Gaps = 10/197 (5%)
Query: 36 SLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQSRS--FRST 93
+LI RR+L +L S S+T+ S S P T S S F++ V S + ST
Sbjct: 2 ALISTRRTLSTLLNKTLS-SSTSYSSSFP-TLSSRSRFAMPLIEKVSSSRTSLGPCYIST 59
Query: 94 SISLLSSRYGETSELSPEIG----PDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYE 149
S Y ++ SP +TIL +GCDY HWL V +F D KP EEMI Y
Sbjct: 60 RPKTSGSGYSPLNDPSPNWSNRPPKETILLDGCDYEHWLIVMEFT-DPKPTEEEMINSYV 118
Query: 150 ETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVN 209
+T L EEAKKKIY+ T+TYTGF A+++EE S K + +PGV++VLPDSY+D N
Sbjct: 119 KTLTSVLGWQ-EEAKKKIYSVCTSTYTGFGALISEELSCKVKALPGVLWVLPDSYLDVPN 177
Query: 210 KQYGGDQYIEGQIIPRP 226
K YGGD Y+EG++IPRP
Sbjct: 178 KDYGGDLYVEGKVIPRP 194
>UniRef100_Q75ID8 Putative chloroplast differentiation and palisade
development-related protein [Oryza sativa]
Length = 180
Score = 122 bits (307), Expect = 2e-26
Identities = 74/170 (43%), Positives = 101/170 (58%), Gaps = 6/170 (3%)
Query: 45 RSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQSRSFRSTSISLLSSRYGE 104
RSLS + S A + P+ + SS + +P G ++ S + S L+
Sbjct: 11 RSLSALLLSSRALQRRFA-PLAAAASSAYLAPWAPPS-RGAKTASSGGSGYSPLNDPSPN 68
Query: 105 TSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAK 164
S P+ +TIL +GCDY HWL V +FP D KP E+M+ Y +T A + S EEAK
Sbjct: 69 WSNRPPK---ETILLDGCDYEHWLIVMEFPTDPKPSEEDMVAAYVKTLAAVVG-SEEEAK 124
Query: 165 KKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGG 214
KKIY+ TTTYTGF A+++EE S K +G+PGV++VLPDSY+D NK YGG
Sbjct: 125 KKIYSVCTTTYTGFGALISEELSYKVKGLPGVLWVLPDSYLDVPNKDYGG 174
>UniRef100_O82169 Hypothetical protein At2g35240 [Arabidopsis thaliana]
Length = 232
Score = 121 bits (304), Expect = 4e-26
Identities = 74/178 (41%), Positives = 99/178 (55%), Gaps = 18/178 (10%)
Query: 85 IQSRSFRST-SISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEE 143
I++R RS S S L S + E+ P LF GCDY HWL V + P ++
Sbjct: 59 IRTRMDRSGGSYSPLKSGSNFSDRPPTEMAP---LFPGCDYEHWLIVMEKPGGENAQKQQ 115
Query: 144 MIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDS 203
MI Y +T AK + S EEA+KKIY S Y GF + EE S K EG+PGV+FVLPDS
Sbjct: 116 MIDCYVQTLAKIVG-SEEEARKKIYNVSCERYFGFGCEIDEETSNKLEGLPGVLFVLPDS 174
Query: 204 YIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGN--PSYNNR 259
Y+DP K YG + ++ G+++PRPP Q R+ +L N RG+ P Y++R
Sbjct: 175 YVDPEFKDYGAELFVNGEVVPRPPERQ-----------RRMVELTNQRGSDKPKYHDR 221
>UniRef100_Q9C7Y2 Plastid protein, putative; 23108-24430 [Arabidopsis thaliana]
Length = 229
Score = 120 bits (302), Expect = 7e-26
Identities = 84/243 (34%), Positives = 119/243 (48%), Gaps = 17/243 (6%)
Query: 27 LSTTTAMASSLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQ 86
++ T A +++ + +R + + S S + S P+ S S N I+
Sbjct: 1 MAKTLARSTASRITKRLISTSGATTPSPSYILSRRSTPV---FSHAVGFISSLNRFTTIR 57
Query: 87 SRSFRST-SISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMI 145
+R RS S S L S + E+ P LF GCDY HWL V D P ++MI
Sbjct: 58 TRMDRSGGSYSPLKSGSNFSDRAPTEMAP---LFPGCDYEHWLIVMDKPGGENATKQQMI 114
Query: 146 RIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYI 205
Y +T AK + S EEAKKKIY S Y GF + EE S K EG+PGV+F+LPDSY+
Sbjct: 115 DCYVQTLAKIIG-SEEEAKKKIYNVSCERYFGFGCEIDEETSNKLEGLPGVLFILPDSYV 173
Query: 206 DPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRD 265
D NK YG + ++ G+I+ RPP Q R+ Q N++ P Y+++ R
Sbjct: 174 DQENKDYGAELFVNGEIVQRPPERQ-------RKIIELTTQRTNDK--PKYHDKTRYVRR 224
Query: 266 GRN 268
N
Sbjct: 225 REN 227
>UniRef100_Q8L949 Plastid protein [Arabidopsis thaliana]
Length = 219
Score = 119 bits (299), Expect = 2e-25
Identities = 82/221 (37%), Positives = 109/221 (49%), Gaps = 31/221 (14%)
Query: 65 ITGSISSHFSLSKSPNVVDGIQ---SR---SFRSTSISLLSSRYGET----------SEL 108
+T ++ S+ +L P + + SR S R SI ++R G T S+
Sbjct: 11 LTRALLSNVTLMAPPRIPSSVHYGGSRLGCSTRFFSIRCGANRSGSTYSPLNSGSNFSDR 70
Query: 109 SP-EIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKI 167
P E+ P LF GCDY HWL V D P ++MI Y +T AK + S EEAKK+I
Sbjct: 71 PPTEMAP---LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVVG-SEEEAKKRI 126
Query: 168 YACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP 227
Y S Y GF + EE S K EG+PGV+FVLPDSY+DP NK YG + ++ G+I+ R P
Sbjct: 127 YNVSCERYLGFGCEIDEETSTKLEGLPGVLFVLPDSYVDPENKDYGAELFVNGEIVQRSP 186
Query: 228 PVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN 268
R R Q + P YN+R R N
Sbjct: 187 ----------ERQRRVEPQPQRXQDRPRYNDRTRYSRRREN 217
>UniRef100_O24657 DAL1 protein [Arabidopsis thaliana]
Length = 219
Score = 119 bits (299), Expect = 2e-25
Identities = 83/221 (37%), Positives = 108/221 (48%), Gaps = 31/221 (14%)
Query: 65 ITGSISSHFSLSKSPNVVDGIQ---SR---SFRSTSISLLSSRYGET----------SEL 108
+T ++ S+ +L P + + SR S R SI ++R G T S+
Sbjct: 11 LTRALLSNVTLMAPPRIPSSVHYGGSRLGCSTRFFSIRCGANRSGSTYSPLNSGSNFSDR 70
Query: 109 SP-EIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKI 167
P E+ P LF GCDY HWL V D P EMI Y +T AK + S EEAKK+I
Sbjct: 71 PPTEMAP---LFPGCDYEHWLIVMDKPGGEGATKHEMIDCYIQTLAKVVG-SEEEAKKRI 126
Query: 168 YACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP 227
Y S Y GF + EE S K EG+PGV+FVLPDSY+DP NK YG + ++ G+I+ R P
Sbjct: 127 YNVSCERYLGFGCEIDEETSTKLEGLPGVLFVLPDSYVDPENKDYGAELFVNGEIVQRSP 186
Query: 228 PVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN 268
R R Q + P YN+R R N
Sbjct: 187 ----------ERQRRVEPQPQRAQDRPRYNDRTRYSRRREN 217
>UniRef100_O22793 Plastid protein [Arabidopsis thaliana]
Length = 219
Score = 119 bits (298), Expect = 2e-25
Identities = 82/221 (37%), Positives = 109/221 (49%), Gaps = 31/221 (14%)
Query: 65 ITGSISSHFSLSKSPNVVDGIQ---SR---SFRSTSISLLSSRYGET----------SEL 108
+T ++ S+ +L P + + SR S R SI ++R G T S+
Sbjct: 11 LTRALLSNVTLMAPPRIPSSVHYGGSRLGCSTRFFSIRCGANRSGSTYSPLNSGSNFSDR 70
Query: 109 SP-EIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKI 167
P E+ P LF GCDY HWL V D P ++MI Y +T AK + S EEAKK+I
Sbjct: 71 PPTEMAP---LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVVG-SEEEAKKRI 126
Query: 168 YACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP 227
Y S Y GF + EE S K EG+PGV+FVLPDSY+DP NK YG + ++ G+I+ R P
Sbjct: 127 YNVSCERYLGFGCEIDEETSTKLEGLPGVLFVLPDSYVDPENKDYGAELFVNGEIVQRSP 186
Query: 228 PVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN 268
R R Q + P YN+R R N
Sbjct: 187 ----------ERQRRVEPQPQRAQDRPRYNDRTRYSRRREN 217
>UniRef100_Q7DLI5 Plastid protein [Arabidopsis thaliana]
Length = 198
Score = 119 bits (297), Expect = 3e-25
Identities = 65/151 (43%), Positives = 81/151 (53%), Gaps = 11/151 (7%)
Query: 118 LFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
LF GCDY HWL V D P ++MI Y +T AK + S EEAKK+IY S Y G
Sbjct: 57 LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVVG-SEEEAKKRIYNVSCERYLG 115
Query: 178 FQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGG 237
F + EE S K EG+PGV+FVLPDSY+DP NK YG + ++ G+I+ R P
Sbjct: 116 FGCEIDEETSTKLEGLPGVLFVLPDSYVDPENKDYGAELFVNGEIVQRSP---------- 165
Query: 238 RRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN 268
R R Q + P YN+R R N
Sbjct: 166 ERQRRVEPQPQRAQDRPRYNDRTRYSRRREN 196
>UniRef100_Q8H656 Putative plastid protein [Oryza sativa]
Length = 227
Score = 116 bits (290), Expect = 2e-24
Identities = 66/152 (43%), Positives = 82/152 (53%), Gaps = 13/152 (8%)
Query: 118 LFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
LF GCDY HWL V D P ++MI Y +T AK L S EEAKKKIY S Y G
Sbjct: 86 LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVLG-SEEEAKKKIYNVSCERYFG 144
Query: 178 FQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGG 237
F + EE S K EG+PGV+FVLPDSY+DP K YG + ++ G+I+ R P Q
Sbjct: 145 FGCEIDEETSNKLEGLPGVLFVLPDSYVDPEYKDYGAELFVNGEIVQRSPERQ------- 197
Query: 238 RRDYRQNNQLPNNRGN-PSYNNRDSMPRDGRN 268
R+ +P + P YN+R R N
Sbjct: 198 ----RRVEPVPQRASDRPRYNDRTRYARRREN 225
>UniRef100_Q5VRK8 Putative DAL1 protein [Oryza sativa]
Length = 165
Score = 116 bits (290), Expect = 2e-24
Identities = 66/152 (43%), Positives = 82/152 (53%), Gaps = 13/152 (8%)
Query: 118 LFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
LF GCDY HWL V D P ++MI Y +T AK L S EEAKKKIY S Y G
Sbjct: 24 LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVLG-SEEEAKKKIYNVSCERYFG 82
Query: 178 FQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGG 237
F + EE S K EG+PGV+FVLPDSY+DP K YG + ++ G+I+ R P Q
Sbjct: 83 FGCEIDEETSNKLEGLPGVLFVLPDSYVDPEYKDYGAELFVNGEIVQRSPERQ------- 135
Query: 238 RRDYRQNNQLPNNRGN-PSYNNRDSMPRDGRN 268
R+ +P + P YN+R R N
Sbjct: 136 ----RRVEPVPQRASDRPRYNDRTRYARRREN 163
>UniRef100_Q7X6N4 OSJNBa0041A02.8 protein [Oryza sativa]
Length = 223
Score = 115 bits (287), Expect = 4e-24
Identities = 62/131 (47%), Positives = 79/131 (59%), Gaps = 5/131 (3%)
Query: 101 RYGETSELSP-EIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNIS 159
R G+ + +P E+ P LF GCDY HWL V D P ++MI Y +T AK + S
Sbjct: 64 RSGQGGDRAPTEMAP---LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVVG-S 119
Query: 160 VEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIE 219
EEAKKKIY S Y GF + EE S K EG+PGV+FVLPDSY+D NK YG + ++
Sbjct: 120 EEEAKKKIYNVSCERYFGFGCEIDEETSNKLEGLPGVLFVLPDSYVDAENKDYGAELFVN 179
Query: 220 GQIIPRPPPVQ 230
G+I+ R P Q
Sbjct: 180 GEIVQRSPERQ 190
>UniRef100_Q6K456 Putative plastid protein [Oryza sativa]
Length = 398
Score = 115 bits (287), Expect = 4e-24
Identities = 106/363 (29%), Positives = 153/363 (41%), Gaps = 51/363 (14%)
Query: 115 DTILFEGCDYNHWLFVCDFPRDNKPPPE----EMIRIYEETCAKGLNISVEEAKKKIYAC 170
+TIL +GCD+ HWL V D P + PE E+I Y +T A+ + S +EA+ KIY+
Sbjct: 73 ETILLDGCDFEHWLVVMDPPPGDPSNPEPTRDEIIDGYIKTLAQIVG-SEDEARHKIYSV 131
Query: 171 STTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQ 230
ST Y F A+++EE S K + +P V +VLPDSY+D NK YGG+ +I G+ +P P
Sbjct: 132 STRHYFAFGALVSEELSYKLKELPKVRWVLPDSYLDVRNKDYGGEPFINGEAVPYDPKY- 190
Query: 231 FGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQ---NHGQASHI 287
++ +NN N R R+ P RN+ +NF ++ ++ Q +
Sbjct: 191 -------HEEWVRNNARANER-----TRRNDRP---RNFDRSRNFERRRENMHNFQNRDV 235
Query: 288 PPQQNIGQ-----QQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYG 342
PP Q Q P + D+ PP A Y P QA GY NY
Sbjct: 236 PPGQGFNSPPPPGQGPVLPRDAPPMPPPPSPPNPGAPPSYQPHAPNPQA--GY---TNYQ 290
Query: 343 QAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRR 402
Y Q+ GP P Q P +G P + G P
Sbjct: 291 GGVPGYQGRAPGYQGGNQEYRGPPPPPPSAYQGNNPGYQGG----GPGYHGGNPPPYQAG 346
Query: 403 NYAPQQNFGPPRQGERRNYVPQQNFGPPG-QGERGNSVPSEGGWDFKPSYMEEFEQGQKG 461
N P Q P G G PG QG+ GN +G ++ +++ + G
Sbjct: 347 NPPPYQAGNPVFAG-----------GAPGYQGQGGNPSYQQGSDNYNAG-APAYKRDEPG 394
Query: 462 NNH 464
N+
Sbjct: 395 RNY 397
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 978,374,102
Number of Sequences: 2790947
Number of extensions: 49428583
Number of successful extensions: 150367
Number of sequences better than 10.0: 4225
Number of HSP's better than 10.0 without gapping: 363
Number of HSP's successfully gapped in prelim test: 4095
Number of HSP's that attempted gapping in prelim test: 116461
Number of HSP's gapped (non-prelim): 15994
length of query: 488
length of database: 848,049,833
effective HSP length: 131
effective length of query: 357
effective length of database: 482,435,776
effective search space: 172229572032
effective search space used: 172229572032
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC134322.18


