

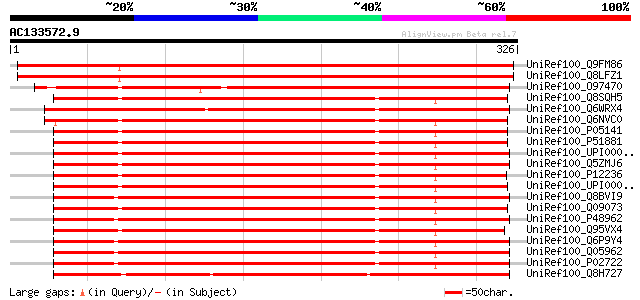
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.9 + phase: 0
(326 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FM86 ADP/ATP translocase-like protein [Arabidopsis t... 503 e-141
UniRef100_Q8LFZ1 ADP/ATP translocase-like protein [Arabidopsis t... 502 e-141
UniRef100_O97470 ADP/ATP translocase [Dictyostelium discoideum] 295 9e-79
UniRef100_Q8SQH5 ADP,ATP carrier protein 2 [Bos taurus] 289 8e-77
UniRef100_Q6WRX4 ADP/ATP translocase [Branchiostoma belcheri tsi... 286 4e-76
UniRef100_Q6NVC0 SLC25A5 protein [Homo sapiens] 286 4e-76
UniRef100_P05141 ADP,ATP carrier protein, fibroblast isoform [Ho... 286 7e-76
UniRef100_P51881 ADP,ATP carrier protein 2 [Mus musculus] 285 1e-75
UniRef100_UPI000024916F UPI000024916F UniRef100 entry 285 2e-75
UniRef100_Q5ZMJ6 Hypothetical protein [Gallus gallus] 285 2e-75
UniRef100_P12236 ADP,ATP carrier protein, liver isoform T2 [Homo... 285 2e-75
UniRef100_UPI000025D98B UPI000025D98B UniRef100 entry 284 2e-75
UniRef100_Q8BVI9 Mus musculus adult male medulla oblongata cDNA,... 284 2e-75
UniRef100_Q09073 ADP,ATP carrier protein 2 [Rattus norvegicus] 284 2e-75
UniRef100_P48962 ADP,ATP carrier protein, heart/skeletal muscle ... 284 2e-75
UniRef100_Q95VX4 ADP-ATP translocator [Ethmostigmus rubripes] 283 3e-75
UniRef100_Q6P9Y4 Solute carrier family 25, member 4 [Rattus norv... 283 4e-75
UniRef100_Q05962 ADP,ATP carrier protein 1 [Rattus norvegicus] 283 6e-75
UniRef100_P02722 ADP,ATP carrier protein, heart isoform T1 [Bos ... 283 6e-75
UniRef100_Q8H727 ADP/ATP translocase [Phytophthora infestans] 282 8e-75
>UniRef100_Q9FM86 ADP/ATP translocase-like protein [Arabidopsis thaliana]
Length = 330
Score = 503 bits (1295), Expect = e-141
Identities = 246/323 (76%), Positives = 285/323 (88%), Gaps = 4/323 (1%)
Query: 6 EDPEEKALNLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNL 65
ED E+ + N + + +FQ+DL+AGAVMGG VHTIVAPIERAKLLLQTQESN+
Sbjct: 6 EDEEDPSRNRRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNI 65
Query: 66 AIVA----SGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS 121
AIV +G+R+FKGMFD I RTVREEGV+SLWRGNGSSVLRYYPSVALNFSLKDLY+S
Sbjct: 66 AIVGDEGHAGKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRS 125
Query: 122 ILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIH 181
ILR +S ++IFSGA ANF+AG+AAGCT+LI+VYPLDIAHTRLAADIG+ E RQFRGIH
Sbjct: 126 ILRNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPEARQFRGIH 185
Query: 182 HFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMV 241
HFL+TI +KDGVRGIYRGLPASLHG++IHRGLYFGGFDT+KE+ SE++KPELALWKRW +
Sbjct: 186 HFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGL 245
Query: 242 AQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNV 301
AQAVTTSAGL SYPLDTVRRR+MMQSGMEHP+Y STLDCW+KIYR+EGL SFYRGA+SN+
Sbjct: 246 AQAVTTSAGLASYPLDTVRRRIMMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNM 305
Query: 302 FRSTGAAAILVLYDEVKKFMNLG 324
FRSTG+AAILV YDEVK+F+N G
Sbjct: 306 FRSTGSAAILVFYDEVKRFLNWG 328
>UniRef100_Q8LFZ1 ADP/ATP translocase-like protein [Arabidopsis thaliana]
Length = 330
Score = 502 bits (1293), Expect = e-141
Identities = 246/323 (76%), Positives = 285/323 (88%), Gaps = 4/323 (1%)
Query: 6 EDPEEKALNLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNL 65
ED E+ + N + + +FQ+DL+AGAVMGG VHTIVAPIERAKLLLQTQESN+
Sbjct: 6 EDEEDPSRNRRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNI 65
Query: 66 AIVA----SGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS 121
AIV +G+R+FKGMFD I RTVREEGV+SLWRGNGSSVLRYYPSVALNFSLKDLY+S
Sbjct: 66 AIVGDEGHAGKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRS 125
Query: 122 ILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIH 181
ILR +S ++IFSGA ANF+AG+AAGCT+LI+VYPLDIAHTRLAADIG+ E RQFRGIH
Sbjct: 126 ILRNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPESRQFRGIH 185
Query: 182 HFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMV 241
HFL+TI +KDGVRGIYRGLPASLHG++IHRGLYFGGFDT+KE+ SE++KPELALWKRW +
Sbjct: 186 HFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGL 245
Query: 242 AQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNV 301
AQAVTTSAGL SYPLDTVRRR+MMQSGMEHP+Y STLDCW+KIYR+EGL SFYRGA+SN+
Sbjct: 246 AQAVTTSAGLASYPLDTVRRRIMMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNM 305
Query: 302 FRSTGAAAILVLYDEVKKFMNLG 324
FRSTG+AAILV YDEVK+F+N G
Sbjct: 306 FRSTGSAAILVFYDEVKRFLNWG 328
>UniRef100_O97470 ADP/ATP translocase [Dictyostelium discoideum]
Length = 309
Score = 295 bits (756), Expect = 9e-79
Identities = 148/307 (48%), Positives = 206/307 (66%), Gaps = 12/307 (3%)
Query: 17 RKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK 76
+K+N SS F +D + G GG TIVAPIER KLLLQ Q ++ I A +++K
Sbjct: 4 QKKNDVSS-----FVKDSLIGGTAGGVSKTIVAPIERVKLLLQVQSASTQIAAD--KQYK 56
Query: 77 GMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS--ILRGGNSNPDNIF 134
G+ DC +R +E+GVISLWRGN ++V+RY+P+ ALNF+ KD YK + NP F
Sbjct: 57 GIVDCFVRVSKEQGVISLWRGNLANVIRYFPTQALNFAFKDKYKKFFVRHTAKENPTKFF 116
Query: 135 SGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVR 194
G N ++G AAG TSL+ VYPLD A TRLAAD+G RQF G+ + +++I+++DG+
Sbjct: 117 IG---NLLSGGAAGATSLLFVYPLDFARTRLAADVGTGSARQFTGLGNCISSIYKRDGLI 173
Query: 195 GIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSY 254
G+YRG S+ G+ ++R +FGG+DT K +L E+ + + W W +AQ VTT AG+VSY
Sbjct: 174 GLYRGFGVSVGGIFVYRAAFFGGYDTAKGILLGENNKKASFWASWGIAQVVTTIAGVVSY 233
Query: 255 PLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLY 314
P DTVRRRMMMQ+G +Y+ST DCW KI EG +F++GA+SN R +G A +LV+Y
Sbjct: 234 PFDTVRRRMMMQAGRADILYSSTWDCWVKIATREGPTAFFKGALSNAIRGSGGALVLVIY 293
Query: 315 DEVKKFM 321
DE++K M
Sbjct: 294 DEIQKLM 300
>UniRef100_Q8SQH5 ADP,ATP carrier protein 2 [Bos taurus]
Length = 297
Score = 289 bits (739), Expect = 8e-77
Identities = 147/295 (49%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 6 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG+RG+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIRGLYQGFNVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KKF
Sbjct: 242 GRKGTDIMYTGTLDCWRKIARDEGAKAFFKGAWSNVLRGMGGAFVLVLYDEIKKF 296
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 16 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_Q6WRX4 ADP/ATP translocase [Branchiostoma belcheri tsingtaunese]
Length = 298
Score = 286 bits (733), Expect = 4e-76
Identities = 143/300 (47%), Positives = 200/300 (66%), Gaps = 4/300 (1%)
Query: 23 SSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCI 82
+S + +F +D +AG + T VAPIER KLLLQ Q+++ + A+G +K+ G+ DC+
Sbjct: 2 ASDQVVSFLKDFLAGGISAAIAKTAVAPIERVKLLLQVQDASKQMQAAGAKKYTGIIDCV 61
Query: 83 IRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFV 142
R +E+G +S WRGN ++V+RY+P+ ALNF+ KD YK + GG + + + N
Sbjct: 62 TRIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLGG-VDKNRFWRYFVGNLA 120
Query: 143 AGAAAGCTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLP 201
+G AAG TSL VYPLD A TRLAADIG+ R + G+ + L ++ DG+ G+YRG
Sbjct: 121 SGGAAGATSLCFVYPLDFARTRLAADIGKGAGERLYSGLGNCLMQTYRSDGLYGLYRGFS 180
Query: 202 ASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRR 261
S+ G++I+R YFG FDT K M+ + K + WM+AQ VTTSAG+VSYP DTVRR
Sbjct: 181 VSVQGIIIYRAAYFGCFDTAKGMMPDPKKTPFVV--SWMIAQVVTTSAGIVSYPFDTVRR 238
Query: 262 RMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
RMMMQSG++ +Y +TLDCWRKI + EG + ++GA SNV R G A +LVLYDE+KK +
Sbjct: 239 RMMMQSGLKELIYKNTLDCWRKIIKDEGGSALFKGAFSNVLRGMGGAFVLVLYDELKKII 298
Score = 33.9 bits (76), Expect = 6.2
Identities = 16/51 (31%), Positives = 28/51 (54%)
Query: 274 YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFMNLG 324
Y +DC +I + +G +SF+RG ++NV R A+ + + K + LG
Sbjct: 54 YTGIIDCVTRIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLG 104
>UniRef100_Q6NVC0 SLC25A5 protein [Homo sapiens]
Length = 323
Score = 286 bits (733), Expect = 4e-76
Identities = 149/307 (48%), Positives = 200/307 (64%), Gaps = 13/307 (4%)
Query: 23 SSSRFN------NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK 76
S+S FN +F +D +AG V T VAPIER KLLLQ Q ++ I A +++K
Sbjct: 20 SASSFNMTDAAVSFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYK 77
Query: 77 GMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSG 136
G+ DC++R +E+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG +
Sbjct: 78 GIIDCVVRIPKEQGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWLY 137
Query: 137 ASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRG 195
+ N +G AAG TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG++G
Sbjct: 138 FAGNLASGGAAGATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKG 197
Query: 196 IYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYP 255
+Y+G S+ G++I+R YFG +DT K ML + + + WM+AQ VT AGL SYP
Sbjct: 198 LYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIVI--SWMIAQTVTAVAGLTSYP 255
Query: 256 LDTVRRRMMMQSGMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVL 313
DTVRRRMMMQSG + +Y TLDCWRKI R EG +F++GA SNV R G A +LVL
Sbjct: 256 FDTVRRRMMMQSGRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVL 315
Query: 314 YDEVKKF 320
YDE+KK+
Sbjct: 316 YDEIKKY 322
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 42 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 98
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 99 NLANVIRYFPTQALNFAFKDKYKQIFLG 126
>UniRef100_P05141 ADP,ATP carrier protein, fibroblast isoform [Homo sapiens]
Length = 297
Score = 286 bits (731), Expect = 7e-76
Identities = 145/295 (49%), Positives = 195/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 6 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG++G+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AGL SYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGIYDTAKGMLPDPKNTHIVI--SWMIAQTVTAVAGLTSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 242 GRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 296
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 16 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_P51881 ADP,ATP carrier protein 2 [Mus musculus]
Length = 297
Score = 285 bits (729), Expect = 1e-75
Identities = 144/295 (48%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 6 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F+G+ L I++ DG++G+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKAGAEREFKGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 242 GRKGTDIMYTGTLDCWRKIARDEGSKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 296
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 16 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_UPI000024916F UPI000024916F UniRef100 entry
Length = 304
Score = 285 bits (728), Expect = 2e-75
Identities = 147/296 (49%), Positives = 195/296 (65%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DCI+R +E
Sbjct: 12 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASQQISAD--KQYKGIVDCIVRIPKE 69
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 70 QGFASFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAAG 129
Query: 149 CTSLILVYPLDIAHTRLAADIGRT-EVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F G+ LA IF+ DG+RG+Y+G S+ G+
Sbjct: 130 ATSLCFVYPLDFARTRLAADVGKAGSTREFSGLADCLAKIFKSDGLRGLYQGFNVSVQGI 189
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AG+VSYP DTVRRRMMMQS
Sbjct: 190 IIYRAAYFGVYDTAKGMLPDPKNTHIMV--SWMIAQTVTAVAGVVSYPFDTVRRRMMMQS 247
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y TLDCWRKI R EG +F++GA+SNV R G A +LVLYDE KK++
Sbjct: 248 GRKGADIMYTGTLDCWRKIARDEGSKAFFKGALSNVLRGMGGAFVLVLYDEFKKYI 303
>UniRef100_Q5ZMJ6 Hypothetical protein [Gallus gallus]
Length = 298
Score = 285 bits (728), Expect = 2e-75
Identities = 143/296 (48%), Positives = 198/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAE--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G+IS WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGIISFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F G+ + IF+ DG+RG+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGVSEREFTGLGDCIVKIFKSDGLRGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT +AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAAAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCW+KI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYKGTIDCWKKIAKDEGSKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 35.0 bits (79), Expect = 2.8
Identities = 24/88 (27%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G+ISF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITAEKQYKGIIDCVVRIPKEQGIISFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>UniRef100_P12236 ADP,ATP carrier protein, liver isoform T2 [Homo sapiens]
Length = 297
Score = 285 bits (728), Expect = 2e-75
Identities = 145/294 (49%), Positives = 196/294 (66%), Gaps = 7/294 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I A +++KG+ DCI+R +E
Sbjct: 6 SFAKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQIAAD--KQYKGIVDCIVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G++ R+FRG+ L I + DG+RG+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKSGTEREFRGLGDCLVKITKSDGIRGLYQGFSVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AG+VSYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGVYDTAKGMLPDPKNTHIVV--SWMIAQTVTAVAGVVSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKK 319
G + +Y T+DCWRKI+R EG +F++GA SNV R G A +LVLYDE+KK
Sbjct: 242 GRKGADIMYTGTVDCWRKIFRDEGGKAFFKGAWSNVLRGMGGAFVLVLYDELKK 295
>UniRef100_UPI000025D98B UPI000025D98B UniRef100 entry
Length = 299
Score = 284 bits (727), Expect = 2e-75
Identities = 145/295 (49%), Positives = 194/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I A ++KG+ DC++R +E
Sbjct: 7 SFMKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQITAE--TQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G IS WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFISFWRGNLANVIRYFPTQALNFAFKDKYKKIFLGGVDQKTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAADIG+ R+F G+ + +A IF+ DG++G+Y G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADIGKGPAEREFTGLGNCIAKIFKTDGIKGLYLGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG FDT K ML + + + WM+AQ VT AGL+SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGCFDTAKGMLPDPKNTHIIV--SWMIAQTVTAVAGLISYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KKF
Sbjct: 243 GRKGADIMYKGTIDCWRKILKDEGGKAFFKGAWSNVIRGMGGAFVLVLYDEIKKF 297
Score = 33.9 bits (76), Expect = 6.2
Identities = 20/74 (27%), Positives = 37/74 (49%), Gaps = 4/74 (5%)
Query: 255 PLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI 310
P++ V+ + +Q + Y +DC +I + +G ISF+RG ++NV R A+
Sbjct: 28 PIERVKLLLQVQHASKQITAETQYKGIIDCVVRIPKEQGFISFWRGNLANVIRYFPTQAL 87
Query: 311 LVLYDEVKKFMNLG 324
+ + K + LG
Sbjct: 88 NFAFKDKYKKIFLG 101
>UniRef100_Q8BVI9 Mus musculus adult male medulla oblongata cDNA, RIKEN full-length
enriched library, clone:6330437A14 product:solute
carrier family 25 (mitochondrial carrier; adenine
nucleotide translocator), member 4, full insert sequence
[Mus musculus]
Length = 298
Score = 284 bits (727), Expect = 2e-75
Identities = 143/296 (48%), Positives = 197/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y TLDCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYTGTLDCWRKIAKDEGANAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 33.5 bits (75), Expect = 8.1
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 17 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>UniRef100_Q09073 ADP,ATP carrier protein 2 [Rattus norvegicus]
Length = 297
Score = 284 bits (727), Expect = 2e-75
Identities = 144/295 (48%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 6 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F+G+ L I++ DG++G+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKAGAEREFKGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 242 GRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 296
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 16 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_P48962 ADP,ATP carrier protein, heart/skeletal muscle isoform T1 [Mus
musculus]
Length = 297
Score = 284 bits (727), Expect = 2e-75
Identities = 143/296 (48%), Positives = 197/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 6 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y TLDCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 242 GRKGADIMYTGTLDCWRKIAKDEGANAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 297
Score = 33.5 bits (75), Expect = 8.1
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 16 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_Q95VX4 ADP-ATP translocator [Ethmostigmus rubripes]
Length = 299
Score = 283 bits (725), Expect = 3e-75
Identities = 143/293 (48%), Positives = 195/293 (65%), Gaps = 7/293 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I +++KGM DC +R +E
Sbjct: 7 SFLKDFIAGGVAAAISKTSVAPIERVKLLLQVQHASKQIAVD--KQYKGMVDCFVRIPQE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G++S WRGN ++V+RY+P+ ALNF+ KD YK I GG + N +G AAG
Sbjct: 65 QGILSYWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFLGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPL A TRLAADIG+ E R+F G+ + +A IF+ DG+ G+YRG S+ G+
Sbjct: 125 ATSLCFVYPLXFARTRLAADIGKGLEQREFTGLGNCIAKIFKSDGLVGLYRGFGVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + W++AQ VTT AG++SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGTYDTAKGMLPDPKNTPIVI--SWLIAQTVTTCAGIISYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
G + +Y +T+DCW KIY+TEG +F++GA SN+ R TG A +LVLYDE+K
Sbjct: 243 GRKKADILYKNTIDCWGKIYKTEGGAAFFKGAFSNILRGTGGAFVLVLYDEIK 295
Score = 33.5 bits (75), Expect = 8.1
Identities = 18/74 (24%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Query: 255 PLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI 310
P++ V+ + +Q + Y +DC+ +I + +G++S++RG ++NV R A+
Sbjct: 28 PIERVKLLLQVQHASKQIAVDKQYKGMVDCFVRIPQEQGILSYWRGNLANVIRYFPTQAL 87
Query: 311 LVLYDEVKKFMNLG 324
+ + K + LG
Sbjct: 88 NFAFKDKYKQIFLG 101
>UniRef100_Q6P9Y4 Solute carrier family 25, member 4 [Rattus norvegicus]
Length = 298
Score = 283 bits (724), Expect = 4e-75
Identities = 142/296 (47%), Positives = 197/296 (65%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYTGTVDCWRKIAKDEGAKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 33.5 bits (75), Expect = 8.1
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 17 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>UniRef100_Q05962 ADP,ATP carrier protein 1 [Rattus norvegicus]
Length = 297
Score = 283 bits (723), Expect = 6e-75
Identities = 142/296 (47%), Positives = 197/296 (65%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 6 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 242 GRKGADIMYTGTVDCWRKIAKDEGRKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 297
Score = 33.5 bits (75), Expect = 8.1
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 16 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_P02722 ADP,ATP carrier protein, heart isoform T1 [Bos taurus]
Length = 297
Score = 283 bits (723), Expect = 6e-75
Identities = 144/296 (48%), Positives = 196/296 (65%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 6 SFLKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 63
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 64 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 123
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F G+ + + IF+ DG+RG+Y+G S+ G+
Sbjct: 124 ATSLCFVYPLDFARTRLAADVGKGAAQREFTGLGNCITKIFKSDGLRGLYQGFNVSVQGI 183
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AGLVSYP DTVRRRMMMQS
Sbjct: 184 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQTVTAVAGLVSYPFDTVRRRMMMQS 241
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KKF+
Sbjct: 242 GRKGADIMYTGTVDCWRKIAKDEGPKAFFKGAWSNVLRGMGGAFVLVLYDEIKKFV 297
Score = 33.5 bits (75), Expect = 8.1
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 16 VAAAISKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 72
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 73 NLANVIRYFPTQALNFAFKDKYKQIFLG 100
>UniRef100_Q8H727 ADP/ATP translocase [Phytophthora infestans]
Length = 310
Score = 282 bits (722), Expect = 8e-75
Identities = 137/293 (46%), Positives = 201/293 (67%), Gaps = 4/293 (1%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F DL AG V GG T+VAPIER KLLLQ Q ++ I + +KG+ DC +R +E
Sbjct: 21 SFLLDLAAGGVAGGISKTVVAPIERVKLLLQVQAASTQIKPEDQ--YKGIVDCFVRVTKE 78
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV SLWRGN ++V+RY+P+ ALNF+ KD +K + G + + + N +G AAG
Sbjct: 79 QGVNSLWRGNLANVIRYFPTQALNFAFKDKFKKLFMDGVTK-EQFWRFFMGNLASGGAAG 137
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
TSL+ VYPLD A TRL AD+G+ + R + G+ + ++TI++ DG+ G+Y+G S+ G++
Sbjct: 138 ATSLLFVYPLDFARTRLGADVGKGKSRMYTGLVNCVSTIYKSDGISGLYQGFGVSVGGII 197
Query: 209 IHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSG 268
++R +FGG+DT++++ + K +W++W+VAQ VT+ AG++SYP DTVRRRMMMQ+G
Sbjct: 198 VYRAAFFGGYDTLRDVALRDPK-NAPVWQKWLVAQTVTSLAGMISYPFDTVRRRMMMQAG 256
Query: 269 MEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
+ +Y STLDC KI + EG +F++GA SN+ R TG A +LVLYDE KK M
Sbjct: 257 RKDILYTSTLDCAMKIAKNEGSGAFFKGAGSNILRGTGGAIVLVLYDEFKKMM 309
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,235,733
Number of Sequences: 2790947
Number of extensions: 20271945
Number of successful extensions: 61833
Number of sequences better than 10.0: 1871
Number of HSP's better than 10.0 without gapping: 1200
Number of HSP's successfully gapped in prelim test: 672
Number of HSP's that attempted gapping in prelim test: 49357
Number of HSP's gapped (non-prelim): 4965
length of query: 326
length of database: 848,049,833
effective HSP length: 127
effective length of query: 199
effective length of database: 493,599,564
effective search space: 98226313236
effective search space used: 98226313236
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133572.9


