

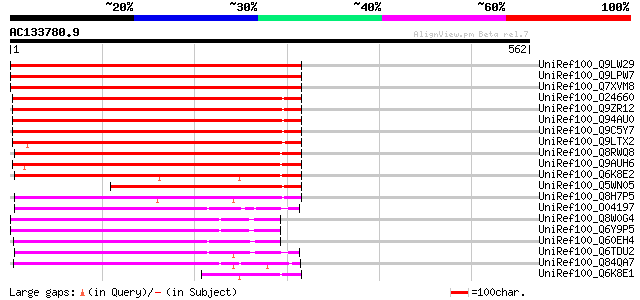
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.9 - phase: 0 /pseudo
(562 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LW29 Transport inhibitor response-like protein [Arab... 505 e-141
UniRef100_Q9LPW7 F13K23.7 protein [Arabidopsis thaliana] 494 e-138
UniRef100_Q7XVM8 OSJNBa0072K14.18 protein [Oryza sativa] 416 e-114
UniRef100_O24660 Transport inhibitor response 1 [Arabidopsis tha... 386 e-106
UniRef100_Q9ZR12 Putative homolog of transport inhibitor respons... 370 e-101
UniRef100_Q94AU0 Putative F-box protein GRR1 protein 1, AtFBL18 ... 369 e-100
UniRef100_Q9C5Y7 GRR1-like protein 1 [Arabidopsis thaliana] 368 e-100
UniRef100_Q9LTX2 Transport inhibitor response 1 protein [Arabido... 307 5e-82
UniRef100_Q8RWQ8 AT4g24390/T22A6_220 [Arabidopsis thaliana] 300 6e-80
UniRef100_Q9AUH6 F-box containing protein TIR1 [Populus tremula ... 299 1e-79
UniRef100_Q6K8E2 Putative F-box containing protein TIR1 [Oryza s... 288 2e-76
UniRef100_Q5WN05 Putative transport inhibitor response TIR1 [Ory... 229 2e-58
UniRef100_Q8H7P5 Putative F-box containing protein TIR1 [Oryza s... 207 5e-52
UniRef100_O04197 Coronatine-insensitive 1 (COI1), AtFBL2 [Arabid... 167 6e-40
UniRef100_Q8W0G4 P0529E05.15 protein [Oryza sativa] 161 5e-38
UniRef100_Q6Y9P5 COI1 [Oryza sativa] 161 5e-38
UniRef100_Q60EH4 Putative LRR-containing F-box protein [Oryza sa... 154 5e-36
UniRef100_Q6TDU2 Coronatine-insensitive 1 [Lycopersicon esculentum] 154 5e-36
UniRef100_Q84QA7 Hypothetical protein OJ1012B02.14 [Oryza sativa] 140 1e-31
UniRef100_Q6K8E1 F-box containing protein TIR1-like [Oryza sativa] 77 1e-12
>UniRef100_Q9LW29 Transport inhibitor response-like protein [Arabidopsis thaliana]
Length = 575
Score = 505 bits (1300), Expect = e-141
Identities = 239/316 (75%), Positives = 284/316 (89%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEHVFD+V SH DRN++SLVCKSWY+IER++RQ+VFIGNCY+I+PERL+ RF
Sbjct: 1 MNYFPDEVIEHVFDFVTSHKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINPERLLRRF 60
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P LKSLTLKGKPHFADF+LVPH WGGFV PWIEALA+++VGLEELRLKRMVV+DESLELL
Sbjct: 61 PCLKSLTLKGKPHFADFNLVPHEWGGFVLPWIEALARSRVGLEELRLKRMVVTDESLELL 120
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
SRSFVNFKSLVLVSCEGFTTDGLA++AANCR LR+LDLQENE++DH+GQWLSCFP++CT+
Sbjct: 121 SRSFVNFKSLVLVSCEGFTTDGLASIAANCRHLRDLDLQENEIDDHRGQWLSCFPDTCTT 180
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LV+LNFACL+G+ NL ALERLV+RSPNLKSL+LNR+VP+DAL R++ APQ++DLG+GS+
Sbjct: 181 LVTLNFACLEGETNLVALERLVARSPNLKSLKLNRAVPLDALARLMACAPQIVDLGVGSY 240
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
+D +S++Y A I KC S+ SLSGFLE AP L+A +PIC NLTSLNLSYAA I G
Sbjct: 241 ENDPDSESYLKLMAVIKKCTSLRSLSGFLEAAPHCLSAFHPICHNLTSLNLSYAAEIHGS 300
Query: 301 ELIKLIRHCGKLQRLW 316
LIKLI+HC KLQRLW
Sbjct: 301 HLIKLIQHCKKLQRLW 316
>UniRef100_Q9LPW7 F13K23.7 protein [Arabidopsis thaliana]
Length = 577
Score = 494 bits (1273), Expect = e-138
Identities = 235/316 (74%), Positives = 281/316 (88%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEHVFD+V SH DRNS+SLVCKSW++IERF+R+ VFIGNCY+I+PERL+ RF
Sbjct: 1 MNYFPDEVIEHVFDFVASHKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINPERLIRRF 60
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P LKSLTLKGKPHFADF+LVPH WGGFV+PWIEALA+++VGLEELRLKRMVV+DESL+LL
Sbjct: 61 PCLKSLTLKGKPHFADFNLVPHEWGGFVHPWIEALARSRVGLEELRLKRMVVTDESLDLL 120
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
SRSF NFKSLVLVSCEGFTTDGLA++AANCR LRELDLQENE++DH+GQWL+CFP+SCT+
Sbjct: 121 SRSFANFKSLVLVSCEGFTTDGLASIAANCRHLRELDLQENEIDDHRGQWLNCFPDSCTT 180
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
L+SLNFACLKG+ N+ ALERLV+RSPNLKSL+LNR+VP+DAL R+++ APQL+DLG+GS+
Sbjct: 181 LMSLNFACLKGETNVAALERLVARSPNLKSLKLNRAVPLDALARLMSCAPQLVDLGVGSY 240
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
++ + +++A I K S+ SLSGFLEVAP L A YPICQNL SLNLSYAA I G
Sbjct: 241 ENEPDPESFAKLMTAIKKYTSLRSLSGFLEVAPLCLPAFYPICQNLISLNLSYAAEIQGN 300
Query: 301 ELIKLIRHCGKLQRLW 316
LIKLI+ C +LQRLW
Sbjct: 301 HLIKLIQLCKRLQRLW 316
>UniRef100_Q7XVM8 OSJNBa0072K14.18 protein [Oryza sativa]
Length = 575
Score = 416 bits (1068), Expect = e-114
Identities = 196/316 (62%), Positives = 249/316 (78%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
M YFP+EV+EH+F ++ + DRN++SLVCK WY IER +R+ VF+GNCY++ R+ RF
Sbjct: 1 MTYFPEEVVEHIFSFLPAQRDRNTVSLVCKVWYEIERLSRRGVFVGNCYAVRAGRVAARF 60
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P++++LT+KGKPHFADF+LVP WGG+ PWIEA A+ GLEELR+KRMVVSDESLELL
Sbjct: 61 PNVRALTVKGKPHFADFNLVPPDWGGYAGPWIEAAARGCHGLEELRMKRMVVSDESLELL 120
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
+RSF F++LVL+SCEGF+TDGLAAVA++C+ LRELDLQENEVED +WLSCFP+SCTS
Sbjct: 121 ARSFPRFRALVLISCEGFSTDGLAAVASHCKLLRELDLQENEVEDRGPRWLSCFPDSCTS 180
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LVSLNFAC+KG++N G+LERLVSRSPNL+SLRLNRSV VD L +IL R P L DLG G+
Sbjct: 181 LVSLNFACIKGEVNAGSLERLVSRSPNLRSLRLNRSVSVDTLAKILLRTPNLEDLGTGNL 240
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
D +++Y + + KCK + SLSGF + +P L+ IYP+C LT LNLSYA +
Sbjct: 241 TDDFQTESYFKLTSALEKCKMLRSLSGFWDASPVCLSFIYPLCAQLTGLNLSYAPTLDAS 300
Query: 301 ELIKLIRHCGKLQRLW 316
+L K+I C KLQRLW
Sbjct: 301 DLTKMISRCVKLQRLW 316
>UniRef100_O24660 Transport inhibitor response 1 [Arabidopsis thaliana]
Length = 594
Score = 386 bits (992), Expect = e-106
Identities = 180/313 (57%), Positives = 241/313 (76%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP+EV+EHVF ++ DRNS+SLVCKSWY IER+ R++VFIGNCY++SP ++ RFP +
Sbjct: 9 FPEEVLEHVFSFIQLDKDRNSVSLVCKSWYEIERWCRRKVFIGNCYAVSPATVIRRFPKV 68
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+S+ LKGKPHFADF+LVP GWGG+VYPWIEA++ + LEE+RLKRMVV+D+ LEL+++S
Sbjct: 69 RSVELKGKPHFADFNLVPDGWGGYVYPWIEAMSSSYTWLEEIRLKRMVVTDDCLELIAKS 128
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F NFK LVL SCEGF+TDGLAA+AA CR+L+ELDL+E++V+D G WLS FP++ TSLVS
Sbjct: 129 FKNFKVLVLSSCEGFSTDGLAAIAATCRNLKELDLRESDVDDVSGHWLSHFPDTYTSLVS 188
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
LN +CL +++ ALERLV+R PNLKSL+LNR+VP++ L +L RAPQL +LG G + +
Sbjct: 189 LNISCLASEVSFSALERLVTRCPNLKSLKLNRAVPLEKLATLLQRAPQLEELGTGGYTAE 248
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ D Y+ + CK + LSGF + P L A+Y +C LT+LNLSYA + +L+
Sbjct: 249 VRPDVYSGLSVALSGCKELRCLSGFWDAVPAYLPAVYSVCSRLTTLNLSYAT-VQSYDLV 307
Query: 304 KLIRHCGKLQRLW 316
KL+ C KLQRLW
Sbjct: 308 KLLCQCPKLQRLW 320
>UniRef100_Q9ZR12 Putative homolog of transport inhibitor response 1 [Arabidopsis
thaliana]
Length = 585
Score = 370 bits (951), Expect = e-101
Identities = 178/313 (56%), Positives = 234/313 (73%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP +V+EH+ ++ S+ DRNS+SLVCKSW+ ER TR+RVF+GNCY++SP + RFP++
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+SLTLKGKPHFAD++LVP GWGG+ +PWIEA+A LEE+R+KRMVV+DE LE ++ S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F +FK LVL SCEGF+TDG+AA+AA CR+LR L+L+E VED G WLS FPES TSLVS
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCRNLRVLELRECIVEDLGGDWLSYFPESSTSLVS 184
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
L+F+CL ++ + LERLVSRSPNLKSL+LN +V +D L +L APQL +LG GSF
Sbjct: 185 LDFSCLDSEVKISDLERLVSRSPNLKSLKLNPAVTLDGLVSLLRCAPQLTELGTGSFAAQ 244
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
L +A++ CK + SLSG +V P L A+Y +C LTSLNLSYA + +L+
Sbjct: 245 LKPEAFSKLSEAFSNCKQLQSLSGLWDVLPEYLPALYSVCPGLTSLNLSYAT-VRMPDLV 303
Query: 304 KLIRHCGKLQRLW 316
+L+R C KLQ+LW
Sbjct: 304 ELLRRCSKLQKLW 316
>UniRef100_Q94AU0 Putative F-box protein GRR1 protein 1, AtFBL18 [Arabidopsis
thaliana]
Length = 585
Score = 369 bits (947), Expect = e-100
Identities = 177/313 (56%), Positives = 234/313 (74%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP +V+EH+ ++ S+ DRNS+SLVCKSW+ ER TR+RVF+GNCY++SP + RFP++
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+SLTLKGKPHFAD++LVP GWGG+ +PWIEA+A LEE+R+KR+VV+DE LE ++ S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRIVVTDECLEKIAAS 124
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F +FK LVL SCEGF+TDG+AA+AA CR+LR L+L+E VED G WLS FPES TSLVS
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCRNLRVLELRECIVEDLGGDWLSYFPESSTSLVS 184
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
L+F+CL ++ + LERLVSRSPNLKSL+LN +V +D L +L APQL +LG GSF
Sbjct: 185 LDFSCLDSEVKISDLERLVSRSPNLKSLKLNPAVTLDGLVSLLRCAPQLTELGTGSFAAQ 244
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
L +A++ CK + SLSG +V P L A+Y +C LTSLNLSYA + +L+
Sbjct: 245 LKPEAFSKLSEAFSNCKQLQSLSGLWDVLPEYLPALYSVCPGLTSLNLSYAT-VRMPDLV 303
Query: 304 KLIRHCGKLQRLW 316
+L+R C KLQ+LW
Sbjct: 304 ELLRRCSKLQKLW 316
>UniRef100_Q9C5Y7 GRR1-like protein 1 [Arabidopsis thaliana]
Length = 585
Score = 368 bits (945), Expect = e-100
Identities = 176/313 (56%), Positives = 234/313 (74%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP +V+EH+ ++ S+ DRNS+SLVCKSW+ ER TR+RVF+GNCY++SP + RFP++
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+SLTLKGKPHFAD++LVP GWGG+ +PWIEA+A LEE+R+KRMVV+DE LE ++ S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F +FK LVL SCEGF+TDG+AA+A+ CR+LR L+L+E VED G WLS FPES TSLVS
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIASTCRNLRVLELRECIVEDLGGDWLSYFPESSTSLVS 184
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
L+F+CL ++ + LERLVSRSPNLKSL+LN +V +D L +L APQL +LG GSF
Sbjct: 185 LDFSCLDSEVKISDLERLVSRSPNLKSLKLNPAVTLDGLVSLLRCAPQLTELGTGSFAAQ 244
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
L +A++ CK + SLSG +V P L A+Y +C LTSLNL+YA + +L+
Sbjct: 245 LKPEAFSKLSEAFSNCKQLQSLSGLWDVLPEYLPALYSVCPGLTSLNLTYAT-VRMPDLV 303
Query: 304 KLIRHCGKLQRLW 316
+L+R C KLQ+LW
Sbjct: 304 ELLRRCSKLQKLW 316
>UniRef100_Q9LTX2 Transport inhibitor response 1 protein [Arabidopsis thaliana]
Length = 619
Score = 307 bits (787), Expect = 5e-82
Identities = 165/320 (51%), Positives = 210/320 (65%), Gaps = 9/320 (2%)
Query: 4 FPDEVIEHVFDYVV----SHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVER 59
FPD V+E+V + V+ S DRN+ SLVCKSW+R+E TR VFIGNCY++SP RL +R
Sbjct: 50 FPDHVLENVLENVLQFLDSRCDRNAASLVCKSWWRVEALTRSEVFIGNCYALSPARLTQR 109
Query: 60 FPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLEL 119
F ++SL LKGKP FADF+L+P WG PW+ +A+ LE++ LKRM V+D+ L L
Sbjct: 110 FKRVRSLVLKGKPRFADFNLMPPDWGANFAPWVSTMAQAYPCLEKVDLKRMFVTDDDLAL 169
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCT 179
L+ SF FK L+LV CEGF T G++ VA CR L+ LDL E+EV D + W+SCFPE T
Sbjct: 170 LADSFPGFKELILVCCEGFGTSGISIVANKCRKLKVLDLIESEVTDDEVDWISCFPEDVT 229
Query: 180 SLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGS 239
L SL F C++ IN ALE LV+RSP LK LRLNR V + L R+L APQL LG GS
Sbjct: 230 CLESLAFDCVEAPINFKALEGLVARSPFLKKLRLNRFVSLVELHRLLLGAPQLTSLGTGS 289
Query: 240 FFHD--LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGI 297
F HD S+ + A CKS+ LSGF E+ P L AI+P+C NLTSLN SYA
Sbjct: 290 FSHDEEPQSEQEPDYAAAFRACKSVVCLSGFRELMPEYLPAIFPVCANLTSLNFSYAN-- 347
Query: 298 LGIELIK-LIRHCGKLQRLW 316
+ ++ K +I +C KLQ W
Sbjct: 348 ISPDMFKPIILNCHKLQVFW 367
Score = 34.7 bits (78), Expect = 7.6
Identities = 46/195 (23%), Positives = 74/195 (37%), Gaps = 31/195 (15%)
Query: 128 KSLVLVSCEGFTTDGLAAVAANCRSLREL-----DLQENEVEDHKGQWLSCFPESCTSLV 182
K V + + +GL AVAA C+ LREL D +E+ L E C L
Sbjct: 362 KLQVFWALDSICDEGLQAVAATCKELRELRIFPFDPREDSEGPVSELGLQAISEGCRKLE 421
Query: 183 SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFH 242
S+ + C R N + ++ + P + R+ D H
Sbjct: 422 SILYFC--------------QRMTNAAVIAMSENCPELTVFRLCIMGRHRPD-------H 460
Query: 243 DLNSDAYAMFKATILKCKSITSL--SGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
F A + CK +T L SG L F Y + + +L++++ AG +
Sbjct: 461 VTGKPMDEGFGAIVKNCKKLTRLAVSGLLTDQAFRYMGEYG--KLVRTLSVAF-AGDSDM 517
Query: 301 ELIKLIRHCGKLQRL 315
L ++ C +LQ+L
Sbjct: 518 ALRHVLEGCPRLQKL 532
>UniRef100_Q8RWQ8 AT4g24390/T22A6_220 [Arabidopsis thaliana]
Length = 623
Score = 300 bits (769), Expect = 6e-80
Identities = 160/313 (51%), Positives = 200/313 (63%), Gaps = 3/313 (0%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
+ V+E+V ++ S DRN++SLVC+SWYR+E TR VFIGNCYS+SP RL+ RF ++S
Sbjct: 56 ENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSLSPARLIHRFKRVRS 115
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP FADF+L+P WG PW+ A AK LE++ LKRM V+D+ L LL+ SF
Sbjct: 116 LVLKGKPRFADFNLMPPNWGAQFSPWVAATAKAYPWLEKVHLKRMFVTDDDLALLAESFP 175
Query: 126 NFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLN 185
FK L LV CEGF T G+A VA CR L+ LDL E+EV D + W+SCFPE T L SL+
Sbjct: 176 GFKELTLVCCEGFGTSGIAIVANKCRQLKVLDLMESEVTDDELDWISCFPEGETHLESLS 235
Query: 186 FACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD-- 243
F C++ IN ALE LV RSP LK LR NR V ++ L R++ RAPQL LG GSF D
Sbjct: 236 FDCVESPINFKALEELVVRSPFLKKLRTNRFVSLEELHRLMVRAPQLTSLGTGSFSPDNV 295
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ + A CKSI LSGF E P L AI +C NLTSLN SY A I L
Sbjct: 296 PQGEQQPDYAAAFRACKSIVCLSGFREFRPEYLLAISSVCANLTSLNFSY-ANISPHMLK 354
Query: 304 KLIRHCGKLQRLW 316
+I +C ++ W
Sbjct: 355 PIISNCHNIRVFW 367
Score = 41.6 bits (96), Expect = 0.062
Identities = 63/243 (25%), Positives = 98/243 (39%), Gaps = 39/243 (16%)
Query: 83 GWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDG 142
G+ F ++ A++ L L +S L+ + + N + V + + +G
Sbjct: 319 GFREFRPEYLLAISSVCANLTSLNFSYANISPHMLKPIISNCHNIR--VFWALDSIRDEG 376
Query: 143 LAAVAANCRSLREL-----DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGA 197
L AVAA C+ LREL D +E+ G L E C L S+ + C ++ GA
Sbjct: 377 LQAVAATCKELRELRIFPFDPREDSEGPVSGVGLQAISEGCRKLESILYFC--QNMTNGA 434
Query: 198 LERLVSRSPNLKSLRL---NRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKA 254
+ + P L RL R P +T P MD G G A
Sbjct: 435 VTAMSENCPQLTVFRLCIMGRHRPDH-----VTGKP--MDDGFG---------------A 472
Query: 255 TILKCKSITSL--SGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKL 312
+ CK +T L SG L FS Y + + +L++++ AG L ++ C KL
Sbjct: 473 IVKNCKKLTRLAVSGLLTDEAFSYIGEYG--KLIRTLSVAF-AGNSDKALRYVLEGCPKL 529
Query: 313 QRL 315
Q+L
Sbjct: 530 QKL 532
>UniRef100_Q9AUH6 F-box containing protein TIR1 [Populus tremula x Populus
tremuloides]
Length = 635
Score = 299 bits (766), Expect = 1e-79
Identities = 156/320 (48%), Positives = 211/320 (65%), Gaps = 8/320 (2%)
Query: 4 FPDEVIEHVFD----YVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVER 59
+PD+V+E+V + ++ S DRN+ SLVC+SWYR+E TR +FIGNCY++SP+R + R
Sbjct: 65 YPDQVLENVLENVLWFLTSRKDRNAASLVCRSWYRVEALTRSDLFIGNCYAVSPKRAMSR 124
Query: 60 FPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLEL 119
F ++S+TLKGKP FADF+L+P WG PW+ A+A LE++ LKRM V+D+ L L
Sbjct: 125 FTRIRSVTLKGKPRFADFNLMPPYWGAHFAPWVSAMAMTYPWLEKVHLKRMSVTDDDLAL 184
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCT 179
L+ SF FK LVLV CEGF T GLA V + CR L+ LDL E++V D + W+SCFP++ T
Sbjct: 185 LAESFSGFKELVLVCCEGFGTSGLAIVVSRCRQLKVLDLIESDVSDDEVDWISCFPDTET 244
Query: 180 SLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGS 239
L SL F C+ I+ LERLV+RSP+LK LRLNR V + L R++ RAP L LG GS
Sbjct: 245 CLESLIFDCVDCPIDFDELERLVARSPSLKKLRLNRYVSIGQLYRLMIRAPHLTHLGTGS 304
Query: 240 F--FHDLNS-DAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAG 296
F D++ + + + CKS+ LSGF E+ P L AI P+C NLTSLN S+ A
Sbjct: 305 FSPSEDVSQVEQGPDYASAFAACKSLVCLSGFREIIPDYLPAINPVCANLTSLNFSF-AD 363
Query: 297 ILGIELIKLIRHCGKLQRLW 316
+ +L +I +C KLQ W
Sbjct: 364 VSAEQLKPIISNCHKLQIFW 383
>UniRef100_Q6K8E2 Putative F-box containing protein TIR1 [Oryza sativa]
Length = 637
Score = 288 bits (738), Expect = 2e-76
Identities = 153/317 (48%), Positives = 207/317 (65%), Gaps = 7/317 (2%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
+ V+E V +++ + DRN+ SLVC+SWYR E TR+ +FIGNCY++SP R VERF +++
Sbjct: 69 ENVLESVLEFLTAARDRNAASLVCRSWYRAEAQTRRELFIGNCYAVSPRRAVERFGGVRA 128
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
+ LKGKP FADFSLVP+GWG +V PW+ AL LE + LKRM VS++ L L+++SF
Sbjct: 129 VVLKGKPRFADFSLVPYGWGAYVSPWVAALGPAYPHLERICLKRMTVSNDDLALIAKSFP 188
Query: 126 NFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE---NEVEDHKGQWLSCFPESCTSLV 182
FK L LV C+GF+T GLAA+A CR LR LDL E +E ED W+S FPES TSL
Sbjct: 189 LFKELSLVCCDGFSTLGLAAIAERCRHLRVLDLIEDYIDEEEDELVDWISKFPESNTSLE 248
Query: 183 SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFH 242
SL F C+ N ALE LV+RSP ++ LR+N V V+ L+R++ RAPQL LG G+F
Sbjct: 249 SLVFDCVSVPFNFEALEALVARSPAMRRLRMNHHVTVEQLRRLMARAPQLTHLGTGAFRS 308
Query: 243 DLNSD---AYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILG 299
+ + + +S+ LSGF +V P L AI+P+C NLTSLN S+ A +
Sbjct: 309 EPGPGGALSVTELATSFAASRSLICLSGFRDVNPEYLPAIHPVCANLTSLNFSF-ANLTA 367
Query: 300 IELIKLIRHCGKLQRLW 316
EL +IR+C +L+ W
Sbjct: 368 EELTPIIRNCVRLRTFW 384
Score = 34.7 bits (78), Expect = 7.6
Identities = 56/211 (26%), Positives = 90/211 (42%), Gaps = 39/211 (18%)
Query: 115 ESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL-QENEVEDHKGQ---- 169
E L + R+ V ++ ++ G +GL AVA C LREL + + ED +G
Sbjct: 368 EELTPIIRNCVRLRTFWVLDTVG--DEGLRAVAETCSDLRELRVFPFDATEDSEGSVSDV 425
Query: 170 WLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRL---NRSVPVDALQRIL 226
L E C L S+ + C + + A+ + +L + RL R P RI
Sbjct: 426 GLQAISEGCRKLESILYFCQR--MTNAAVIAMSKNCSDLVTFRLCIMGRHRP----DRI- 478
Query: 227 TRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSIT--SLSGFLEVAPFSLAAIYPICQ 284
T P MD G G A ++ CK +T S+SG L F+ Y +
Sbjct: 479 TGEP--MDDGFG---------------AIVMNCKKLTRLSVSGLLTDKAFAYIGKYG--K 519
Query: 285 NLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ +L++++ AG + L + C +LQ+L
Sbjct: 520 LIKTLSVAF-AGNSDMSLQSVFEGCTRLQKL 549
>UniRef100_Q5WN05 Putative transport inhibitor response TIR1 [Oryza sativa]
Length = 462
Score = 229 bits (583), Expect = 2e-58
Identities = 117/207 (56%), Positives = 151/207 (72%), Gaps = 1/207 (0%)
Query: 110 MVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQ 169
MVV+DE LE+++ SF NF+ L LVSC+GF+T GLAA+AA CR LRELDLQENE+ED
Sbjct: 1 MVVTDECLEMIAASFRNFQVLRLVSCDGFSTAGLAAIAAGCRHLRELDLQENEIEDCSIH 60
Query: 170 WLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRA 229
WLS FPES TSLV+LNF+CL+G++N+ LERLV+R NLK+L+LN ++P+D L +L +A
Sbjct: 61 WLSLFPESFTSLVTLNFSCLEGEVNITVLERLVTRCHNLKTLKLNNAIPLDKLASLLHKA 120
Query: 230 PQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSL 289
PQL++LG G F D +SD +A +A CKS+ LSG + P L A Y +C+ LTSL
Sbjct: 121 PQLVELGTGKFSADYHSDLFAKLEAAFGGCKSLRRLSGAWDAVPDYLPAFYCVCEGLTSL 180
Query: 290 NLSYAAGILGIELIKLIRHCGKLQRLW 316
NLSYA + G ELIK I C LQ+LW
Sbjct: 181 NLSYAT-VRGPELIKFISRCRNLQQLW 206
>UniRef100_Q8H7P5 Putative F-box containing protein TIR1 [Oryza sativa]
Length = 603
Score = 207 bits (528), Expect = 5e-52
Identities = 125/324 (38%), Positives = 185/324 (56%), Gaps = 14/324 (4%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D V+E V ++ S DR + SLVC+SW R E TR V + N + SP R+ RFP +
Sbjct: 29 DNVLETVLQFLDSARDRCAASLVCRSWSRAESATRASVAVRNLLAASPARVARRFPAARR 88
Query: 66 LTLKGKPHFADFSLVPHGWGGFVY-PWIEALAKNKV-GLEELRLKRMVVSDESLELLSRS 123
+ LKG+P FADF+L+P GW G + PW A+A L L LKR+ V+D+ L+L+SRS
Sbjct: 89 VLLKGRPRFADFNLLPPGWAGADFRPWAAAVAAAAFPALASLFLKRITVTDDDLDLVSRS 148
Query: 124 F-VNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL---QENEVEDHKGQWLSCFPESCT 179
+F+ L L+ C+GF++ GLA++A++CR LR LD+ + N+ +D W++ FP T
Sbjct: 149 LPASFRDLSLLLCDGFSSAGLASIASHCRGLRVLDVVDCEMNDDDDEVVDWVAAFPPGTT 208
Query: 180 SLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGS 239
L SL+F C ++ ALE LV+RSP L L +N V + L+R++ P+L LG G+
Sbjct: 209 DLESLSFECYVRPVSFAALEALVARSPRLTRLGVNEHVSLGQLRRLMANTPRLTHLGTGA 268
Query: 240 FF-----HDLNSDAYAMFK--ATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLS 292
F D+ D M A+ + ++ SLSGF E P L I + NLT+L+ S
Sbjct: 269 FRPGDGPEDVGLDIEQMASAFASAGRTNTLVSLSGFREFEPEYLPTIAAVSGNLTNLDFS 328
Query: 293 YAAGILGIELIKLIRHCGKLQRLW 316
Y + + + I C L+RL+
Sbjct: 329 YCP-VTPDQFLPFIGQCHNLERLY 351
>UniRef100_O04197 Coronatine-insensitive 1 (COI1), AtFBL2 [Arabidopsis thaliana]
Length = 592
Score = 167 bits (424), Expect = 6e-40
Identities = 100/311 (32%), Positives = 166/311 (53%), Gaps = 14/311 (4%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D+VIE V Y+ DR+S SLVC+ W++I+ TR+ V + CY+ +P+RL RFP+L+S
Sbjct: 18 DDVIEQVMTYITDPKDRDSASLVCRRWFKIDSETREHVTMALCYTATPDRLSRRFPNLRS 77
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP A F+L+P WGG+V PW+ ++ N L+ + +RM+VSD L+ L+++
Sbjct: 78 LKLKGKPRAAMFNLIPENWGGYVTPWVTEISNNLRQLKSVHFRRMIVSDLDLDRLAKARA 137
Query: 126 -NFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSL 184
+ ++L L C GFTTDGL ++ +CR ++ L ++E+ + G+WL + TSL L
Sbjct: 138 DDLETLKLDKCSGFTTDGLLSIVTHCRKIKTLLMEESSFSEKDGKWLHELAQHNTSLEVL 197
Query: 185 NFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
NF + I+ LE + +L S+++ + L A L + GS D
Sbjct: 198 NFYMTEFAKISPKDLETIARNCRSLVSVKVG-DFEILELVGFFKAAANLEEFCGGSLNED 256
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ M ++ + + L G + P + ++P + L+L YA L+
Sbjct: 257 IGMPEKYM---NLVFPRKLCRL-GLSYMGPNEMPILFPFAAQIRKLDLLYA-------LL 305
Query: 304 KLIRHCGKLQR 314
+ HC +Q+
Sbjct: 306 ETEDHCTLIQK 316
>UniRef100_Q8W0G4 P0529E05.15 protein [Oryza sativa]
Length = 630
Score = 161 bits (407), Expect = 5e-38
Identities = 102/293 (34%), Positives = 147/293 (49%), Gaps = 7/293 (2%)
Query: 2 NYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFP 61
++ PDE + V +V DR + S VC+ W+RI+ TR+ V + CY+ P RL ERFP
Sbjct: 54 DWVPDEALHLVMGHVEDPRDREAASRVCRRWHRIDALTRKHVTVAFCYAARPARLRERFP 113
Query: 62 DLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLS 121
L+SL+LKGKP A + L+P WG + PWI+ LA L+ L L+RM V+D + L
Sbjct: 114 RLESLSLKGKPRAAMYGLIPDDWGAYAAPWIDELAAPLECLKALHLRRMTVTDADIAALV 173
Query: 122 RSFVN-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
R+ + + L L C GF+TD L VA +CRSLR L L+E + D G+WL + +
Sbjct: 174 RARGHMLQELKLDKCIGFSTDALRLVARSCRSLRTLFLEECHITDKGGEWLHELAVNNSV 233
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LV+LNF + + LE L +L SL+++ D + T A L D G+F
Sbjct: 234 LVTLNFYMTELKVAPADLELLAKNCKSLISLKMSECDLSDLISFFQT-ANALQDFAGGAF 292
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSY 293
+ Y K C G + + I+P L L+L Y
Sbjct: 293 YEVGELTKYEKVKFPPRLC-----FLGLTYMGTNEMPVIFPFSMKLKKLDLQY 340
>UniRef100_Q6Y9P5 COI1 [Oryza sativa]
Length = 595
Score = 161 bits (407), Expect = 5e-38
Identities = 102/293 (34%), Positives = 147/293 (49%), Gaps = 7/293 (2%)
Query: 2 NYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFP 61
++ PDE + V +V DR + S VC+ W+RI+ TR+ V + CY+ P RL ERFP
Sbjct: 19 DWVPDEALHLVMGHVEDPRDREAASRVCRRWHRIDALTRKHVTVAFCYAARPARLRERFP 78
Query: 62 DLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLS 121
L+SL+LKGKP A + L+P WG + PWI+ LA L+ L L+RM V+D + L
Sbjct: 79 RLESLSLKGKPRAAMYGLIPDDWGAYAAPWIDELAAPLECLKALHLRRMTVTDADIAALV 138
Query: 122 RSFVN-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
R+ + + L L C GF+TD L VA +CRSLR L L+E + D G+WL + +
Sbjct: 139 RARGHMLQELKLDKCIGFSTDALRLVARSCRSLRTLFLEECHITDKGGEWLHELAVNNSV 198
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LV+LNF + + LE L +L SL+++ D + T A L D G+F
Sbjct: 199 LVTLNFYMTELKVAPADLELLAKNCKSLISLKMSECDLSDLISFFQT-ANALQDFAGGAF 257
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSY 293
+ Y K C G + + I+P L L+L Y
Sbjct: 258 YEVGELTKYEKVKFPPRLC-----FLGLTYMGTNEMPVIFPFSMKLKKLDLQY 305
>UniRef100_Q60EH4 Putative LRR-containing F-box protein [Oryza sativa]
Length = 597
Score = 154 bits (390), Expect = 5e-36
Identities = 98/290 (33%), Positives = 143/290 (48%), Gaps = 7/290 (2%)
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLK 64
P+E + V YV DR ++SLVC+ W+RI+ TR+ V + CY+ SP L+ RFP L+
Sbjct: 24 PEEALHLVLGYVDDPRDREAVSLVCRRWHRIDALTRKHVTVPFCYAASPAHLLARFPRLE 83
Query: 65 SLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSF 124
SL +KGKP A + L+P WG + PW+ LA L+ L L+RMVV+D+ L L R+
Sbjct: 84 SLAVKGKPRAAMYGLIPEDWGAYARPWVAELAAPLECLKALHLRRMVVTDDDLAALVRAR 143
Query: 125 VN-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
+ + L L C GF+TD L VA +CRSLR L L+E + D+ +WL + L +
Sbjct: 144 GHMLQELKLDKCSGFSTDALRLVARSCRSLRTLFLEECSIADNGTEWLHDLAVNNPVLET 203
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
LNF + + LE L + +L SL+++ L A L + G+F
Sbjct: 204 LNFHMTELTVVPADLELLAKKCKSLISLKIS-DCDFSDLIGFFRMAASLQEFAGGAFIEQ 262
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSY 293
Y K C G + + I+P L L+L Y
Sbjct: 263 GELTKYGNVKFPSRLCS-----LGLTYMGTNEMPIIFPFSALLKKLDLQY 307
>UniRef100_Q6TDU2 Coronatine-insensitive 1 [Lycopersicon esculentum]
Length = 603
Score = 154 bits (390), Expect = 5e-36
Identities = 102/321 (31%), Positives = 162/321 (49%), Gaps = 25/321 (7%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D V E V Y+ DR+++SLVCK W++I+ TR+ + + CY+ PE+L RFP L+S
Sbjct: 15 DTVWECVIPYIQESRDRDAVSLVCKRWWQIDAITRKHITMALCYTAKPEQLSRRFPHLES 74
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL-SRSF 124
+ LKGKP A F+L+P WGG+V PW+ + K+ L+ L +RM+V D LELL +R
Sbjct: 75 VKLKGKPRAAMFNLIPEDWGGYVTPWVMEITKSFSKLKALHFRRMIVRDSDLELLANRRG 134
Query: 125 VNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSL 184
+ L L C GF+TDGL ++ +C++LR L ++E+ + + G+W + T L +L
Sbjct: 135 RVLQVLKLDKCSGFSTDGLLHISRSCKNLRTLLMEESYIIEKDGEWAHELALNNTVLENL 194
Query: 185 NFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF--- 240
NF + LE + +L S++++ + L A L + G G+F
Sbjct: 195 NFYMTDLLQVRAEDLELIARNCKSLVSMKIS-ECEITNLLGFFRAAAALEEFGGGAFNDQ 253
Query: 241 -------FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSY 293
++ +S YA C+ G + ++ ++PI L L+L Y
Sbjct: 254 PELVVENGYNEHSGKYAALVFPPRLCQ-----LGLTYLGRNEMSILFPIASRLRKLDLLY 308
Query: 294 AAGILGIELIKLIRHCGKLQR 314
A L+ HC LQR
Sbjct: 309 A-------LLDTAAHCFLLQR 322
>UniRef100_Q84QA7 Hypothetical protein OJ1012B02.14 [Oryza sativa]
Length = 589
Score = 140 bits (353), Expect = 1e-31
Identities = 105/337 (31%), Positives = 163/337 (48%), Gaps = 30/337 (8%)
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLK 64
PD + V +V DR+++SLVC+ W R++ +R+ V + YS +P+RL RFP L+
Sbjct: 19 PDVALGLVMGFVEDPWDRDAISLVCRHWCRVDALSRKHVTVAMAYSTTPDRLFRRFPCLE 78
Query: 65 SLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSF 124
SL LK KP A F+L+P WGG PWI L+ + L+ L L+RM+VSD+ L++L R+
Sbjct: 79 SLKLKAKPRAAMFNLIPEDWGGSASPWIRQLSASFHFLKALHLRRMIVSDDDLDVLVRAK 138
Query: 125 VN-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEV-EDHKGQWLSCFPESCTSLV 182
+ S L C GF+T LA VA C+ L L L+++ + E +W+ + + L
Sbjct: 139 AHMLSSFKLDRCSGFSTSSLALVARTCKKLETLFLEDSIIAEKENDEWIRELATNNSVLE 198
Query: 183 SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF-- 240
+LNF + L LV LK L+++ +D + T A L D GSF
Sbjct: 199 TLNFFLTDLRASPAYLTLLVRNCRRLKVLKISECFMLDLVDLFRT-AEILQDFAGGSFDD 257
Query: 241 ----FHDLNSDAYAMFKATILKCKSITSLSGFLEVA-PFSLA-----------------A 278
N + Y F ++L+ + + ++V P+ A
Sbjct: 258 QGQVEESRNYENY-YFPPSLLRLSLLYMGTKEMQVLFPYGAALKKLDLQFTFLSTEDHCQ 316
Query: 279 IYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ C NL L + G G+E++ + C KLQRL
Sbjct: 317 LVQRCPNLEILEVRDVIGDRGLEVV--AQTCKKLQRL 351
>UniRef100_Q6K8E1 F-box containing protein TIR1-like [Oryza sativa]
Length = 364
Score = 77.4 bits (189), Expect = 1e-12
Identities = 43/112 (38%), Positives = 65/112 (57%), Gaps = 4/112 (3%)
Query: 208 LKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSD---AYAMFKATILKCKSITS 264
++ LR+N V V+ L+R++ RAPQL LG G+F + + + +S+
Sbjct: 1 MRRLRMNHHVTVEQLRRLMARAPQLTHLGTGAFRSEPGPGGALSVTELATSFAASRSLIC 60
Query: 265 LSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
LSGF +V P L AI+P+C NLTSLN S+ A + EL +IR+C +L+ W
Sbjct: 61 LSGFRDVNPEYLPAIHPVCANLTSLNFSF-ANLTAEELTPIIRNCVRLRTFW 111
Score = 34.7 bits (78), Expect = 7.6
Identities = 56/211 (26%), Positives = 90/211 (42%), Gaps = 39/211 (18%)
Query: 115 ESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL-QENEVEDHKGQ---- 169
E L + R+ V ++ ++ G +GL AVA C LREL + + ED +G
Sbjct: 95 EELTPIIRNCVRLRTFWVLDTVG--DEGLRAVAETCSDLRELRVFPFDATEDSEGSVSDV 152
Query: 170 WLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRL---NRSVPVDALQRIL 226
L E C L S+ + C + + A+ + +L + RL R P RI
Sbjct: 153 GLQAISEGCRKLESILYFCQR--MTNAAVIAMSKNCSDLVTFRLCIMGRHRP----DRI- 205
Query: 227 TRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSIT--SLSGFLEVAPFSLAAIYPICQ 284
T P MD G G A ++ CK +T S+SG L F+ Y +
Sbjct: 206 TGEP--MDDGFG---------------AIVMNCKKLTRLSVSGLLTDKAFAYIGKYG--K 246
Query: 285 NLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ +L++++ AG + L + C +LQ+L
Sbjct: 247 LIKTLSVAF-AGNSDMSLQSVFEGCTRLQKL 276
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.340 0.146 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 826,914,587
Number of Sequences: 2790947
Number of extensions: 30488928
Number of successful extensions: 97187
Number of sequences better than 10.0: 312
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 236
Number of HSP's that attempted gapping in prelim test: 96228
Number of HSP's gapped (non-prelim): 947
length of query: 562
length of database: 848,049,833
effective HSP length: 133
effective length of query: 429
effective length of database: 476,853,882
effective search space: 204570315378
effective search space used: 204570315378
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC133780.9


