

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.6 - phase: 0
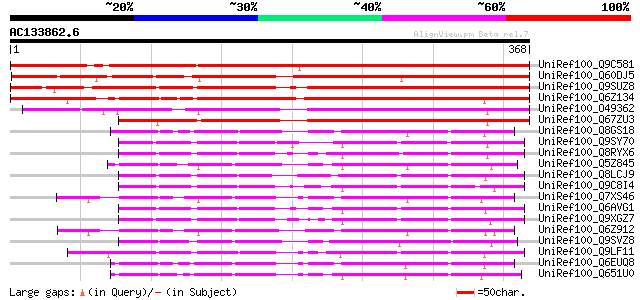
(368 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C581 Hypothetical protein At5g22070 [Arabidopsis tha... 459 e-128
UniRef100_Q60DJ5 Hypothetical protein OSJNBb0088L13.7 [Oryza sat... 367 e-100
UniRef100_Q9SUZ8 Hypothetical protein F4F15.170 [Arabidopsis tha... 360 3e-98
UniRef100_Q6Z134 Hypothetical protein P0455F03.29 [Oryza sativa] 309 7e-83
UniRef100_O49362 Hypothetical protein F10M6.70 [Arabidopsis thal... 275 1e-72
UniRef100_Q67ZU3 Hypothetical protein At5g25330 [Arabidopsis tha... 265 2e-69
UniRef100_Q8GS18 Hypothetical protein [Arabidopsis thaliana] 159 2e-37
UniRef100_Q9SY70 F14N23.16 [Arabidopsis thaliana] 149 9e-35
UniRef100_Q8RYX6 P0648C09.24 protein [Oryza sativa] 149 2e-34
UniRef100_Q5Z845 Hypothetical protein P0468G03.24 [Oryza sativa] 149 2e-34
UniRef100_Q8LCJ9 Hypothetical protein [Arabidopsis thaliana] 148 3e-34
UniRef100_Q9C8I4 Hypothetical protein F19C24.28 [Arabidopsis tha... 147 5e-34
UniRef100_Q7XS46 OSJNBa0035M09.18 protein [Oryza sativa] 146 1e-33
UniRef100_Q6AVG1 Expressed protein [Oryza sativa] 145 2e-33
UniRef100_Q9XGZ7 T1N24.19 protein [Arabidopsis thaliana] 144 3e-33
UniRef100_Q6Z912 Hypothetical protein P0494D11.22 [Oryza sativa] 143 9e-33
UniRef100_Q9SVZ8 Hypothetical protein AT4g25870 [Arabidopsis tha... 142 2e-32
UniRef100_Q9LF11 Hypothetical protein T21H19_90 [Arabidopsis tha... 141 3e-32
UniRef100_Q6EUQ8 Hypothetical protein OJ1077_E05.12 [Oryza sativa] 141 3e-32
UniRef100_Q651U0 Hypothetical protein OSJNBa0051O02.41 [Oryza sa... 140 4e-32
>UniRef100_Q9C581 Hypothetical protein At5g22070 [Arabidopsis thaliana]
Length = 362
Score = 459 bits (1180), Expect = e-128
Identities = 229/371 (61%), Positives = 280/371 (74%), Gaps = 12/371 (3%)
Query: 1 MFSNPYLLTFSLLLSLP-ILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPS 59
MFSN ++L FSLLL LP + FF+AP + P P I +DE+DD +LF A +
Sbjct: 1 MFSNQFVLFFSLLLCLPFVFFFVAPHVFPSPPGESLIPISDEIDDRSLFIAAAGST---- 56
Query: 60 SSSHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNL 119
S SH S + NP KIAFLFLTN+DLHF P+W+ FF L+NVYVH+DP VN+
Sbjct: 57 SLSHLSS----GNPNPKLKIAFLFLTNSDLHFAPIWDRFFSGHSKSLYNVYVHADPFVNV 112
Query: 120 TLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLH 179
T + + + F ++K+T RASPTLISATRRLLA+A LDD +N YF VLSQYCIPLH
Sbjct: 113 TRPGNGSVFENAF-IANAKRTARASPTLISATRRLLATAFLDDPANTYFAVLSQYCIPLH 171
Query: 180 SFDYIYKSLFLSPTFDLTDSESTQF--GVRLKYKSFIEIINNGPRLWKRYTARGRYAMMP 237
SF+Y+Y SLF S FD +D + G+R+ Y+SF+E+I++ PRLWKRYTARGRYAMMP
Sbjct: 172 SFNYVYSSLFESSIFDKSDPDPNPNPRGIRILYRSFMELISDEPRLWKRYTARGRYAMMP 231
Query: 238 EVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDS 297
EVPFEKFRVGSQFF +TR+HAL+ +KDR LWRKFK+PCYR DECYPEEHYFPTLL+M+D
Sbjct: 232 EVPFEKFRVGSQFFVMTRRHALLTIKDRILWRKFKLPCYRSDECYPEEHYFPTLLNMKDP 291
Query: 298 DGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFARKFVPDCLEPL 357
DG TGYTLT VNWTGTV GHP+TY+P+EV PELI RLR+S +S S+ FARKF PDCL+PL
Sbjct: 292 DGCTGYTLTRVNWTGTVKGHPYTYKPKEVVPELIQRLRRSNHSSSYFFARKFTPDCLKPL 351
Query: 358 MGIAKSVIFKD 368
+ IA SVIF+D
Sbjct: 352 LAIADSVIFRD 362
>UniRef100_Q60DJ5 Hypothetical protein OSJNBb0088L13.7 [Oryza sativa]
Length = 362
Score = 367 bits (942), Expect = e-100
Identities = 199/376 (52%), Positives = 254/376 (66%), Gaps = 30/376 (7%)
Query: 2 FSNPYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSS- 60
+++P++L+ LL+S+P++F LAP +LPP P I DE DD+ LF AI S+ PSS
Sbjct: 8 YASPFVLSVLLLVSIPVIFLLAPRLLPPKTLPA-IPDADESDDLALFRRAILSSSSPSSA 66
Query: 61 ----SSHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPR 116
SS S FF + P K+AFLFLTN+DL F+PLW FF+ LFN+YVH+DP
Sbjct: 67 TPTPSSAASYFFR---RRPAPKVAFLFLTNSDLVFSPLWEKFFRGH-HHLFNLYVHADPF 122
Query: 117 VNLTLLRSSNNYNPIFK--FISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQY 174
LT+ + P F+ F+ +K T RASPTLISA RRL+A+A+LDD SN +F +LSQ
Sbjct: 123 SALTMPPT-----PSFRGRFVPAKATQRASPTLISAARRLIATALLDDPSNQFFALLSQS 177
Query: 175 CIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA 234
CIPLH F +Y +L S G +++SFIEI+++ + RY ARG
Sbjct: 178 CIPLHPFPTLYNTLL-----------SDNAGPHGRHRSFIEIMDDAYMIHDRYYARGDDV 226
Query: 235 MMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCY--RDDECYPEEHYFPTLL 292
M+PEVP+++FR GSQFF LTRKHA++VV+D LWRKFK+PC R D CYPEEHYFPTLL
Sbjct: 227 MLPEVPYDQFRFGSQFFVLTRKHAIMVVRDMKLWRKFKLPCLIKRRDSCYPEEHYFPTLL 286
Query: 293 SMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFARKFVPD 352
M+D +G TGYTLT VNWT V GHPHTY+P EVS LI LRKS + S++FARKF P+
Sbjct: 287 DMQDPEGCTGYTLTRVNWTDQVEGHPHTYRPGEVSASLIKELRKSNGTYSYMFARKFAPE 346
Query: 353 CLEPLMGIAKSVIFKD 368
CLEPLM IA SVI +D
Sbjct: 347 CLEPLMEIADSVILRD 362
>UniRef100_Q9SUZ8 Hypothetical protein F4F15.170 [Arabidopsis thaliana]
Length = 346
Score = 360 bits (924), Expect = 3e-98
Identities = 198/372 (53%), Positives = 258/372 (69%), Gaps = 30/372 (8%)
Query: 1 MFSNPYLL-TFSLLLSLPILFFLAPAILPPH---PSPIPISPTDELDDINLFNTAISHST 56
MFS+ L+ + SL LSL +LFFL IL PH S IS D+LDD++LF+ A+
Sbjct: 1 MFSSSTLVYSLSLFLSLSLLFFLF--ILSPHILLSSSTLISTADDLDDLSLFHRAVV--- 55
Query: 57 PPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPR 116
SSS++ ++ S NP KIAFLFLTN+DL F PLW FFQ L+NVY+H+DP
Sbjct: 56 --SSSTNNNRRLISLSPNPPPKIAFLFLTNSDLTFLPLWESFFQGHQD-LYNVYIHADPT 112
Query: 117 VNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCI 176
+++ L S++ N KFI +++T RASPTLISA RRLLA+AILDD +N YF ++SQ+CI
Sbjct: 113 SSVSPLLDSSSINA--KFIPARRTARASPTLISAERRLLANAILDDPNNLYFALISQHCI 170
Query: 177 PLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMM 236
PLHSF YI+ LF S+ Q +SFIEI+++ P L KRY ARG AM+
Sbjct: 171 PLHSFSYIHNHLF---------SDHHQ-------QSFIEILSDEPFLLKRYNARGDDAML 214
Query: 237 PEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMED 296
PE+ ++ FRVGSQFF L ++HAL+V+K+R LWRKFK+PC + CYPEEHYFPTLLS+ED
Sbjct: 215 PEIQYQDFRVGSQFFVLAKRHALMVIKERKLWRKFKLPCLDVESCYPEEHYFPTLLSLED 274
Query: 297 SDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFARKFVPDCLEP 356
G + +TLT VNWTG+V GHPHTY E+SP+LI LR+S +S + FARKF P+ L+P
Sbjct: 275 PQGCSHFTLTRVNWTGSVGGHPHTYDASEISPQLIHSLRRSNSSLDYFFARKFTPESLQP 334
Query: 357 LMGIAKSVIFKD 368
LM IA +VIF+D
Sbjct: 335 LMEIADAVIFRD 346
>UniRef100_Q6Z134 Hypothetical protein P0455F03.29 [Oryza sativa]
Length = 359
Score = 309 bits (792), Expect = 7e-83
Identities = 171/378 (45%), Positives = 234/378 (61%), Gaps = 29/378 (7%)
Query: 1 MFSNPYLLTFSLLLSLPILFFLAPAILPPHPSPIPISPT--------DELDDINLFNTAI 52
M S+P +LT L SL L LAP P + P P E D+ LF A
Sbjct: 1 MSSSPLVLTVLLFASLTWLVVLAPRSSSPPATATPSPPVVGDGVGGGGEDGDLALFRRAT 60
Query: 53 SHSTPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVH 112
+++ ++ P K+AFLFLTN++L F PLW FF+ +L NVYVH
Sbjct: 61 LDGGEGAAA--------MAVAEP--KVAFLFLTNSELTFAPLWERFFEGHGERL-NVYVH 109
Query: 113 SDPRVNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLS 172
+DP L ++ + ++ +F+++ T RA TLI+A RRLLA+A++DDA+NAYF +LS
Sbjct: 110 ADPAARL-MMPPTRSFKG--RFVAAGPTKRADATLIAAARRLLAAALVDDAANAYFALLS 166
Query: 173 QYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGR 232
Q+CIP+HSF +++ +LF P + + + S+IE+++ P++ RY ARG
Sbjct: 167 QHCIPVHSFRHLHATLFPPPAAAAAAARRQR-----RLPSYIEVLDGEPQMASRYAARGE 221
Query: 233 YAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLL 292
AM+PEVPF++FRVGSQFFTL R+HA +VV +R LW KF+ PC + CYPEEHYFPTLL
Sbjct: 222 GAMLPEVPFDRFRVGSQFFTLARRHAALVVGERRLWDKFRQPCLDQNACYPEEHYFPTLL 281
Query: 293 SMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLR--KSTNSESFLFARKFV 350
M D GV YTLT+VNW G+V+GHPHTY EVS EL+ LR K + ++FARKF
Sbjct: 282 DMADPAGVARYTLTHVNWAGSVHGHPHTYTAAEVSAELVADLRRPKKNTTHDYMFARKFS 341
Query: 351 PDCLEPLMGIAKSVIFKD 368
PDCL PLM IA +++F D
Sbjct: 342 PDCLAPLMDIADAILFND 359
>UniRef100_O49362 Hypothetical protein F10M6.70 [Arabidopsis thaliana]
Length = 384
Score = 275 bits (703), Expect = 1e-72
Identities = 167/401 (41%), Positives = 220/401 (54%), Gaps = 70/401 (17%)
Query: 10 FSLLLSLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPS---- 65
F LL +L + +A ISP +++F+ S + P SSSS P+
Sbjct: 7 FPLLCALFLCLPVAVIFTVQRELTCVISPEFGFRSLSVFSL-YSRNVPDSSSSSPAVRIR 65
Query: 66 --------KFFHLSSK-NP------TFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVY 110
LSS+ NP T KIAF++LT + L F PLW +FF L+NVY
Sbjct: 66 QPIPKEDDPLLRLSSRVNPNLPPGSTRKIAFMYLTTSPLPFAPLWEMFFDGISKNLYNVY 125
Query: 111 VHSDPRVNLTLLRSSNNYNPIF------KFISSKKTYRASPTLISATRRLLASAILDDAS 164
VH+DP + Y+P F + I SK + R +PTL +A RRLLA A+LDD
Sbjct: 126 VHADP---------TREYDPPFSGVFLNRVIHSKPSLRHTPTLTAAARRLLAHALLDDPL 176
Query: 165 NAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLW 224
N F V+S C+P+ SFD+ YK+L S KSFIEI+ + P +
Sbjct: 177 NYMFAVISPSCVPIRSFDFTYKTLVSSR------------------KSFIEILKDEPWQF 218
Query: 225 KRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPE 284
R+TA GR+AM+PEV E+FR+GSQF+ L R+HA VV +DR +W KF C R+D CYPE
Sbjct: 219 DRWTAIGRHAMLPEVKLEEFRIGSQFWVLKRRHARVVARDRRIWVKFNQTCVREDSCYPE 278
Query: 285 EHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKS------- 337
E YFPTLL+M D G TLT+V+WT GHP Y+PEEV PEL+LRLRK+
Sbjct: 279 ESYFPTLLNMRDPRGCVPATLTHVDWTVNDGGHPRMYEPEEVVPELVLRLRKTRPRYGED 338
Query: 338 ----------TNSESFLFARKFVPDCLEPLMGIAKSVIFKD 368
+ FLFARKF P LEPL+G+A++V+F D
Sbjct: 339 GINGSEWSKVERMDPFLFARKFSPQALEPLLGMARTVLFND 379
>UniRef100_Q67ZU3 Hypothetical protein At5g25330 [Arabidopsis thaliana]
Length = 366
Score = 265 bits (677), Expect = 2e-69
Identities = 148/302 (49%), Positives = 182/302 (60%), Gaps = 31/302 (10%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTP--SKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFI 135
K+AF+FLT L PLW LFF + L+NVYVH DP N I
Sbjct: 81 KLAFMFLTTNSLPLAPLWELFFNQSSHHKSLYNVYVHVDPTQKHKPGSHGTFQNRIIP-- 138
Query: 136 SSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFD 195
SSK YR +PTLISA RRLLA A+L+D SN FI+LS CIP HSF++ YK+L S
Sbjct: 139 SSKPAYRHTPTLISAARRLLAHALLEDPSNYMFILLSPSCIPSHSFNFTYKTLVSST--- 195
Query: 196 LTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTR 255
KSFIEI+ + P ++R+ ARG YAM PEVP E+FR+GSQF+TLTR
Sbjct: 196 ---------------KSFIEILKDEPGWYERWAARGPYAMFPEVPPEEFRIGSQFWTLTR 240
Query: 256 KHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVN 315
HAL+VV D +W KF C R+D CYPEEHYFPTLL+M D G T+T+V+W+ +
Sbjct: 241 AHALMVVSDVEIWSKFNKSCVREDICYPEEHYFPTLLNMRDPQGCVSATVTHVDWSVNDH 300
Query: 316 GHPHTYQPEEVSPELILRLRKS---------TNSESFLFARKFVPDCLEPLMGIAKSVIF 366
GHP TY+P EV ELI +LR + T + FLFARKF P + LM I +SVIF
Sbjct: 301 GHPRTYKPLEVRAELIQKLRSARPRYGDGNRTRKDPFLFARKFSPAGINQLMNITRSVIF 360
Query: 367 KD 368
D
Sbjct: 361 ND 362
>UniRef100_Q8GS18 Hypothetical protein [Arabidopsis thaliana]
Length = 376
Score = 159 bits (401), Expect = 2e-37
Identities = 107/314 (34%), Positives = 164/314 (52%), Gaps = 57/314 (18%)
Query: 72 SKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPI 131
SK K+AF+FLT L F PLW +FF+ +K F+VYVH+ + + + +S+ + +
Sbjct: 83 SKTANPKLAFMFLTPGTLPFEPLWEMFFRGHENK-FSVYVHASKK---SPVHTSSYF--V 136
Query: 132 FKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
+ I S K +++ A RRLLA A++D N +FI+LS C+PL F+YIY L +
Sbjct: 137 GRDIHSHKVAWGQISMVDAERRLLAHALVDP-DNQHFILLSDSCVPLFDFNYIYNHLIFA 195
Query: 192 PTFDLTDSESTQFGVRLKYKSFIEIINN-GPRLWKRYTARGRYAMMPEVPFEKFRVGSQF 250
SFI+ + GP RY+ M+PEV + FR GSQ+
Sbjct: 196 NL------------------SFIDCFEDPGPHGSGRYSQH----MLPEVEKKDFRKGSQW 233
Query: 251 FTLTRKHALVVVKDRTLWRKFKVPCYRDDE---CYPEEHYFPTLLSMEDSDGVTGYTLTN 307
F++ R+HA+VV+ D + KFK+ C + E CY +EHYFPTL +M D DG+ +++T+
Sbjct: 234 FSMKRRHAIVVMADSLYYTKFKLYCRPNMEGRNCYADEHYFPTLFNMIDPDGIANWSVTH 293
Query: 308 VNWTGTVNGHPHTYQPEEVSPELILRLR-----------------------KSTNSESFL 344
V+W+ HP Y +++P LI +++ K +L
Sbjct: 294 VDWS-EGKWHPKLYNARDITPYLIRKIKSIQLAYHVTSDLKKVTTVKPCLWKGEQRPCYL 352
Query: 345 FARKFVPDCLEPLM 358
FARKF P+ L+ LM
Sbjct: 353 FARKFNPETLDRLM 366
>UniRef100_Q9SY70 F14N23.16 [Arabidopsis thaliana]
Length = 412
Score = 149 bits (377), Expect = 9e-35
Identities = 101/302 (33%), Positives = 156/302 (51%), Gaps = 48/302 (15%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
K+AF+FLT L PLW FF+ K +VYVH+ P ++ + R S Y+ + I S
Sbjct: 143 KVAFMFLTRGPLPMLPLWEKFFKGN-EKYLSVYVHTPPGYDMNVSRDSPFYD---RQIPS 198
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
++ SP L A +RLLA+A+LD SN F++LS+ C+P+++F +Y L ++ +
Sbjct: 199 QRVEWGSPLLTDAEKRLLANALLD-FSNERFVLLSESCVPVYNFSTVYTYL-INSAYSFV 256
Query: 198 DS--ESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTL 253
DS E T++G RGRY+ M+P++ +R GSQ+F +
Sbjct: 257 DSYDEPTRYG------------------------RGRYSRKMLPDIKLHHWRKGSQWFEV 292
Query: 254 TRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGT 313
RK A+ ++ D + FK C CYP+EHY PT L+M ++T V+W +
Sbjct: 293 NRKIAIYIISDSKYYSLFKQFC--RPACYPDEHYIPTFLNMFHGSMNANRSVTWVDW--S 348
Query: 314 VNG-HPHTYQPEEVSPELILRLRKS---------TNSESFLFARKFVPDCLEPLMGIAKS 363
+ G HP TY ++ + +RK+ S FLFARKF P L PLM ++ +
Sbjct: 349 IGGPHPATYAAANITEGFLQSIRKNETDCLYNEEPTSLCFLFARKFSPSALAPLMNLSST 408
Query: 364 VI 365
V+
Sbjct: 409 VL 410
>UniRef100_Q8RYX6 P0648C09.24 protein [Oryza sativa]
Length = 481
Score = 149 bits (375), Expect = 2e-34
Identities = 109/300 (36%), Positives = 157/300 (52%), Gaps = 43/300 (14%)
Query: 78 KIAFLFLTNTD-LHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFIS 136
K+AF+FL L PLW FF+ LF++YVH+ P + L + S Y + I
Sbjct: 110 KVAFMFLAGRGVLPLAPLWERFFRGHEG-LFSIYVHAPPGMVLNVSDDSPFYG---RQIP 165
Query: 137 SKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDL 196
S++T S TL+ A +RLLA+A+LD SN F++LS+ CIP+ SF Y L
Sbjct: 166 SQETSWGSITLMDAEKRLLANALLD-FSNDRFVLLSESCIPVQSFPVAYGYL-------- 216
Query: 197 TDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLT 254
T S SF+E+ + K T RGRY+ M P++ ++R GSQ+F L
Sbjct: 217 TGSRH----------SFVEVYYH-----KGKTCRGRYSRRMEPDITLPQWRKGSQWFELR 261
Query: 255 RKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTV 314
R A+ + D + F+ C CYP+EHY PT ++M + T+T V+W+
Sbjct: 262 RDLAVAALTDARYYPLFRRHCR--PSCYPDEHYLPTFVAMLHGADNSNRTVTYVDWS-RG 318
Query: 315 NGHPHTYQPEEVSPELILRLRKS---------TNSESFLFARKFVPDCLEPLMGIAKSVI 365
HP TY +V+PELIL +R+S + FLFARKF D LEPL+ I+ +V+
Sbjct: 319 GAHPATYTAGDVTPELILSIRRSEVPCMYNSRPTTACFLFARKFSADALEPLLNISSTVM 378
>UniRef100_Q5Z845 Hypothetical protein P0468G03.24 [Oryza sativa]
Length = 372
Score = 149 bits (375), Expect = 2e-34
Identities = 105/322 (32%), Positives = 163/322 (50%), Gaps = 67/322 (20%)
Query: 70 LSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYN 129
+ SKNP KIAF+FLT + L F LW FF + + +YVH+ + + +
Sbjct: 80 VQSKNP--KIAFMFLTPSSLPFEKLWEKFFMGHEDR-YTIYVHASRERPV-------HAS 129
Query: 130 PIF--KFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKS 187
PIF + I S+K + ++I A RRLLA+A L D N +F++LS+ C+PLH+FDY+Y
Sbjct: 130 PIFNGRDIRSEKVVWGTISMIDAERRLLANA-LQDPDNQHFVLLSESCVPLHNFDYVYSY 188
Query: 188 LFLSPTFDLTDSESTQFGVRLKYKSFIEIINN-GPRLWKRYTARGRYA--MMPEVPFEKF 244
L + SF++ ++ GP GRY+ M+PE+ +
Sbjct: 189 LMETNI------------------SFVDCFDDPGPH------GAGRYSDHMLPEIVKRDW 224
Query: 245 RVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDE---CYPEEHYFPTLLSMEDSDGVT 301
R G+Q+FT+ R+HA++++ D + KFK C +E CY +EHY PTL +M D G+
Sbjct: 225 RKGAQWFTVKRQHAVLILSDFLYYAKFKRYCKPGNEWHNCYSDEHYLPTLFNMVDPTGIA 284
Query: 302 GYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNS--------------------- 340
+++T+V+W+ HP Y+ + S EL+ + S
Sbjct: 285 NWSVTHVDWS-EGKWHPKAYRAVDTSFELLKNISSIDESIHVTSNAKHQVMRRPCLWNGM 343
Query: 341 --ESFLFARKFVPDCLEPLMGI 360
+LFARKF P+ L+ LM I
Sbjct: 344 KRPCYLFARKFYPEALDNLMNI 365
>UniRef100_Q8LCJ9 Hypothetical protein [Arabidopsis thaliana]
Length = 381
Score = 148 bits (373), Expect = 3e-34
Identities = 99/296 (33%), Positives = 152/296 (50%), Gaps = 37/296 (12%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
K+AF+FLT L F PLW FF+ +++YVH+ P SS Y + I S
Sbjct: 113 KMAFMFLTKGPLPFAPLWERFFKGHEG-FYSIYVHTLPNYRSDFPSSSVFYR---RQIPS 168
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
+ ++ A RRLLA+A+LD SN +F++LS+ CIPL F+++Y+
Sbjct: 169 QHVAWGEMSMCDAERRLLANALLD-ISNEWFVLLSEACIPLRGFNFVYR----------- 216
Query: 198 DSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKH 257
+ R +Y + +GP RY+ YAM PEV ++R GSQ+F + R
Sbjct: 217 ------YVSRSRYSFMGSVDEDGPYGRGRYS----YAMGPEVSLNEWRKGSQWFEINRAL 266
Query: 258 ALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGH 317
A+ +V+D + KFK C CY +EHYFPT+LS+ D + TLT +W+ H
Sbjct: 267 AVDIVEDMVYYNKFKEFC--RPPCYVDEHYFPTMLSIGYPDFLANRTLTWTDWS-RGGAH 323
Query: 318 PHTYQPEEVSPELILRLRK--------STNSESFLFARKFVPDCLEPLMGIAKSVI 365
P T+ +++ + I +L + + +LFARKF P L+PL+ +A V+
Sbjct: 324 PATFGKADITEKFIKKLSRGKACFYNDQPSQVCYLFARKFAPSALKPLLKLAPKVL 379
>UniRef100_Q9C8I4 Hypothetical protein F19C24.28 [Arabidopsis thaliana]
Length = 399
Score = 147 bits (371), Expect = 5e-34
Identities = 107/301 (35%), Positives = 153/301 (50%), Gaps = 47/301 (15%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
K+AF+FL L F PLW F + L+++YVHS P RSS Y ++I S
Sbjct: 117 KLAFMFLAKGPLPFAPLWEKFCKGHEG-LYSIYVHSLPSYKSDFSRSSVFYR---RYIPS 172
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
+ ++ A RRLLA+A+LD SN +F++LS+ CIPL F +IY +
Sbjct: 173 QAVAWGEMSMGEAERRLLANALLD-ISNEWFVLLSESCIPLRGFSFIYSYV--------- 222
Query: 198 DSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLTR 255
SES +Y GP RGRY M PE+ ++R GSQ+F + R
Sbjct: 223 -SES-------RYSFMGAADEEGP------DGRGRYRTEMEPEITLSQWRKGSQWFEINR 268
Query: 256 KHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVN 315
K A+ +V+D T + KFK C CY +EHYFPT+LSM+ + TLT +W+
Sbjct: 269 KLAVEIVQDTTYYPKFKEFC--RPPCYVDEHYFPTMLSMKHRVLLANRTLTWTDWS-RGG 325
Query: 316 GHPHTYQPEEVSPELILRLRKSTNSES-----------FLFARKFVPDCLEPLMGIAKSV 364
HP T+ +V+ L+K T ++S +LFARKF P LEPL+ +A +
Sbjct: 326 AHPATFGKADVTESF---LKKLTGAKSCLYNDHQSQICYLFARKFAPSALEPLLQLAPKI 382
Query: 365 I 365
+
Sbjct: 383 L 383
>UniRef100_Q7XS46 OSJNBa0035M09.18 protein [Oryza sativa]
Length = 382
Score = 146 bits (368), Expect = 1e-33
Identities = 110/358 (30%), Positives = 166/358 (45%), Gaps = 78/358 (21%)
Query: 34 IPISPTDELDDINLFNTAISH---STPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLH 90
+P P EL D + + STPP S K+AF+FLT L
Sbjct: 61 LPAEPVRELTDQERASQVVFKQILSTPPVKSKRS-------------KVAFMFLTPGTLP 107
Query: 91 FTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIF--KFISSKKTYRASPTLI 148
F LW FF+ + + +YVH+ + +P+F + I S+K +++
Sbjct: 108 FERLWEKFFEGHEGR-YTIYVHASRE-------KPEHASPLFIDRDIRSEKVVWGKISMV 159
Query: 149 SATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRL 208
A RRLLA+A L+D N +F++LS C+PLH+FDY+Y L G +
Sbjct: 160 DAERRLLANA-LEDVDNQHFVLLSDSCVPLHNFDYVYNYLI---------------GTNI 203
Query: 209 KYKSFIE-IINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTL 267
SFI+ + GP RY+ M+PEV FR GSQ+F++ R+HAL+++ D
Sbjct: 204 ---SFIDSFYDPGPHGNFRYSKH----MLPEVRESDFRKGSQWFSVKRQHALMIIADSLY 256
Query: 268 WRKFKVPCYRDDE----CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQP 323
+ KFK+ C E CY +EHY PTL M D +G+ +++T+V+W+ HP Y+
Sbjct: 257 YTKFKLHCKPGMEDGRNCYADEHYLPTLFHMIDPNGIANWSVTHVDWS-EGKWHPKAYRA 315
Query: 324 EEVSPELILR-----------------------LRKSTNSESFLFARKFVPDCLEPLM 358
+V+ EL+ L +LFARKF P+ + LM
Sbjct: 316 NDVTYELLKNITSIDMSYHITSDSKKVVTQRPCLWNGVKRPCYLFARKFYPESINRLM 373
>UniRef100_Q6AVG1 Expressed protein [Oryza sativa]
Length = 398
Score = 145 bits (366), Expect = 2e-33
Identities = 102/299 (34%), Positives = 157/299 (52%), Gaps = 43/299 (14%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
K+AF+FLT L PLW FF+ L++VYVH+ P S Y + I S
Sbjct: 129 KVAFMFLTRGPLPLAPLWERFFRGHDG-LYSVYVHALPSYRANFTTDSVFYR---RQIPS 184
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
K T+ A RRLLA+A+LD SN +F+++S+ CIP+ +F+ Y+ L
Sbjct: 185 KVAEWGEMTMCDAERRLLANALLD-ISNEWFVLVSESCIPIFNFNTTYRYL--------- 234
Query: 198 DSESTQFGVRLKYKSFIEIINN-GPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLT 254
S+Q SF+ ++ GP RGRY M PEV ++R GSQ+F +
Sbjct: 235 -QNSSQ--------SFVMAFDDPGPY------GRGRYNWNMTPEVELTQWRKGSQWFEVN 279
Query: 255 RKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTV 314
R+ A+ +V+D + KFK C CY +EHYFPT+L++E + ++T V+W+
Sbjct: 280 RELAIEIVRDTLYYPKFKEFC--RPHCYVDEHYFPTMLTIEAPQSLANRSITWVDWS-RG 336
Query: 315 NGHPHTYQPEEVSPELILRLRK--------STNSESFLFARKFVPDCLEPLMGIAKSVI 365
HP T+ +++ E + R+++ ++ FLFARKF P LEPL+ +A +V+
Sbjct: 337 GAHPATFGRGDITEEFLRRVQEGRTCLYNGQNSTMCFLFARKFAPSALEPLLELAPTVL 395
>UniRef100_Q9XGZ7 T1N24.19 protein [Arabidopsis thaliana]
Length = 436
Score = 144 bits (364), Expect = 3e-33
Identities = 107/298 (35%), Positives = 155/298 (51%), Gaps = 41/298 (13%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
KIAF+FLT L PLW + KL++VY+HS + SS Y + I S
Sbjct: 168 KIAFMFLTMGPLPLAPLWERLLKGH-EKLYSVYIHSPVSSSAKFPASSVFYR---RHIPS 223
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
+ T+ A RRLLA+A+LD SN +F++LS+ CIPL +F IY + T
Sbjct: 224 QVAEWGRMTMCDAERRLLANALLD-ISNEWFVLLSESCIPLFNFTTIYTYM--------T 274
Query: 198 DSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLTR 255
SE + G SF + P + RGRY M PEV +++R GSQ+F + R
Sbjct: 275 KSEHSFMG------SF-----DDPGAY----GRGRYHGNMAPEVFIDQWRKGSQWFEINR 319
Query: 256 KHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVN 315
+ A+ +VKD + KFK C CY +EHYFPT+L++E + ++T V+W+
Sbjct: 320 ELAVSIVKDTLYYPKFKEFC--QPACYVDEHYFPTMLTIEKPAALANRSVTWVDWS-RGG 376
Query: 316 GHPHTYQPEEVSPELILRLRKSTN--------SESFLFARKFVPDCLEPLMGIAKSVI 365
HP T+ ++++ E R+ K N S +LFARKF P LEPL+ IA ++
Sbjct: 377 AHPATFGAQDINEEFFARILKGDNCTYNGGYTSMCYLFARKFSPSALEPLVQIAPKLL 434
>UniRef100_Q6Z912 Hypothetical protein P0494D11.22 [Oryza sativa]
Length = 376
Score = 143 bits (360), Expect = 9e-33
Identities = 114/357 (31%), Positives = 165/357 (45%), Gaps = 78/357 (21%)
Query: 35 PISPTDELDDINLFNTAISH---STPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLHF 91
P P EL D + STPP S +P KIAF+FLT L F
Sbjct: 56 PAEPARELTDEETAARVVFRQILSTPPFPSRNP-------------KIAFMFLTPGKLPF 102
Query: 92 TPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIF--KFISSKKTYRASPTLIS 149
LW LFF+ + + +YVH+ + +P+F + I S K +++
Sbjct: 103 EKLWELFFKGHEGR-YTIYVHASRE-------KPEHVSPVFVGRDIHSDKVGWGMISMVD 154
Query: 150 ATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLK 209
A RRLLA A L+D N F++LS C+PLH+FDY+Y L S
Sbjct: 155 AERRLLAKA-LEDTDNQLFVLLSDSCVPLHNFDYVYDFLMGS------------------ 195
Query: 210 YKSFIEIINN-GPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLW 268
SF++ ++ GP RY+ M+PEV FR GSQ+F + R+HA+VVV D +
Sbjct: 196 RHSFLDCFDDPGPHGVFRYSKH----MLPEVREIDFRKGSQWFAIKRQHAMVVVADSLYY 251
Query: 269 RKFKVPCYRDDE----CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPE 324
KF+ C E CY +EHY PTL M D G+ +++T V+W+ HP +++ +
Sbjct: 252 TKFRRFCKPGMEEGRNCYADEHYLPTLFLMMDPAGIANWSVTYVDWS-EGKWHPRSFRAK 310
Query: 325 EVSPELILRLRK------STNSES-----------------FLFARKFVPDCLEPLM 358
+V+ EL+ + T+ E +LFARKF P+ L LM
Sbjct: 311 DVTYELLKNMTSVDISYHITSDEKKELLQRPCLWNGLKRPCYLFARKFYPETLNNLM 367
>UniRef100_Q9SVZ8 Hypothetical protein AT4g25870 [Arabidopsis thaliana]
Length = 389
Score = 142 bits (357), Expect = 2e-32
Identities = 100/312 (32%), Positives = 152/312 (48%), Gaps = 59/312 (18%)
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
KIAFLFLT L F LW+ FF+ K F++Y+H + + R ++ + I S
Sbjct: 99 KIAFLFLTPGTLPFEKLWDEFFKGHEGK-FSIYIHPSKERPVHISRHFSD-----REIHS 152
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
+ +++ A +RLL SA L+D N +F+++S+ CIPLH+FDY Y+ L S
Sbjct: 153 DEVTWGRISMVDAEKRLLVSA-LEDPDNQHFVLVSESCIPLHTFDYTYRYLLYSNV---- 207
Query: 198 DSESTQFGVRLKYKSFIE-IINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRK 256
SFIE ++ GP T R M+PE+ E FR G+Q+FT+ R+
Sbjct: 208 --------------SFIESFVDPGPH----GTGRHMEHMLPEIAKEDFRKGAQWFTMKRQ 249
Query: 257 HALVVVKDRTLWRKFKVPC----YRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTG 312
HA++V+ D + KF+ C D C +EHY PT +M D G++ +++T V+W+
Sbjct: 250 HAIIVMADGLYYSKFREYCGPGIEADKNCIADEHYLPTFFNMIDPMGISNWSVTFVDWSE 309
Query: 313 TVNGHPHTYQPEEVSPELILRLRKSTNS------------------------ESFLFARK 348
HP TY E+S E + + S +LFARK
Sbjct: 310 R-RWHPKTYGGNEISLEFMKNVTSEDMSVHVTSVGKHGDELHWPCTWNGIKRPCYLFARK 368
Query: 349 FVPDCLEPLMGI 360
F PD L+ L+ +
Sbjct: 369 FHPDTLDTLVNL 380
>UniRef100_Q9LF11 Hypothetical protein T21H19_90 [Arabidopsis thaliana]
Length = 329
Score = 141 bits (356), Expect = 3e-32
Identities = 110/341 (32%), Positives = 167/341 (48%), Gaps = 48/341 (14%)
Query: 42 LDDINLFNTAISHSTPPSSSSHPSKFF--HLSSKNPTFKIAFLFLTNTDLHFTPLWNLFF 99
+ D LF A S S+P SS S S H + K+AF+F+T L LW FF
Sbjct: 16 MSDQELFTKASSLSSPTSSLSSSSWLGRRHNNDGKMAVKVAFMFMTGGRLPLAGLWEKFF 75
Query: 100 QTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAI 159
+ +++YVH++P + +S Y+ + I S+ Y + +++ A +RLLA+A+
Sbjct: 76 EGHEG-FYSIYVHTNPSFQDSFPETSVFYS---RRIPSQPVYWGTSSMVDAEKRLLANAL 131
Query: 160 LDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINN 219
LD+ SN F++LS CIPL++F IY L G L SFI ++
Sbjct: 132 LDE-SNQRFVLLSDSCIPLYNFTTIYDYLT---------------GTNL---SFIGSFDD 172
Query: 220 GPRLWKRYTARGRY--AMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYR 277
R + RGRY M P + +R GSQ+F TR+ AL +++D ++ F C
Sbjct: 173 -----PRKSGRGRYNHTMYPHINITHWRKGSQWFETTRELALHIIEDTVYYKIFDQHC-- 225
Query: 278 DDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLR-- 335
CY +EHY PTL+ M + TLT V+W+ HP + +++ E + R+R
Sbjct: 226 KPPCYMDEHYIPTLVHMLHGEMSANRTLTWVDWS-KAGPHPGRFIWPDITDEFLNRIRFK 284
Query: 336 -----------KSTNSESFLFARKFVPDCLEPLMGIAKSVI 365
T S+ FLFARKF + LEPL+ I+ V+
Sbjct: 285 EECVYFGRGGENVTTSKCFLFARKFTAETLEPLLRISPIVL 325
>UniRef100_Q6EUQ8 Hypothetical protein OJ1077_E05.12 [Oryza sativa]
Length = 390
Score = 141 bits (355), Expect = 3e-32
Identities = 98/313 (31%), Positives = 159/313 (50%), Gaps = 58/313 (18%)
Query: 72 SKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPI 131
SKNP KIA +FLT L F LW F Q + +++Y+H+ + SS+ + +
Sbjct: 102 SKNP--KIALMFLTPGSLPFEKLWEKFLQGHEDR-YSIYIHASRE---RPVHSSSLF--V 153
Query: 132 FKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
+ I S+K +++ A +RLLA+A L+D N +F++LS C+PLH+FDYIY L
Sbjct: 154 GREIHSEKVVWGRISMVDAEKRLLANA-LEDVDNQFFVLLSDSCVPLHTFDYIYNFLM-- 210
Query: 192 PTFDLTDSESTQFGVRLKYKSFIE-IINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQF 250
G + SFI+ ++ GP RY+ M+PE+ FR G+Q+
Sbjct: 211 -------------GTNV---SFIDCFLDPGPHGSGRYSVE----MLPEIEQRDFRKGAQW 250
Query: 251 FTLTRKHALVVVKDRTLWRKFKVPCYRDD--ECYPEEHYFPTLLSMEDSDGVTGYTLTNV 308
F +TR+HAL+++ D + KF++ C + C +EHY PTL +M D G++ +++T+V
Sbjct: 251 FAVTRRHALLILADHLYYNKFELYCKPAEGRNCIADEHYLPTLFNMVDPGGISNWSVTHV 310
Query: 309 NWTGTVNGHPHTYQPEEVSPELILRLR-----------------------KSTNSESFLF 345
+W+ HP +Y+ +V+ L+ + T +LF
Sbjct: 311 DWS-EGKWHPRSYRAIDVTYALLKNITAIKENFRITSDDKKVVTMTPCMWNGTKRPCYLF 369
Query: 346 ARKFVPDCLEPLM 358
ARKF P+ L L+
Sbjct: 370 ARKFYPEALNNLL 382
>UniRef100_Q651U0 Hypothetical protein OSJNBa0051O02.41 [Oryza sativa]
Length = 378
Score = 140 bits (354), Expect = 4e-32
Identities = 100/320 (31%), Positives = 161/320 (50%), Gaps = 62/320 (19%)
Query: 72 SKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPI 131
SKNP KIA +FLT L F LW F Q + +++YVH+ + +S+ + +
Sbjct: 89 SKNP--KIALMFLTPGTLPFEKLWEKFLQGQEGR-YSIYVHASREKPV---HTSSLF--V 140
Query: 132 FKFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
+ I S +++ A +RLLA+A L D N +F++LS C+PLH+FDY+Y L
Sbjct: 141 GRDIHSDAVVWGKISMVDAEKRLLANA-LADVDNQFFVLLSDSCVPLHTFDYVYNYLM-- 197
Query: 192 PTFDLTDSESTQFGVRLKYKSFIEIINN-GPRLWKRYTARGRYA--MMPEVPFEKFRVGS 248
G + SFI+ + GP GRY+ M+PE+ + FR G+
Sbjct: 198 -------------GTNI---SFIDCFRDPGPH------GNGRYSPEMLPEIEEKDFRKGA 235
Query: 249 QFFTLTRKHALVVVKDRTLWRKFKVPCYRDD--ECYPEEHYFPTLLSMEDSDGVTGYTLT 306
Q+F +TR+HAL+++ D ++KFK+ C D C +EHY PTL +M D G+ +++T
Sbjct: 236 QWFAITRRHALLILADSLYYKKFKLYCKPADGRNCIADEHYLPTLFNMVDPGGIANWSVT 295
Query: 307 NVNWTGTVNGHPHTYQPEEVSPELILR-----------------------LRKSTNSESF 343
+V+W+ HP +Y+ +V+ +L+ L + +
Sbjct: 296 HVDWS-EGKWHPRSYRAADVTYDLLKNITAVDENFHVTSDDKKLMTQKPCLWNGSKRPCY 354
Query: 344 LFARKFVPDCLEPLMGIAKS 363
LFARKF P+ L+ L+ + S
Sbjct: 355 LFARKFYPETLDNLLKLFTS 374
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 647,556,156
Number of Sequences: 2790947
Number of extensions: 28365514
Number of successful extensions: 74143
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 73872
Number of HSP's gapped (non-prelim): 84
length of query: 368
length of database: 848,049,833
effective HSP length: 129
effective length of query: 239
effective length of database: 488,017,670
effective search space: 116636223130
effective search space used: 116636223130
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133862.6


