

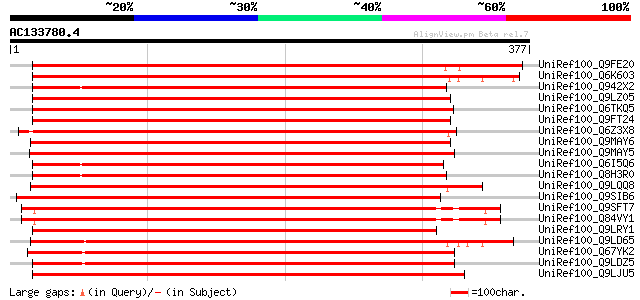
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.4 - phase: 1
(377 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidops... 456 e-127
UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa] 446 e-124
UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [... 429 e-119
UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana] 426 e-118
UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis] 426 e-118
UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica ... 423 e-117
UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa] 420 e-116
UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra] 420 e-116
UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra] 418 e-115
UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza... 415 e-115
UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa] 412 e-114
UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVI... 407 e-112
UniRef100_Q9SIB6 Hypothetical protein At2g28590 [Arabidopsis tha... 404 e-111
UniRef100_Q9SFT7 Hypothetical protein T1B9.28 [Arabidopsis thali... 403 e-111
UniRef100_Q84VY1 At3g07070 [Arabidopsis thaliana] 403 e-111
UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidops... 397 e-109
UniRef100_Q9LD65 Hypothetical protein P0462H08.22 (EST C22619(S1... 395 e-108
UniRef100_Q67YK2 Hypothetical protein At1g20650 [Arabidopsis tha... 392 e-108
UniRef100_Q9LDZ5 F2D10.13 [Arabidopsis thaliana] 392 e-107
UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidops... 388 e-106
>UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidopsis thaliana]
Length = 456
Score = 456 bits (1173), Expect = e-127
Identities = 228/381 (59%), Positives = 280/381 (72%), Gaps = 25/381 (6%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A F FRELA AT NF +TF+G+GGFG VYKG+L STGQ VAVK+LD G QG +EFLV
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYE+MP+GSLE HLHDL PD E LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL +LH +A P VIYRD KSSNILLDEGF+PKLSDFGLAK GPTGD+S+V+TRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVV LELITGR+A D H +++LV WARPLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F KL DP L+G +P L AL +A MC++E RP D+V AL YL+++ Y P
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
Query: 316 -----------KASEIVSPGE-------------MEHDESPNETTMILVKDSLREQALAE 351
+ + +++ + E ++SP ET IL +D RE+A+AE
Sbjct: 371 SKDDSRRNRDERGARLITRNDDGGGSGSKFDLEGSEKEDSPRETARILNRDINRERAVAE 430
Query: 352 AKQWGETWREKRKQSGQNSPE 372
AK WGE+ REKR+QS Q + E
Sbjct: 431 AKMWGESLREKRRQSEQGTSE 451
>UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa]
Length = 526
Score = 446 bits (1146), Expect = e-124
Identities = 234/412 (56%), Positives = 281/412 (67%), Gaps = 58/412 (14%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
AQ FTFRELATAT+NFR E F+G+GGFG VYKG+L STGQ VA+K+L+ G QG +EFLV
Sbjct: 107 AQTFTFRELATATRNFRPECFLGEGGFGRVYKGRLESTGQVVAIKQLNRDGLQGNREFLV 166
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHH NLV++IGYCA+GDQRLLVYEYM GSLE HLHDL PD E LDWNTRM+I
Sbjct: 167 EVLMLSLLHHQNLVNLIGYCADGDQRLLVYEYMHFGSLEDHLHDLPPDKEALDWNTRMKI 226
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL YLH +A P VIYRD KSSNILLDE F+PKLSDFGLAK GP GD+S+V+TRV
Sbjct: 227 AAGAAKGLEYLHDKANPPVIYRDFKSSNILLDESFHPKLSDFGLAKLGPVGDKSHVSTRV 286
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVVLLELITGRRA D TR H +++LV WARPLF
Sbjct: 287 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVLLELITGRRAIDSTRPHGEQNLVSWARPLFN 346
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ K+ DP L+G YP+ GL AL +A MC++ + RP D+V AL YL+S+ Y P
Sbjct: 347 DRRKLPKMADPRLEGRYPMRGLYQALAVASMCIQSEAASRPLIADVVTALSYLASQSYDP 406
Query: 316 KAS-------------------------EIVSPG----EMEHDESPNETTMILVKD---- 342
A+ E S G + ++SP E IL KD
Sbjct: 407 NAAHASRKPGGDQRSKVGENGRVVSRNDEASSSGHKSPNKDREDSPKEPPGILNKDFDRE 466
Query: 343 ----------------------SLREQALAEAKQWGETWREKRK--QSGQNS 370
RE+ +AEAK WGE WR+KR+ ++GQ S
Sbjct: 467 RMVAEAKMWGDRERMVAEAKMWGDRERMVAEAKMWGENWRDKRRAIENGQGS 518
>UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [Oryza sativa]
Length = 497
Score = 429 bits (1103), Expect = e-119
Identities = 208/302 (68%), Positives = 247/302 (80%), Gaps = 2/302 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
AQ FTFRELA ATKNFR + +G+GGFG VYKG+L TGQAVAVK+LD G QG +EFLV
Sbjct: 74 AQTFTFRELAAATKNFRQDCLLGEGGFGRVYKGRL-ETGQAVAVKQLDRNGLQGNREFLV 132
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHH NLV++IGYCA+GDQRLLVYE+MP+GSLE HLHDL PD EPLDWNTRM+I
Sbjct: 133 EVLMLSLLHHTNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEPLDWNTRMKI 192
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL YLH +A P VIYRD KSSNILL EGF+PKLSDFGLAK GP GD+++V+TRV
Sbjct: 193 AAGAAKGLEYLHDKASPPVIYRDFKSSNILLGEGFHPKLSDFGLAKLGPVGDKTHVSTRV 252
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVV LELITGR+A D T+ +++LV WARPLF+
Sbjct: 253 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPQGEQNLVAWARPLFK 312
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F K+ DP LQG +P+ GL AL +A MCL+E RP GD+V AL YL+S+ Y P
Sbjct: 313 DRRKFPKMADPMLQGRFPMRGLYQALAVAAMCLQEQATTRPHIGDVVTALSYLASQTYDP 372
Query: 316 KA 317
A
Sbjct: 373 NA 374
>UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana]
Length = 395
Score = 426 bits (1096), Expect = e-118
Identities = 203/305 (66%), Positives = 247/305 (80%), Gaps = 1/305 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
AQ FTF ELATAT+NFR E IG+GGFG VYKG L ST Q A+K+LD G QG +EFLV
Sbjct: 58 AQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGNREFLV 117
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYEYMP+GSLE HLHD+ P +PLDWNTRM+I
Sbjct: 118 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPGKQPLDWNTRMKI 177
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL YLH + P VIYRDLK SNILLD+ ++PKLSDFGLAK GP GD+S+V+TRV
Sbjct: 178 AAGAAKGLEYLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSHVSTRV 237
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRA-HDKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVVLLE+ITGR+A D +R+ +++LV WARPLF+
Sbjct: 238 MGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDSSRSTGEQNLVAWARPLFK 297
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F ++ DP LQG YP GL AL +A MC++E P LRP D+V AL YL+S+K+ P
Sbjct: 298 DRRKFSQMADPMLQGQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLASQKFDP 357
Query: 316 KASEI 320
A +
Sbjct: 358 LAQPV 362
>UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis]
Length = 376
Score = 426 bits (1095), Expect = e-118
Identities = 203/307 (66%), Positives = 248/307 (80%), Gaps = 1/307 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A FTFRELA ATKNFR E +G+GGFG VYKG+L ST + VA+K+LD G QG +EFLV
Sbjct: 56 AHTFTFRELAAATKNFRAECLLGEGGFGRVYKGRLESTNKIVAIKQLDRNGLQGNREFLV 115
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYEYM +GSLE HLHDL PD + LDWNTRM+I
Sbjct: 116 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMALGSLEDHLHDLPPDKKRLDWNTRMKI 175
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL YLH +A P VIYRDLK SNILL EG++PKLSDFGLAK GP GD+++V+TRV
Sbjct: 176 AAGAAKGLEYLHDKASPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPVGDKTHVSTRV 235
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKH-LVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVVLLE+ITGR+A D ++A +H LV WARPLF+
Sbjct: 236 MGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSKAAGEHNLVAWARPLFK 295
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F ++ DP L G YP+ GL AL +A MC++E P +RP D+V AL YL+S+KY P
Sbjct: 296 DRRKFSQMADPMLHGQYPLRGLYQALAVAAMCVQEQPNMRPLIADVVTALTYLASQKYDP 355
Query: 316 KASEIVS 322
+ + S
Sbjct: 356 ETQPVQS 362
>UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica napus]
Length = 376
Score = 423 bits (1087), Expect = e-117
Identities = 200/305 (65%), Positives = 246/305 (80%), Gaps = 1/305 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
AQ FTF ELATAT+NFR E IG+GGFG VYKG L STGQ A+K+LD G QG +EFLV
Sbjct: 55 AQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTGQTAAIKQLDHNGLQGNREFLV 114
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYEYMP+GSLE HLHD+ P +PLDWNTRM+I
Sbjct: 115 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPSKQPLDWNTRMKI 174
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL YLH + P VIYRDLK SNILL + ++PKLSDFGLAK GP GD+S+V+TRV
Sbjct: 175 AAGAAKGLEYLHDKTMPPVIYRDLKCSNILLGDDYFPKLSDFGLAKLGPVGDKSHVSTRV 234
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVVLLE+ITGR+A D +R +++LV WARPLF+
Sbjct: 235 MGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSRCTGEQNLVAWARPLFK 294
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F ++ DP +QG YP GL AL +A MC++E P LRP D+V AL YL+S+++ P
Sbjct: 295 DRRKFSQMADPMIQGQYPPRGLYQALAVAAMCVQEQPNLRPVIADVVTALTYLASQRFDP 354
Query: 316 KASEI 320
+ +
Sbjct: 355 MSQPV 359
>UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa]
Length = 390
Score = 420 bits (1080), Expect = e-116
Identities = 208/325 (64%), Positives = 255/325 (78%), Gaps = 9/325 (2%)
Query: 7 TAENTDISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTT 66
++EN I+ A+ FTFRELA AT NFR + +G+GGFG VYKG L + Q VA+K+LD
Sbjct: 64 SSENRRIA--ARTFTFRELAAATSNFRVDCLLGEGGFGRVYKGYLETVDQVVAIKQLDRN 121
Query: 67 GFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNE 126
G QG +EFLVEVLMLS+LHHPNLV++IGYCA+GDQRLLVYEYMP+GSLE HLHD P
Sbjct: 122 GLQGNREFLVEVLMLSMLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDPPPGKS 181
Query: 127 PLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPT 186
LDWNTRM+IA GAA+GL YLH +A P VIYRDLK SNILL EG++PKLSDFGLAK GP
Sbjct: 182 RLDWNTRMKIAAGAAKGLEYLHDKANPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPI 241
Query: 187 GDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKH 245
GD+S+V+TRVMGT+GYCAPEYA TG+LT++SD+YSFGVVLLE+ITGRRA D TR A +++
Sbjct: 242 GDKSHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRRAIDNTRAAGEQN 301
Query: 246 LVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
LV WARPLF+D+ F ++ DP L G YP GL AL +A MC++E P +RP GD+V AL
Sbjct: 302 LVAWARPLFKDRRKFPQMADPALHGQYPSRGLYQALAVAAMCVQEQPTMRPLIGDVVTAL 361
Query: 306 DYLSSKKYVPKA------SEIVSPG 324
YL+S+ Y P+A S ++SPG
Sbjct: 362 AYLASQTYDPEAHGVHHTSRLMSPG 386
>UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra]
Length = 405
Score = 420 bits (1079), Expect = e-116
Identities = 198/306 (64%), Positives = 250/306 (80%), Gaps = 1/306 (0%)
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
+AQ FTF ELA AT NFR + F+G+GGFG VYKG L QAVA+K+LD G QG +EF+
Sbjct: 82 QAQTFTFEELAAATSNFRSDCFLGEGGFGKVYKGYLDKINQAVAIKQLDRNGVQGIREFV 141
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
VEV+ LSL HPNLV +IG+CAEGDQRLLVYEYMP+GSLE+HLHD+ P+ +PLDWNTRM+
Sbjct: 142 VEVVTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHLHDIPPNRQPLDWNTRMK 201
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
IA GAA+GL YLH+E +P VIYRDLK SNILL EG++PKLSDFGLAK GP+GD+++V+TR
Sbjct: 202 IAAGAAKGLEYLHNEMKPPVIYRDLKCSNILLGEGYHPKLSDFGLAKVGPSGDKTHVSTR 261
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLF 254
VMGT+GYCAP+YA TG+LT +SDIYSFGVVLLELITGR+A D+ + +++LV WARP+F
Sbjct: 262 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDQRKERGEQNLVAWARPMF 321
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYV 314
+D+ NF +VDP LQG YPI GL AL +A MC++E P +RP+ D+V+AL+YL+S KY
Sbjct: 322 KDRRNFSCMVDPLLQGQYPIRGLYQALAIAAMCVQEQPNMRPAVSDLVMALNYLASHKYD 381
Query: 315 PKASEI 320
P+ +
Sbjct: 382 PQVHSV 387
>UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra]
Length = 406
Score = 418 bits (1074), Expect = e-115
Identities = 198/310 (63%), Positives = 246/310 (78%), Gaps = 1/310 (0%)
Query: 15 NKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEF 74
N AQ FTF EL AT NFR + F+G+GGFG VYKG L Q VA+K+LD G QG +EF
Sbjct: 81 NPAQTFTFEELVAATDNFRSDCFLGEGGFGKVYKGYLEKINQVVAIKQLDQNGLQGIREF 140
Query: 75 LVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRM 134
+VEVL LSL +PNLV +IG+CAEGDQRLLVYEYMP+GSLE+HLHD+ P+ +PLDWN RM
Sbjct: 141 VVEVLTLSLADNPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHLHDIPPNRQPLDWNARM 200
Query: 135 RIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVAT 194
+IA GAA+GL YLH+E P VIYRDLK SNILL EG++PKLSDFGLAK GP+GD ++V+T
Sbjct: 201 KIAAGAAKGLEYLHNEMAPPVIYRDLKCSNILLGEGYHPKLSDFGLAKVGPSGDHTHVST 260
Query: 195 RVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPL 253
RVMGT+GYCAP+YA TG+LT +SD+YSFGVVLLELITGR+A D+T+ +++LV WARP+
Sbjct: 261 RVMGTYGYCAPDYAMTGQLTFKSDVYSFGVVLLELITGRKAIDQTKERSEQNLVAWARPM 320
Query: 254 FRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKY 313
F+D+ NF +VDP LQG YPI GL AL +A MC++E P +RP+ D+VLAL+YL+S KY
Sbjct: 321 FKDRRNFSGMVDPFLQGQYPIKGLYQALAIAAMCVQEQPNMRPAVSDVVLALNYLASHKY 380
Query: 314 VPKASEIVSP 323
P+ P
Sbjct: 381 DPQIHPFKDP 390
>UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza sativa]
Length = 491
Score = 415 bits (1067), Expect = e-115
Identities = 201/300 (67%), Positives = 242/300 (80%), Gaps = 2/300 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A FTFRELA ATKNFR + +G+GGFG VYKG L GQAVAVK+LD G QG +EFLV
Sbjct: 65 AHTFTFRELAAATKNFRQDCLLGEGGFGRVYKGHL-ENGQAVAVKQLDRNGLQGNREFLV 123
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHH NLV++IGYCA+GDQRLLVYE+MP+GSLE HLHD+ PD EPLDWNTRM+I
Sbjct: 124 EVLMLSLLHHDNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDIPPDKEPLDWNTRMKI 183
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A GAA+GL +LH +A P VIYRD KSSNILL EG++PKLSDFGLAK GP GD+++V+TRV
Sbjct: 184 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLGEGYHPKLSDFGLAKLGPVGDKTHVSTRV 243
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVV LELITGR+A D T+ +++LV WARPLF+
Sbjct: 244 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPLGEQNLVAWARPLFK 303
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F K+ DP L G +P+ GL AL +A MCL+E RP GD+V AL YL+S+ Y P
Sbjct: 304 DRRKFPKMADPLLAGRFPMRGLYQALAVAAMCLQEQAATRPFIGDVVTALSYLASQTYDP 363
>UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa]
Length = 479
Score = 412 bits (1059), Expect = e-114
Identities = 203/302 (67%), Positives = 242/302 (79%), Gaps = 2/302 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A+IFTFRELA ATKNFR + +G+GGFG VYKG++ GQ +AVK+LD G QG +EFLV
Sbjct: 64 AKIFTFRELAVATKNFRKDCLLGEGGFGRVYKGQM-ENGQVIAVKQLDRNGLQGNREFLV 122
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV +IGYCA+GDQRLLVYEYM +GSLE+HLHD P +PLDWN RM+I
Sbjct: 123 EVLMLSLLHHPNLVRLIGYCADGDQRLLVYEYMLLGSLENHLHDRPPGKKPLDWNARMKI 182
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
AVGAA+GL YLH +A P VIYRD KSSNILL E +YPKLSDFGLAK GP GD+++V+TRV
Sbjct: 183 AVGAAKGLEYLHDKANPPVIYRDFKSSNILLGEDYYPKLSDFGLAKLGPVGDKTHVSTRV 242
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDWARPLFR 255
MGT+GYCAPEYA TG+LT++SD+YSFGVV LELITGR+A D T+ A +++LV WARPLFR
Sbjct: 243 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDHTQPAGEQNLVAWARPLFR 302
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F ++ DP LQG YP GL AL +A MCL+E+ RP DIV AL YL+S Y P
Sbjct: 303 DRRKFCQMADPSLQGCYPKRGLYQALAVASMCLQENATSRPLIADIVTALSYLASNHYDP 362
Query: 316 KA 317
A
Sbjct: 363 NA 364
>UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVII [Arabidopsis
thaliana]
Length = 423
Score = 407 bits (1046), Expect = e-112
Identities = 203/331 (61%), Positives = 247/331 (74%), Gaps = 3/331 (0%)
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
KAQ FTF+ELA AT NFR + F+G+GGFG V+KG + Q VA+K+LD G QG +EF+
Sbjct: 87 KAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFV 146
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
VEVL LSL HPNLV +IG+CAEGDQRLLVYEYMP GSLE HLH L +PLDWNTRM+
Sbjct: 147 VEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMK 206
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
IA GAARGL YLH P VIYRDLK SNILL E + PKLSDFGLAK GP+GD+++V+TR
Sbjct: 207 IAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTR 266
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARPLF 254
VMGT+GYCAP+YA TG+LT +SDIYSFGVVLLELITGR+A D T+ D++LV WARPLF
Sbjct: 267 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLF 326
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYV 314
+D+ NF K+VDP LQG YP+ GL AL ++ MC++E P +RP D+VLAL++L+S KY
Sbjct: 327 KDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKYD 386
Query: 315 PK--ASEIVSPGEMEHDESPNETTMILVKDS 343
P +S D E LVK++
Sbjct: 387 PNSPSSSSGKNPSFHRDRDDEEKRPHLVKET 417
>UniRef100_Q9SIB6 Hypothetical protein At2g28590 [Arabidopsis thaliana]
Length = 424
Score = 404 bits (1039), Expect = e-111
Identities = 193/309 (62%), Positives = 242/309 (77%), Gaps = 1/309 (0%)
Query: 6 TTAENTDISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDT 65
T E+ I KAQ FTF EL+ +T NF+ + F+G+GGFG VYKG + Q VA+K+LD
Sbjct: 72 TNVEDEVIVKKAQTFTFEELSVSTGNFKSDCFLGEGGFGKVYKGFIEKINQVVAIKQLDR 131
Query: 66 TGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDN 125
G QG +EF+VEVL LSL HPNLV +IG+CAEG QRLLVYEYMP+GSL++HLHDL
Sbjct: 132 NGAQGIREFVVEVLTLSLADHPNLVKLIGFCAEGVQRLLVYEYMPLGSLDNHLHDLPSGK 191
Query: 126 EPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGP 185
PL WNTRM+IA GAARGL YLH +P VIYRDLK SNIL+DEG++ KLSDFGLAK GP
Sbjct: 192 NPLAWNTRMKIAAGAARGLEYLHDTMKPPVIYRDLKCSNILIDEGYHAKLSDFGLAKVGP 251
Query: 186 TGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHD-K 244
G +++V+TRVMGT+GYCAP+YA TG+LT +SD+YSFGVVLLELITGR+AYD TR + +
Sbjct: 252 RGSETHVSTRVMGTYGYCAPDYALTGQLTFKSDVYSFGVVLLELITGRKAYDNTRTRNHQ 311
Query: 245 HLVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLA 304
LV+WA PLF+D+ NF+K+VDP L+G YP+ GL AL +A MC++E P +RP D+V+A
Sbjct: 312 SLVEWANPLFKDRKNFKKMVDPLLEGDYPVRGLYQALAIAAMCVQEQPSMRPVIADVVMA 371
Query: 305 LDYLSSKKY 313
LD+L+S KY
Sbjct: 372 LDHLASSKY 380
>UniRef100_Q9SFT7 Hypothetical protein T1B9.28 [Arabidopsis thaliana]
Length = 414
Score = 403 bits (1036), Expect = e-111
Identities = 209/355 (58%), Positives = 260/355 (72%), Gaps = 14/355 (3%)
Query: 9 ENTDISNK--AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTT 66
E+ +++N AQ F+FRELATATKNFR E IG+GGFG VYKGKL TG VAVK+LD
Sbjct: 54 EDKEVTNNIAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRN 113
Query: 67 GFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNE 126
G QG KEF+VEVLMLSLLHH +LV++IGYCA+GDQRLLVYEYM GSLE HL DL PD
Sbjct: 114 GLQGNKEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQI 173
Query: 127 PLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPT 186
PLDW+TR+RIA+GAA GL YLH +A P VIYRDLK++NILLD F KLSDFGLAK GP
Sbjct: 174 PLDWDTRIRIALGAAMGLEYLHDKANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPV 233
Query: 187 GDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHD-KH 245
GD+ +V++RVMGT+GYCAPEY TG+LT +SD+YSFGVVLLELITGRR D TR D ++
Sbjct: 234 GDKQHVSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDTTRPKDEQN 293
Query: 246 LVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
LV WA+P+F++ F +L DP L+G +P L A+ +A MCL+E+ +RP D+V AL
Sbjct: 294 LVTWAQPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTAL 353
Query: 306 DYLSSKKYVPKASEIVSPGEMEHDESPNETTMILVKDSL----REQALAEAKQWG 356
+L + P S V +D+ P + V+DS+ RE+A+AEA +WG
Sbjct: 354 GFLGT---APDGSISVP----HYDDPPQPSDETSVEDSVAAEERERAVAEAMEWG 401
>UniRef100_Q84VY1 At3g07070 [Arabidopsis thaliana]
Length = 414
Score = 403 bits (1035), Expect = e-111
Identities = 209/355 (58%), Positives = 259/355 (72%), Gaps = 14/355 (3%)
Query: 9 ENTDISNK--AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTT 66
E+ +++N AQ F+FRELATATKNFR E IG+GGFG VYKGKL TG VAVK+LD
Sbjct: 54 EDKEVTNNIAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRN 113
Query: 67 GFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNE 126
G QG KEF+VEVLMLSLLHH +LV++IGYCA+GDQRLLVYEYM GSLE HL DL PD
Sbjct: 114 GLQGNKEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQI 173
Query: 127 PLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPT 186
PLDW+TR+RIA+GAA GL YLH A P VIYRDLK++NILLD F KLSDFGLAK GP
Sbjct: 174 PLDWDTRIRIALGAAMGLEYLHDRANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPV 233
Query: 187 GDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHD-KH 245
GD+ +V++RVMGT+GYCAPEY TG+LT +SD+YSFGVVLLELITGRR D TR D ++
Sbjct: 234 GDKQHVSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDTTRPKDEQN 293
Query: 246 LVDWARPLFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
LV WA+P+F++ F +L DP L+G +P L A+ +A MCL+E+ +RP D+V AL
Sbjct: 294 LVTWAQPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTAL 353
Query: 306 DYLSSKKYVPKASEIVSPGEMEHDESPNETTMILVKDSL----REQALAEAKQWG 356
+L + P S V +D+ P + V+DS+ RE+A+AEA +WG
Sbjct: 354 GFLGT---APDGSISVP----HYDDPPQPSDETSVEDSVAAEERERAVAEAMEWG 401
>UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 381
Score = 397 bits (1019), Expect = e-109
Identities = 193/295 (65%), Positives = 234/295 (78%), Gaps = 1/295 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A+IFTFRELATATKNFR E IG+GGFG VYKGKL + Q VAVK+LD G QG++EFLV
Sbjct: 50 ARIFTFRELATATKNFRQECLIGEGGFGRVYKGKLENPAQVVAVKQLDRNGLQGQREFLV 109
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHH NLV++IGYCA+GDQRLLVYEYMP+GSLE HL DL P +PLDWNTR++I
Sbjct: 110 EVLMLSLLHHRNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLLDLEPGQKPLDWNTRIKI 169
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
A+GAA+G+ YLH EA+P VIYRDLKSSNILLD + KLSDFGLAK GP GD +V++RV
Sbjct: 170 ALGAAKGIEYLHDEADPPVIYRDLKSSNILLDPEYVAKLSDFGLAKLGPVGDTLHVSSRV 229
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDWARPLFR 255
MGT+GYCAPEY TG LT +SD+YSFGVVLLELI+GRR D R +H+++LV WA P+FR
Sbjct: 230 MGTYGYCAPEYQRTGYLTNKSDVYSFGVVLLELISGRRVIDTMRPSHEQNLVTWALPIFR 289
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSS 310
D + +L DP L+G YP L A+ +A MCL E+P +RP D++ AL +L +
Sbjct: 290 DPTRYWQLADPLLRGDYPEKSLNQAIAVAAMCLHEEPTVRPLMSDVITALSFLGA 344
>UniRef100_Q9LD65 Hypothetical protein P0462H08.22 (EST C22619(S11214) corresponds to
a region of the predicted gene) [Oryza sativa]
Length = 407
Score = 395 bits (1014), Expect = e-108
Identities = 215/394 (54%), Positives = 259/394 (65%), Gaps = 44/394 (11%)
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
+A IFT RELA AT NF E +G+GGFG+VYK L Q VAVK+LD G QG +EFL
Sbjct: 12 EATIFTLRELADATNNFSTECLLGRGGFGSVYKAFLNDR-QVVAVKQLDLNGLQGNREFL 70
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
VEVLMLSLLHHPNLV + GYC +GDQRLL+YEYMP+GSLE LHDL P EPLDW TRM+
Sbjct: 71 VEVLMLSLLHHPNLVKLFGYCVDGDQRLLIYEYMPLGSLEDRLHDLRPGQEPLDWTTRMK 130
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
IA AA GL YLH EA P+VIYRD+K SNILL EG+ KLSDFGLAK GP GD+++V TR
Sbjct: 131 IAADAAAGLEYLHDEAIPAVIYRDIKPSNILLGEGYNAKLSDFGLAKLGPVGDKTHVTTR 190
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKH-LVDWARPLF 254
VMGTHGYCAPEY +TGKLT++SDIYSFGVV LELITGRRA D R D+ LV WARPLF
Sbjct: 191 VMGTHGYCAPEYLSTGKLTIKSDIYSFGVVFLELITGRRALDSNRPPDEQDLVAWARPLF 250
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYV 314
+D+ F K+ DP L GH+P GL AL +A MCL+E + RPS ++ +AL YL+S+ +
Sbjct: 251 KDQRKFPKMADPSLHGHFPKRGLFQALAIAAMCLQEKAKNRPSIREVAVALSYLASQTHE 310
Query: 315 PK--ASEIVSPG---------EMEHDES----------PNETTMILVKD----------- 342
+ A+ PG ++ D S P T +V++
Sbjct: 311 SQNTAARHTLPGPSVPRVLDNQINQDTSLPSQHGVHMPPLAGTDHMVQEVKENCRSSSHR 370
Query: 343 ----------SLREQALAEAKQWGETWREKRKQS 366
+ RE+ALAEA W E WR + K S
Sbjct: 371 PGRGRVTPNGADRERALAEANVWVEAWRRQEKTS 404
>UniRef100_Q67YK2 Hypothetical protein At1g20650 [Arabidopsis thaliana]
Length = 381
Score = 392 bits (1008), Expect = e-108
Identities = 195/311 (62%), Positives = 241/311 (76%), Gaps = 2/311 (0%)
Query: 14 SNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKE 73
S A+ FTF+ELA AT+NFR+ +G+GGFG VYKG+L S GQ VA+K+L+ G QG +E
Sbjct: 60 SGGARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDS-GQVVAIKQLNPDGLQGNRE 118
Query: 74 FLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTR 133
F+VEVLMLSLLHHPNLV++IGYC GDQRLLVYEYMPMGSLE HL DL + EPL WNTR
Sbjct: 119 FIVEVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSWNTR 178
Query: 134 MRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVA 193
M+IAVGAARG+ YLH A P VIYRDLKS+NILLD+ F PKLSDFGLAK GP GD+++V+
Sbjct: 179 MKIAVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRTHVS 238
Query: 194 TRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYD-ETRAHDKHLVDWARP 252
TRVMGT+GYCAPEYA +GKLT++SDIY FGVVLLELITGR+A D + +++LV W+RP
Sbjct: 239 TRVMGTYGYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTWSRP 298
Query: 253 LFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKK 312
+D+ F LVDP L+G YP L A+ + MCL E+ RP GDIV+AL+YL+++
Sbjct: 299 YLKDQKKFGHLVDPSLRGKYPRRCLNYAIAIIAMCLNEEAHYRPFIGDIVVALEYLAAQS 358
Query: 313 YVPKASEIVSP 323
+A + SP
Sbjct: 359 RSHEARNVSSP 369
>UniRef100_Q9LDZ5 F2D10.13 [Arabidopsis thaliana]
Length = 381
Score = 392 bits (1006), Expect = e-107
Identities = 194/308 (62%), Positives = 240/308 (76%), Gaps = 2/308 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A+ FTF+ELA AT+NFR+ +G+GGFG VYKG+L S GQ VA+K+L+ G QG +EF+V
Sbjct: 63 ARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDS-GQVVAIKQLNPDGLQGNREFIV 121
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYC GDQRLLVYEYMPMGSLE HL DL + EPL WNTRM+I
Sbjct: 122 EVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSWNTRMKI 181
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
AVGAARG+ YLH A P VIYRDLKS+NILLD+ F PKLSDFGLAK GP GD+++V+TRV
Sbjct: 182 AVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRTHVSTRV 241
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYD-ETRAHDKHLVDWARPLFR 255
MGT+GYCAPEYA +GKLT++SDIY FGVVLLELITGR+A D + +++LV W+RP +
Sbjct: 242 MGTYGYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTWSRPYLK 301
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYVP 315
D+ F LVDP L+G YP L A+ + MCL E+ RP GDIV+AL+YL+++
Sbjct: 302 DQKKFGHLVDPSLRGKYPRRCLNYAIAIIAMCLNEEAHYRPFIGDIVVALEYLAAQSRSH 361
Query: 316 KASEIVSP 323
+A + SP
Sbjct: 362 EARNVSSP 369
>UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 377
Score = 388 bits (996), Expect = e-106
Identities = 190/316 (60%), Positives = 238/316 (75%), Gaps = 2/316 (0%)
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A IFTFREL ATKNF + +G+GGFG VYKG++ + Q VAVK+LD G+QG +EFLV
Sbjct: 58 AHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNREFLV 117
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNE-PLDWNTRMR 135
EV+MLSLLHH NLV+++GYCA+GDQR+LVYEYM GSLE HL +L + + PLDW+TRM+
Sbjct: 118 EVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWDTRMK 177
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
+A GAARGL YLH A+P VIYRD K+SNILLDE F PKLSDFGLAK GPTG +++V+TR
Sbjct: 178 VAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGETHVSTR 237
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETR-AHDKHLVDWARPLF 254
VMGT+GYCAPEYA TG+LT++SD+YSFGVV LE+ITGRR D T+ +++LV WA PLF
Sbjct: 238 VMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWASPLF 297
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKKYV 314
+D+ F + DP L+G YPI GL AL +A MCL+E+ RP D+V AL+YL+ K
Sbjct: 298 KDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLAVTKTE 357
Query: 315 PKASEIVSPGEMEHDE 330
+ E E DE
Sbjct: 358 EDGQTVEGEEEEEEDE 373
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 641,182,975
Number of Sequences: 2790947
Number of extensions: 27634424
Number of successful extensions: 108080
Number of sequences better than 10.0: 18065
Number of HSP's better than 10.0 without gapping: 8380
Number of HSP's successfully gapped in prelim test: 9687
Number of HSP's that attempted gapping in prelim test: 66323
Number of HSP's gapped (non-prelim): 20682
length of query: 377
length of database: 848,049,833
effective HSP length: 129
effective length of query: 248
effective length of database: 488,017,670
effective search space: 121028382160
effective search space used: 121028382160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133780.4


