

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.2 - phase: 0
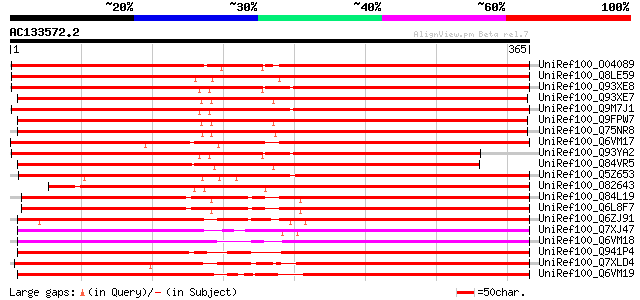
(365 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04089 Zinc transporter 4, chloroplast precursor [Arab... 530 e-149
UniRef100_Q8LE59 Fe(II) transport protein 3, chloroplast precurs... 524 e-147
UniRef100_Q93XE8 ZIP-like zinc transporter [Thlaspi caerulescens] 523 e-147
UniRef100_Q93XE7 ZIP-like zinc transporter [Thlaspi caerulescens] 519 e-146
UniRef100_Q9M7J1 Zn and Cd transporter ZNT1 [Thlaspi caerulescens] 518 e-145
UniRef100_Q9FPW7 Putative Zn transporter ZNT4 [Thlaspi caerulesc... 516 e-145
UniRef100_Q75NR8 Zn/Cd transporter homolog [Thlaspi japonicum] 515 e-145
UniRef100_Q6VM17 Metal transport protein [Medicago truncatula] 483 e-135
UniRef100_Q93YA2 Putative Zn and Cd transporter [Thlaspi caerule... 462 e-129
UniRef100_Q84VR5 Putative Zn transporter [Thlaspi caerulescens] 456 e-127
UniRef100_Q5Z653 Putative ZIP-like zinc transporter [Oryza sativa] 431 e-119
UniRef100_O82643 Zinc transporter 9 [Arabidopsis thaliana] 405 e-112
UniRef100_Q84L19 Putative ZIP-like zinc transporter [Oryza sativa] 383 e-105
UniRef100_Q6L8F7 Iron regulated transporter-like protein [Oryza ... 383 e-105
UniRef100_Q6ZJ91 Putative iron transporter Fe2 [Oryza sativa] 318 1e-85
UniRef100_Q7XJ47 Putative zinc transporter [Oryza sativa] 311 1e-83
UniRef100_Q6VM18 Metal transport protein [Medicago truncatula] 311 2e-83
UniRef100_Q941P4 Zinc transporter protein ZIP1 [Glycine max] 305 1e-81
UniRef100_Q7XLD4 OSJNBa0070C17.15 protein [Oryza sativa] 305 2e-81
UniRef100_Q6VM19 Metal transport protein [Medicago truncatula] 304 3e-81
>UniRef100_O04089 Zinc transporter 4, chloroplast precursor [Arabidopsis thaliana]
Length = 374
Score = 530 bits (1364), Expect = e-149
Identities = 272/375 (72%), Positives = 309/375 (81%), Gaps = 17/375 (4%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T C ESDLCRD+ AAF+LKF+A+ASILLAG AG+AIPLIG++RR+L+T+GNLFVAA
Sbjct: 6 TKILCDAGESDLCRDDSAAFLLKFVAIASILLAGAAGVAIPLIGRNRRFLQTEGNLFVAA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL++PCLP+FPWSKFPF GFFAM+AAL TLL+DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALSNPCLPDFPWSKFPFPGFFAMVAALATLLVDFMGTQ 125
Query: 122 YYERKQGMNRAVDEQARVGTSEEGNV---------GKVFGEEESGGMHIVGMHAHAAHHR 172
YYERKQ N+A E A G+ E V KVFGEE+ GG+HIVG+ AHAAHHR
Sbjct: 126 YYERKQERNQAATEAA-AGSEEIAVVPVVGERVTDNKVFGEEDGGGIHIVGIRAHAAHHR 184
Query: 173 HNHP--HGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLS 230
H+H HG CDG HGH H H ++ E RHVVVSQ+LELGIVSHS+IIGLS
Sbjct: 185 HSHSNSHG-TCDGHA----HGHSHGHMHGNSDVENGARHVVVSQILELGIVSHSIIIGLS 239
Query: 231 LGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIG 290
LGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQF+ S TIMACFFALTTP+G+GIG
Sbjct: 240 LGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPLGIGIG 299
Query: 291 TGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCM 350
T +AS +N +SPGAL+ EGILD+LSAGILVYMALVDLIAADFLSKRMSCN RLQ+VSY M
Sbjct: 300 TAVASSFNSHSPGALVTEGILDSLSAGILVYMALVDLIAADFLSKRMSCNLRLQVVSYVM 359
Query: 351 LFLGAGLMSSLAIWA 365
LFLGAGLMS+LAIWA
Sbjct: 360 LFLGAGLMSALAIWA 374
>UniRef100_Q8LE59 Fe(II) transport protein 3, chloroplast precursor [Arabidopsis
thaliana]
Length = 389
Score = 524 bits (1350), Expect = e-147
Identities = 268/384 (69%), Positives = 308/384 (79%), Gaps = 20/384 (5%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
+N C +ESDLCRD+ AAF+LKF+A+ASILLAG AG+ IPLIG++RR+L+TDGNLFV A
Sbjct: 6 SNVLCNASESDLCRDDSAAFLLKFVAIASILLAGAAGVTIPLIGRNRRFLQTDGNLFVTA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL +PCLP+FPWSKFPF GFFAM+AAL TL +DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALKNPCLPDFPWSKFPFPGFFAMIAALITLFVDFMGTQ 125
Query: 122 YYERKQGM--NRAVDEQARVGT--------SEEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
YYERKQ + +V+ R + E N GKVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 126 YYERKQEREASESVEPFGREQSPGIVVPMIGEGTNDGKVFGEEDSGGIHIVGIHAHAAHH 185
Query: 172 RHNHPHG-DACDGGGIVK---------EHGHDHSHALIAANEETDVRHVVVSQVLELGIV 221
RH+HP G D+C+G + HGH H H + RH+VVSQVLELGIV
Sbjct: 186 RHSHPPGHDSCEGHSKIDIGHAHAHGHGHGHGHGHVHGGLDAVNGARHIVVSQVLELGIV 245
Query: 222 SHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFAL 281
SHS+IIGLSLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQF+ S TIMACFFAL
Sbjct: 246 SHSIIIGLSLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFAL 305
Query: 282 TTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNF 341
TTPIG+GIGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAADFLS +M CNF
Sbjct: 306 TTPIGIGIGTAVASSFNSHSVGALVTEGILDSLSAGILVYMALVDLIAADFLSTKMRCNF 365
Query: 342 RLQLVSYCMLFLGAGLMSSLAIWA 365
RLQ+VSY MLFLGAGLMSSLAIWA
Sbjct: 366 RLQIVSYVMLFLGAGLMSSLAIWA 389
>UniRef100_Q93XE8 ZIP-like zinc transporter [Thlaspi caerulescens]
Length = 408
Score = 523 bits (1347), Expect = e-147
Identities = 270/376 (71%), Positives = 304/376 (80%), Gaps = 15/376 (3%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T C ESDLCRD+ AAF+LKF+A+ASILLAG AG+AIPLIGK+RR+L+T+GNLFVAA
Sbjct: 36 TKIFCDAGESDLCRDDAAAFLLKFVAIASILLAGAAGVAIPLIGKNRRFLQTEGNLFVAA 95
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ ATEAL +PCLP++PWSKFPF GFFAM+AAL TLL+DF+GTQ
Sbjct: 96 KAFAAGVILATGFVHMLAGATEALTNPCLPDYPWSKFPFPGFFAMVAALITLLVDFMGTQ 155
Query: 122 YYERKQGMNRA--------VDEQARV--GTSEEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
YYE KQ N V+E + V E GN K FGEE+ GGMHIVG+ AHAAHH
Sbjct: 156 YYESKQQRNEVAGGGEAAVVEETSSVLPVVVERGNDSKAFGEEDGGGMHIVGIRAHAAHH 215
Query: 172 RHNHP--HGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGL 229
H+H HG CDG + HGH H H + + E RHVVVSQ+LELGIVSHS+IIGL
Sbjct: 216 NHSHSNAHG-TCDGHAHGQSHGHVHVHG--SHDVENGARHVVVSQILELGIVSHSIIIGL 272
Query: 230 SLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGI 289
SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S IMACFFALT PIG+GI
Sbjct: 273 SLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSAIIMACFFALTAPIGIGI 332
Query: 290 GTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYC 349
GT +AS +N +SPGAL+ EGILD+LSAGIL YMALVDLIAADFLSKRMSCN RLQ+VSY
Sbjct: 333 GTAVASSFNSHSPGALVTEGILDSLSAGILTYMALVDLIAADFLSKRMSCNVRLQVVSYV 392
Query: 350 MLFLGAGLMSSLAIWA 365
MLFLGAGLMS+LAIWA
Sbjct: 393 MLFLGAGLMSALAIWA 408
>UniRef100_Q93XE7 ZIP-like zinc transporter [Thlaspi caerulescens]
Length = 422
Score = 519 bits (1336), Expect = e-146
Identities = 267/376 (71%), Positives = 300/376 (79%), Gaps = 17/376 (4%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DLCRD+ AAF+LK +A+ASI LAG AG+ IPLIG++RR+L+TDG+LFVAAKAFA
Sbjct: 46 CNAGEPDLCRDDSAAFLLKLVAIASIFLAGAAGVVIPLIGRNRRFLQTDGSLFVAAKAFA 105
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGFVHML+ TEAL +PCLPEFPW KFPF GFFAM+AAL TLL+DF+GTQYYE+
Sbjct: 106 AGVILATGFVHMLAGGTEALTNPCLPEFPWKKFPFPGFFAMVAALITLLVDFMGTQYYEK 165
Query: 126 KQGMNRAV--DEQARVG-----------TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHR 172
KQ DE G T+EEGN KVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 166 KQEREATTRSDELPSSGPEQSPGIVVPVTAEEGNDEKVFGEEDSGGIHIVGIHAHAAHHT 225
Query: 173 HNHPHGDA-CDGG---GIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIG 228
HNH G + CDG I HGH H H+ RHVVVSQVLELGIVSHS+IIG
Sbjct: 226 HNHTQGQSSCDGHRKIDIGHAHGHGHGHSHGGLELGNGARHVVVSQVLELGIVSHSIIIG 285
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVG 288
+SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S TIMACFFALTTPI +G
Sbjct: 286 ISLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSATIMACFFALTTPISIG 345
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAADFLSKRMSCNFRLQ+VSY
Sbjct: 346 IGTVVASSFNAHSVGALVTEGILDSLSAGILVYMALVDLIAADFLSKRMSCNFRLQIVSY 405
Query: 349 CMLFLGAGLMSSLAIW 364
+LFLG+GLMSSLAIW
Sbjct: 406 LLLFLGSGLMSSLAIW 421
>UniRef100_Q9M7J1 Zn and Cd transporter ZNT1 [Thlaspi caerulescens]
Length = 378
Score = 518 bits (1333), Expect = e-145
Identities = 266/375 (70%), Positives = 301/375 (79%), Gaps = 13/375 (3%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T C ESDLCRD+ AAF+LKF+A+ASILLAG AG+AIPLIGK+RR+L+T+GNLFVAA
Sbjct: 6 TKILCDAGESDLCRDDAAAFLLKFVAIASILLAGAAGVAIPLIGKNRRFLQTEGNLFVAA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL +PCLP++PWSKFPF GFFAM+AAL TLL+DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALTNPCLPDYPWSKFPFPGFFAMVAALITLLVDFMGTQ 125
Query: 122 YYERKQGMNRA--------VDEQARV--GTSEEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
YYE KQ N V+E + V E GN K FGEE+ GGMHIVG+ AHAAHH
Sbjct: 126 YYESKQQRNEVAGGGEAAVVEETSSVLPVVVERGNDSKAFGEEDGGGMHIVGIRAHAAHH 185
Query: 172 RHNHPHGDAC-DGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLS 230
H+H + DG + HGH H H + + E RHVVVSQ+LELGIVSHS+IIGLS
Sbjct: 186 NHSHSNAHGTFDGHAHGQSHGHVHVHG--SHDVENGARHVVVSQILELGIVSHSIIIGLS 243
Query: 231 LGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIG 290
LGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S IMACFFALT PIG+GIG
Sbjct: 244 LGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSAIIMACFFALTAPIGIGIG 303
Query: 291 TGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCM 350
T +AS +N +SPGAL+ EGILD+LSAGIL YMALVDLIAADFLSKRMSCN RLQ+VSY M
Sbjct: 304 TAVASSFNSHSPGALVTEGILDSLSAGILTYMALVDLIAADFLSKRMSCNVRLQVVSYVM 363
Query: 351 LFLGAGLMSSLAIWA 365
LFLGAGLMS+LAIWA
Sbjct: 364 LFLGAGLMSALAIWA 378
>UniRef100_Q9FPW7 Putative Zn transporter ZNT4 [Thlaspi caerulescens]
Length = 386
Score = 516 bits (1328), Expect = e-145
Identities = 266/376 (70%), Positives = 299/376 (78%), Gaps = 17/376 (4%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DLCRD+ AAF+LK +A+ASI LAG AG+ IPLIG++RR+L+TDG+LFVAAKAFA
Sbjct: 10 CNAGEPDLCRDDSAAFLLKLVAIASIFLAGAAGVVIPLIGRNRRFLQTDGSLFVAAKAFA 69
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGFVHML+ TEAL +PCLPEFPW KFPF FFAM+AAL TLL+DF+GTQYYE+
Sbjct: 70 AGVILATGFVHMLAGGTEALTNPCLPEFPWKKFPFPRFFAMVAALITLLVDFMGTQYYEK 129
Query: 126 KQGMNRAV--DEQARVG-----------TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHR 172
KQ DE G T+EEGN KVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 130 KQEREATTRSDELPSSGPEQSPGIVVPVTAEEGNDEKVFGEEDSGGIHIVGIHAHAAHHT 189
Query: 173 HNHPHGDA-CDGG---GIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIG 228
HNH G + CDG I HGH H H+ RHVVVSQVLELGIVSHS+IIG
Sbjct: 190 HNHTQGQSSCDGHRKIDIGHAHGHGHGHSHGGLELGNGARHVVVSQVLELGIVSHSIIIG 249
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVG 288
+SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S TIMACFFALTTPI +G
Sbjct: 250 ISLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSATIMACFFALTTPISIG 309
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAADFLSKRMSCNFRLQ+VSY
Sbjct: 310 IGTVVASSFNAHSVGALVTEGILDSLSAGILVYMALVDLIAADFLSKRMSCNFRLQIVSY 369
Query: 349 CMLFLGAGLMSSLAIW 364
+LFLG+GLMSSLAIW
Sbjct: 370 LLLFLGSGLMSSLAIW 385
>UniRef100_Q75NR8 Zn/Cd transporter homolog [Thlaspi japonicum]
Length = 423
Score = 515 bits (1326), Expect = e-145
Identities = 265/376 (70%), Positives = 297/376 (78%), Gaps = 17/376 (4%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DLCRD+ AAF+LK +A+ASI LAG AG+AIPLIG++RR+L+TDG+LFVAAKAFA
Sbjct: 47 CDAGEPDLCRDDSAAFLLKLVAIASIFLAGAAGVAIPLIGRNRRFLQTDGSLFVAAKAFA 106
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGFVHML+ TEAL +PCLPEFPW KFPF GFFAM+AAL TLL+DF+GTQYYE+
Sbjct: 107 AGVILATGFVHMLAGGTEALTNPCLPEFPWKKFPFPGFFAMVAALITLLVDFMGTQYYEK 166
Query: 126 KQGMNRAVD--EQARVG-----------TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHR 172
KQ EQ G EEGN KVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 167 KQEREATTHSGEQPSSGPEQSLGIVVPVAGEEGNDEKVFGEEDSGGIHIVGIHAHAAHHT 226
Query: 173 HNHPHGDA-CDGGG---IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIG 228
HNH G + CDG I HGH H H+ RHVVVSQVLELGIVSHS+IIG
Sbjct: 227 HNHTQGQSSCDGHSKIDIGHAHGHGHGHSHGGLELGNGARHVVVSQVLELGIVSHSIIIG 286
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVG 288
+SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK TIMACFFALTTPI +G
Sbjct: 287 ISLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKPATIMACFFALTTPISIG 346
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAADFLSK MSCNFRLQ+VSY
Sbjct: 347 IGTAVASSFNAHSVGALVTEGILDSLSAGILVYMALVDLIAADFLSKMMSCNFRLQIVSY 406
Query: 349 CMLFLGAGLMSSLAIW 364
+LFLG+GLMSSLAIW
Sbjct: 407 LLLFLGSGLMSSLAIW 422
>UniRef100_Q6VM17 Metal transport protein [Medicago truncatula]
Length = 374
Score = 483 bits (1243), Expect = e-135
Identities = 255/377 (67%), Positives = 294/377 (77%), Gaps = 23/377 (6%)
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVA 60
MTNS+C ESDLCRDE AA +LKF+AMASIL+AG +GIA+PL+G R LR+DG + A
Sbjct: 9 MTNSSCESGESDLCRDESAALILKFVAMASILVAGFSGIAVPLLGNRRGLLRSDGEILPA 68
Query: 61 AKAFAAGVILATGFVHMLSDATEALNSPCLPEFP--WSKFPFTGFFAMMAALFTLLLDFV 118
AKAFAAGVILATGFVHML DA +ALN CL + WS+FPFTGFFAMM+AL TLL+DFV
Sbjct: 69 AKAFAAGVILATGFVHMLQDAWKALNHSCLKSYSHVWSEFPFTGFFAMMSALLTLLVDFV 128
Query: 119 GTQYYERKQGMNRAVDEQARVGTSEEG--------NVGKVFGEEESGGMHIVGMHAHAAH 170
TQYYE + + D RV + EG + +V GE GGMHIVGMHAHA+
Sbjct: 129 ATQYYESQH--QKTHDRHGRVVGNGEGLEEELLGSGIVEVQGETFGGGMHIVGMHAHASQ 186
Query: 171 HRHNHP-HGDACDGGGIVKEHGHDHSHALIAANE-ETDVRHVVVSQVLELGIVSHSVIIG 228
H H+H HGD HGH HSH+ + ++ VRHVVVSQVLELGIVSHS+IIG
Sbjct: 187 HGHSHQNHGDG---------HGHGHSHSFGEHDGVDSSVRHVVVSQVLELGIVSHSLIIG 237
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVG 288
LSLGVSQSPC +RPLIAALSFHQFFEGFALGGCIS+A+FK SS TIMACFFALTTP+GV
Sbjct: 238 LSLGVSQSPCTMRPLIAALSFHQFFEGFALGGCISEARFKTSSATIMACFFALTTPLGVA 297
Query: 289 IGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSY 348
IGT +AS +NPYSPGALI EGILD+LSAGILVYMALVDLIAADFLSK+M C+ RLQ+VS+
Sbjct: 298 IGTLVASNFNPYSPGALITEGILDSLSAGILVYMALVDLIAADFLSKKMRCSLRLQIVSF 357
Query: 349 CMLFLGAGLMSSLAIWA 365
C+LFLGAG MSSLA+WA
Sbjct: 358 CLLFLGAGSMSSLALWA 374
>UniRef100_Q93YA2 Putative Zn and Cd transporter [Thlaspi caerulescens]
Length = 344
Score = 462 bits (1190), Expect = e-129
Identities = 238/342 (69%), Positives = 271/342 (78%), Gaps = 15/342 (4%)
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T C ESDLCRD+ AAF+LKF+A+ASIL+AG AG+AIPLIGK+RR+L+T+GNLFVAA
Sbjct: 6 TKILCDAGESDLCRDDAAAFLLKFVAIASILIAGAAGVAIPLIGKNRRFLQTEGNLFVAA 65
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQ 121
KAFAAGVILATGFVHML+ TEAL +PCLP++PWSKFPF GFFAM+AAL TLL+DF+GTQ
Sbjct: 66 KAFAAGVILATGFVHMLAGGTEALTNPCLPDYPWSKFPFPGFFAMVAALITLLVDFMGTQ 125
Query: 122 YYERKQGMNRA--------VDEQARV--GTSEEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
YYE KQ N V+E + V E GN K FGEE+ GGMHIVG+ AHAAHH
Sbjct: 126 YYESKQQRNEVAGGGEAAVVEETSSVLPVVVERGNDSKAFGEEDGGGMHIVGIRAHAAHH 185
Query: 172 RHNHP--HGDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGL 229
H+H HG CDG + HGH H H + + E RHVVVSQ+LELGIVSHS+IIGL
Sbjct: 186 NHSHSNAHG-TCDGHAHGQSHGHVHVHG--SHDVENGARHVVVSQILELGIVSHSIIIGL 242
Query: 230 SLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGI 289
SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S IMACFFALT PIG+GI
Sbjct: 243 SLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSAIIMACFFALTAPIGIGI 302
Query: 290 GTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAAD 331
GT +AS +N +SPGAL+ EGILD+LSAGIL YMALVDLIAAD
Sbjct: 303 GTAVASSFNSHSPGALVTEGILDSLSAGILTYMALVDLIAAD 344
>UniRef100_Q84VR5 Putative Zn transporter [Thlaspi caerulescens]
Length = 350
Score = 456 bits (1172), Expect = e-127
Identities = 236/343 (68%), Positives = 266/343 (76%), Gaps = 19/343 (5%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DLCRD+ AAF+LK +A+ASI LAG AG+AIPLIG++RR+L+TDG+LFVAAKAFA
Sbjct: 9 CNAGEPDLCRDDSAAFLLKLVAIASIFLAGAAGVAIPLIGRNRRFLQTDGSLFVAAKAFA 68
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGFVHML+ TEAL +PCLPEFPW KFPF GFFAM+AAL TLL+DF+GTQYYE+
Sbjct: 69 AGVILATGFVHMLAGGTEALTNPCLPEFPWKKFPFPGFFAMVAALITLLVDFMGTQYYEK 128
Query: 126 KQGMNRAVDEQARVGTS--------------EEGNVGKVFGEEESGGMHIVGMHAHAAHH 171
KQ A + S EEGN KVFGEE+SGG+HIVG+HAHAAHH
Sbjct: 129 KQE-REATTHSGELPPSGPEQSPGIVVPVAAEEGNDEKVFGEEDSGGIHIVGIHAHAAHH 187
Query: 172 RHNHPHGDA-CDGG---GIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVII 227
HNH G + CDG I HGH H H+ RHVVVSQVLELGIVSHS+II
Sbjct: 188 THNHTQGQSSCDGHRKIDIGHAHGHGHGHSHGGLELGNGARHVVVSQVLELGIVSHSIII 247
Query: 228 GLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGV 287
G+SLGVSQSPC IRPLIAALSFHQFFEGFALGGCISQAQFK S TIMACFFALTTPI +
Sbjct: 248 GISLGVSQSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSATIMACFFALTTPISI 307
Query: 288 GIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAA 330
GIGT +AS +N +S GAL+ EGILD+LSAGILVYMALVDLIAA
Sbjct: 308 GIGTAVASSFNAHSVGALVTEGILDSLSAGILVYMALVDLIAA 350
>UniRef100_Q5Z653 Putative ZIP-like zinc transporter [Oryza sativa]
Length = 422
Score = 431 bits (1109), Expect = e-119
Identities = 230/376 (61%), Positives = 272/376 (72%), Gaps = 20/376 (5%)
Query: 7 GGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYL-------RTDGNLFV 59
GG + + CRDE AA LK +A+A+IL+AG AG+AIPL+G+ RR + G LFV
Sbjct: 50 GGDKDEECRDEAAALRLKMVAVAAILIAGAAGVAIPLVGRRRRGGGGGGGGGASSGGLFV 109
Query: 60 AAKAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVG 119
AKAFAAGVILATGFVHML DA AL++PCLP PW +FPF GF AM+AAL TL++DFVG
Sbjct: 110 LAKAFAAGVILATGFVHMLHDAEHALSNPCLPHSPWRRFPFPGFVAMLAALATLVVDFVG 169
Query: 120 TQYYERKQGMNRAVD--EQARVGTSEEGN---VGKVFGEEESGG----MHIVGMHAHAAH 170
T +YERK A E+A E+G VG G + GG MHIVG+HAHAA
Sbjct: 170 THFYERKHRQEEAAAAAEEAAAALLEDGGALPVGDGEGRDGRGGKRDAMHIVGIHAHAAA 229
Query: 171 HRHNHPH-GDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGL 229
HRH+H H AC GG + H H H H E RHVVVSQ+LELGIVSHSVIIGL
Sbjct: 230 HRHSHAHVHGACHGGAVNDAHAHGHGHG---HEEGPSARHVVVSQILELGIVSHSVIIGL 286
Query: 230 SLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGI 289
SLGVSQSPC I+PL+AALSFHQFFEGFALGGCIS+AQ K S +MA FFA+TTP G+ +
Sbjct: 287 SLGVSQSPCTIKPLVAALSFHQFFEGFALGGCISEAQLKNFSAFLMAFFFAITTPAGITV 346
Query: 290 GTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYC 349
G +AS YNP SP AL+ EGILD++SAGIL+YMALVDLIAADFLS++MSCN RLQ+ SY
Sbjct: 347 GAAVASFYNPNSPRALVVEGILDSMSAGILIYMALVDLIAADFLSRKMSCNPRLQVGSYI 406
Query: 350 MLFLGAGLMSSLAIWA 365
LFLGA M++LA+WA
Sbjct: 407 ALFLGAMAMAALALWA 422
>UniRef100_O82643 Zinc transporter 9 [Arabidopsis thaliana]
Length = 344
Score = 405 bits (1042), Expect = e-112
Identities = 214/347 (61%), Positives = 259/347 (73%), Gaps = 12/347 (3%)
Query: 28 MASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGVILATGFVHMLSDATEALNS 87
MASIL++G AG++IPL+G L +G L AKAFAAGVILATGFVHMLS ++AL+
Sbjct: 1 MASILISGAAGVSIPLVGT---LLPLNGGLMRGAKAFAAGVILATGFVHMLSGGSKALSD 57
Query: 88 PCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQG--MNRAVDE---QARVGTS 142
PCLPEFPW FPF FFAM+AAL TLL DF+ T YYERKQ MN++V+ Q V +
Sbjct: 58 PCLPEFPWKMFPFPEFFAMVAALLTLLADFMITGYYERKQEKMMNQSVESLGTQVSVMSD 117
Query: 143 EEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHG----DACDGGGIVKEHGHDHSHAL 198
G + +E+ G +HIVGM AHA HHRH+ G +A V HGH HSH
Sbjct: 118 PGLESGFLRDQEDGGALHIVGMRAHAEHHRHSLSMGAEGFEALSKRSGVSGHGHGHSHGH 177
Query: 199 IAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFAL 258
++ VRHVVVSQ+LE+GIVSHS+IIG+SLGVS SPC IRPL+ ALSFHQFFEGFAL
Sbjct: 178 GDVGLDSGVRHVVVSQILEMGIVSHSIIIGISLGVSHSPCTIRPLLLALSFHQFFEGFAL 237
Query: 259 GGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGI 318
GGC+++A+ + +MA FFA+TTPIGV +GT IAS YN YS AL+AEG+LD+LSAGI
Sbjct: 238 GGCVAEARLTPRGSAMMAFFFAITTPIGVAVGTAIASSYNSYSVAALVAEGVLDSLSAGI 297
Query: 319 LVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
LVYMALVDLIAADFLSK+MS +FR+Q+VSYC LFLGAG+MS+LAIWA
Sbjct: 298 LVYMALVDLIAADFLSKKMSVDFRVQVVSYCFLFLGAGMMSALAIWA 344
>UniRef100_Q84L19 Putative ZIP-like zinc transporter [Oryza sativa]
Length = 384
Score = 383 bits (984), Expect = e-105
Identities = 209/368 (56%), Positives = 256/368 (68%), Gaps = 25/368 (6%)
Query: 9 AESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGV 68
AE CRD+ AA LK +AMA+IL+AG+ G+ +PL G+ RR LRTD FVAAKAFAAGV
Sbjct: 31 AEGAGCRDDAAALRLKGVAMATILVAGVVGVGLPLAGRKRRALRTDSAAFVAAKAFAAGV 90
Query: 69 ILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQG 128
ILATGFVHML DA AL+SPCLP PW FPF GF AM AAL TL+LDF+ T++YE G
Sbjct: 91 ILATGFVHMLHDAEHALSSPCLPAHPWRSFPFPGFVAMSAALATLVLDFLATRFYE---G 147
Query: 129 MNRAVDEQARVG--------TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDA 180
+RA E+ + ++ + ++ V E+ + H+ H H+HPHG
Sbjct: 148 NHRAETERVKAAAAAALAASSASDDDITVVTVTEDDNDNKAPLLQPHS--HSHSHPHGHG 205
Query: 181 CDGGGIVKEHGHDHSHALIAANE---ETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSP 237
HGH+ + + E VR VVVSQ+LE+GIVSHSVIIGLSLGVS+SP
Sbjct: 206 ---------HGHELAQPEGSGGEGEVPAQVRSVVVSQILEMGIVSHSVIIGLSLGVSRSP 256
Query: 238 CAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVY 297
C IRPL+AALSFHQFFEGFALGGCI+QAQFK S IMACFFA+TTP G+ G G+AS Y
Sbjct: 257 CTIRPLVAALSFHQFFEGFALGGCIAQAQFKTLSAAIMACFFAITTPAGIAAGAGVASFY 316
Query: 298 NPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGL 357
NP SP AL+ EGILD++SAGIL+YM+ VDLIAADFL +M+ + R Q+++Y LFLGA
Sbjct: 317 NPNSPRALVVEGILDSVSAGILIYMSQVDLIAADFLGGKMTGSTRQQVMAYIALFLGALS 376
Query: 358 MSSLAIWA 365
MSSLAIWA
Sbjct: 377 MSSLAIWA 384
>UniRef100_Q6L8F7 Iron regulated transporter-like protein [Oryza sativa]
Length = 384
Score = 383 bits (983), Expect = e-105
Identities = 209/368 (56%), Positives = 256/368 (68%), Gaps = 25/368 (6%)
Query: 9 AESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFAAGV 68
AE CRD+ AA LK +AMA+IL+AG+ G+ +PL G+ RR LRTD FVAAKAFAAGV
Sbjct: 31 AEGAGCRDDAAALRLKGVAMATILVAGVVGVGLPLAGRKRRALRTDSAAFVAAKAFAAGV 90
Query: 69 ILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQG 128
ILATGFVHML DA AL+SPCLP PW FPF GF AM AAL TL+LDF+ T++YE G
Sbjct: 91 ILATGFVHMLHDAEHALSSPCLPAHPWRSFPFPGFVAMSAALATLVLDFLATRFYE---G 147
Query: 129 MNRAVDEQARVG--------TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDA 180
+RA E+ + ++ + ++ V E+ + H+ H H+HPHG
Sbjct: 148 KHRAETERVKAAAAAALAASSASDDDITVVTVTEDDNDNKAPLLQPHS--HSHSHPHGHG 205
Query: 181 CDGGGIVKEHGHDHSHALIAANE---ETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSP 237
HGH+ + + E VR VVVSQ+LE+GIVSHSVIIGLSLGVS+SP
Sbjct: 206 ---------HGHELAQPEGSGGEGEVPAQVRSVVVSQILEMGIVSHSVIIGLSLGVSRSP 256
Query: 238 CAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVY 297
C IRPL+AALSFHQFFEGFALGGCI+QAQFK S IMACFFA+TTP G+ G G+AS Y
Sbjct: 257 CTIRPLVAALSFHQFFEGFALGGCIAQAQFKTLSAAIMACFFAITTPAGIAAGAGVASFY 316
Query: 298 NPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGL 357
N SP AL+ EGILD++SAGIL+YM+LVDLIAADFL +M+ + R Q+++Y LFLGA
Sbjct: 317 NANSPRALVVEGILDSVSAGILIYMSLVDLIAADFLGGKMTGSTRQQVMAYIALFLGALS 376
Query: 358 MSSLAIWA 365
MSSLAIWA
Sbjct: 377 MSSLAIWA 384
>UniRef100_Q6ZJ91 Putative iron transporter Fe2 [Oryza sativa]
Length = 396
Score = 318 bits (816), Expect = 1e-85
Identities = 185/372 (49%), Positives = 231/372 (61%), Gaps = 26/372 (6%)
Query: 6 CGGAESDLCRDEPA--AFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKA 63
CG A + E A A LK +A+ASIL AG AG+ +P++G+ LR DG++F A KA
Sbjct: 39 CGNAAAAAVAGEDARGALRLKLVAIASILAAGAAGVLVPVLGRSFAALRPDGDVFFAVKA 98
Query: 64 FAAGVILATGFVHMLSDATEALNSPCLP-EFPWSKFPFTGFFAMMAALFTLLLDFVGTQY 122
FAAGVILATG VH+L A +AL SPC FPF G AM AA+ T+++D V Y
Sbjct: 99 FAAGVILATGMVHILPAAFDALASPCGGGRGGGGGFPFAGLVAMAAAMATMMIDSVAAGY 158
Query: 123 YERKQGMN-RAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDAC 181
Y R R VD+ A + + G EE G H +H H H H H HG
Sbjct: 159 YRRSHFKKPRPVDDPA--------DAARAAGVEEGGAEHAGHVHVHT-HATHGHAHGHVH 209
Query: 182 DGGGIVKEHGHDHSH--ALIAANEETD------VRHVVVSQVLELGIVSHSVIIGLSLGV 233
G HGH HSH A AA D +RH VVSQVLELGI+ HSVIIG+SLG
Sbjct: 210 SHG-----HGHGHSHGSAPAAATSPEDASVAETIRHRVVSQVLELGILVHSVIIGVSLGA 264
Query: 234 SQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGI 293
S P +IRPL+ ALSFHQFFEG LGGCI QA FKA +T IMA FF+LT P+G+ +G I
Sbjct: 265 SLRPSSIRPLVGALSFHQFFEGIGLGGCIVQANFKAKATVIMATFFSLTAPVGIALGIAI 324
Query: 294 ASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFL 353
+S Y+ +S AL+ EG+ ++ +AGIL+YM+LVDL+AADF + ++ N +LQL Y LFL
Sbjct: 325 SSSYSKHSSTALVVEGVFNSAAAGILIYMSLVDLLAADFNNPKLQTNTKLQLAVYLALFL 384
Query: 354 GAGLMSSLAIWA 365
GAG+MS LAIWA
Sbjct: 385 GAGMMSLLAIWA 396
>UniRef100_Q7XJ47 Putative zinc transporter [Oryza sativa]
Length = 390
Score = 311 bits (798), Expect = 1e-83
Identities = 181/378 (47%), Positives = 228/378 (59%), Gaps = 37/378 (9%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
CG ++ RD A LK A SIL+ G G +P +G+H LR DG++F KAFA
Sbjct: 32 CGKEDAAAGRDRARARGLKIAAFFSILVCGALGCGLPSLGRHVPALRPDGDVFFLVKAFA 91
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEF-PWSKFPFTGFFAMMAALFTLLLDFVGTQYYE 124
AGVILATGF+H+L DA + L CLP PW +FPF GF AM+ A+ TL++D + T Y+
Sbjct: 92 AGVILATGFIHILPDAFDNLTDDCLPAGGPWKEFPFAGFGAMVGAIGTLVVDTLATGYFT 151
Query: 125 RKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGG 184
R A A V EE+ A H H+H H DGG
Sbjct: 152 RALSKKDAATAAA------------VADEEKQSA-------AATQQHNHHHNHHVVGDGG 192
Query: 185 GIVKEH------------GHDH-SHALIAA----NEETDVRHVVVSQVLELGIVSHSVII 227
G +EH GH H S AL+AA ++ET +RH V+SQVLELGIV HSVII
Sbjct: 193 GGGEEHEGQVHVHTHATHGHAHGSSALVAAVGEDDKETTLRHRVISQVLELGIVVHSVII 252
Query: 228 GLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGV 287
G+SLG SQ+P I+PL+ ALSFHQ FEG LGGCI QA+FK S M FF LTTP+G+
Sbjct: 253 GISLGASQNPETIKPLVVALSFHQMFEGMGLGGCIVQAKFKVRSIVTMVLFFCLTTPVGI 312
Query: 288 GIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVS 347
+G GI+SVYN SP AL+ EGIL++++AGIL+YMALVDL+A DF++ R+ +LQL
Sbjct: 313 AVGVGISSVYNESSPTALVVEGILNSVAAGILIYMALVDLLAEDFMNPRVQSKGKLQLGI 372
Query: 348 YCMLFLGAGLMSSLAIWA 365
+ GAGLMS LA WA
Sbjct: 373 NLAMLAGAGLMSMLAKWA 390
>UniRef100_Q6VM18 Metal transport protein [Medicago truncatula]
Length = 372
Score = 311 bits (797), Expect = 2e-83
Identities = 173/361 (47%), Positives = 218/361 (59%), Gaps = 36/361 (9%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E D RD A K A+ SIL+A G+ +PL+GK L + ++F KAFA
Sbjct: 47 CTCDEEDEERDRSKALRYKIAALVSILVASAIGVCLPLLGKVIPALSPEKDIFFIIKAFA 106
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVIL+TGF+H+L DA E L SPCL E PW FPFTGF AM A+ TL++D T Y++
Sbjct: 107 AGVILSTGFIHVLPDAFENLTSPCLNEHPWGDFPFTGFVAMCTAMGTLMVDTYATAYFQN 166
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
+++ E H H H H
Sbjct: 167 HYSKRAPAQVESQTTPDVENE-----------------------EHTHVHAHAS------ 197
Query: 186 IVKEHGHDHSHALIAANEETDV-RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLI 244
H H+H I+ ++ +++ RH V+SQVLELGI+ HSVIIG+SLG S+SP IRPL+
Sbjct: 198 ------HSHAHGHISFDQSSELLRHRVISQVLELGIIGHSVIIGISLGASESPKTIRPLV 251
Query: 245 AALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGA 304
AAL+FHQFFEG LG CI+QA FK+ S TIM FFALTTP+G+GIG GI++VY+ SP A
Sbjct: 252 AALTFHQFFEGMGLGSCITQANFKSLSITIMGLFFALTTPVGIGIGLGISNVYDENSPTA 311
Query: 305 LIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIW 364
I EGI +A SAGIL+YMALVDL+AADF++ RM N RLQL S L LGAG MS +A W
Sbjct: 312 FIFEGIFNAASAGILIYMALVDLLAADFMNPRMQKNGRLQLGSNISLLLGAGCMSLIAKW 371
Query: 365 A 365
A
Sbjct: 372 A 372
>UniRef100_Q941P4 Zinc transporter protein ZIP1 [Glycine max]
Length = 354
Score = 305 bits (781), Expect = 1e-81
Identities = 175/361 (48%), Positives = 221/361 (60%), Gaps = 37/361 (10%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C D RD+ A K A+ SIL+AG G+ IPL+GK L + + F KAFA
Sbjct: 30 CTCDREDEERDKSKALRYKIAALVSILVAGAIGVCIPLLGKVISALSPEKDTFFIIKAFA 89
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVIL+TGF+H+L DA E L SPCL E PW +FPFTGF AM A+ TL++D T Y+++
Sbjct: 90 AGVILSTGFIHVLPDAFENLTSPCLKEHPWGEFPFTGFVAMCTAMGTLMVDTYATAYFKK 149
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
+ + DE V E+ESG V +H HA H
Sbjct: 150 H---HHSQDEATDV-------------EKESGHEGHVHLHTHATH--------------- 178
Query: 186 IVKEHGHDHSHALIAANEETDV-RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLI 244
GH H H ++ +++ RH V+SQVLE+GI+ HS+IIG+SLG S+SP IRPL+
Sbjct: 179 -----GHAHGHVPTDDDQSSELLRHRVISQVLEVGIIVHSIIIGISLGASESPKTIRPLM 233
Query: 245 AALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGA 304
AAL FHQFFEG LG CI+QA FK S T+M FALTTP+G+GIG GI VY+ SP A
Sbjct: 234 AALIFHQFFEGMGLGSCITQANFKKLSITLMGLVFALTTPMGIGIGIGITKVYDENSPTA 293
Query: 305 LIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIW 364
LI EGI +A SAGIL+YMALVDL+AADF++ RM + L+L + L LGAG MS LA W
Sbjct: 294 LIVEGIFNAASAGILIYMALVDLLAADFMNPRMQKSGSLRLGANLSLLLGAGCMSLLAKW 353
Query: 365 A 365
A
Sbjct: 354 A 354
>UniRef100_Q7XLD4 OSJNBa0070C17.15 protein [Oryza sativa]
Length = 364
Score = 305 bits (780), Expect = 2e-81
Identities = 177/365 (48%), Positives = 222/365 (60%), Gaps = 27/365 (7%)
Query: 4 STCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKA 63
S C A + D A LK IA+ASIL AG AG+ +P+IG+ LR DG++F A KA
Sbjct: 24 SACDCANTTDGADRQGAMKLKLIAIASILAAGAAGVLVPVIGRSMAALRPDGDIFFAVKA 83
Query: 64 FAAGVILATGFVHMLSDATEALNSPCLPEFPWSK--FPFTGFFAMMAALFTLLLDFVGTQ 121
FAAGVILATG VH+L A +AL SPCL + FPF G +M AA+ T+++D +
Sbjct: 84 FAAGVILATGMVHILPAAFDALTSPCLKRGGGDRNPFPFAGLVSMSAAVSTMVVDSLAAG 143
Query: 122 YYERKQGMN-RAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDA 180
YY R Q R VD NV K G+E + + H H H H HGD
Sbjct: 144 YYHRSQFRKARPVDNI---------NVHKHAGDERAEHAQHINAHTHGGH---THSHGDI 191
Query: 181 CDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAI 240
G E G + +RH VVSQVLELGI+ HSVIIG+SLG S P I
Sbjct: 192 VVCGS--PEEG----------SVAESIRHKVVSQVLELGILVHSVIIGVSLGASVRPSTI 239
Query: 241 RPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPY 300
RPL+ ALSFHQFFEG LGGCI QA FK +T IMA FF+LT P+G+ +G I+S YN +
Sbjct: 240 RPLVGALSFHQFFEGVGLGGCIVQANFKVRATVIMAIFFSLTAPVGIVLGIAISSSYNVH 299
Query: 301 SPGALIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSS 360
S A + EG+ ++ SAGIL+YM+LVDL+A DF + ++ N +LQL++Y LFLGAGLMS
Sbjct: 300 SSTAFVVEGVFNSASAGILIYMSLVDLLATDFNNPKLQINTKLQLMAYLALFLGAGLMSM 359
Query: 361 LAIWA 365
LAIWA
Sbjct: 360 LAIWA 364
>UniRef100_Q6VM19 Metal transport protein [Medicago truncatula]
Length = 359
Score = 304 bits (778), Expect = 3e-81
Identities = 174/361 (48%), Positives = 222/361 (61%), Gaps = 30/361 (8%)
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C E DL RD+P A K A+ SIL+A G+ IPL+GK L + ++F KAFA
Sbjct: 28 CTCDEEDLDRDKPKALRYKIAALVSILVASGIGVCIPLLGKVIPALSPEKDIFFIIKAFA 87
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVILATGF+H+L DA E L SP L + PW FPFTGF AM A+ TL++D T Y++
Sbjct: 88 AGVILATGFIHVLPDAFENLTSPRLKKHPWGDFPFTGFVAMCTAMGTLMVDTYATAYFQN 147
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
+ V E ++ G M + H HA+H H HPH + G
Sbjct: 148 HYSKKAPAQVENEVSPDVE--------KDHEGHMDV---HTHASHG-HAHPHMSSVSSGP 195
Query: 186 IVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIA 245
+ +RH V++QVLELGI+ HSVIIG+SLG S+SP IRPL+A
Sbjct: 196 STEL-----------------LRHRVITQVLELGIIVHSVIIGISLGASESPKTIRPLVA 238
Query: 246 ALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGAL 305
AL+FHQFFEG LG CI+QA FK+ S TIM FFALTTP+G+ IG GI+S Y+ SP AL
Sbjct: 239 ALTFHQFFEGMGLGSCITQANFKSLSITIMGLFFALTTPVGIAIGIGISSGYDENSPTAL 298
Query: 306 IAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQL-VSYCMLFLGAGLMSSLAIW 364
I EGI +A S+GIL+YMALVDL+AADF++ RM N L+L + +L LG+GLMS +A W
Sbjct: 299 IVEGIFNAASSGILIYMALVDLLAADFMNPRMQKNGILRLGCNISLLLLGSGLMSLIAKW 358
Query: 365 A 365
A
Sbjct: 359 A 359
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 584,665,882
Number of Sequences: 2790947
Number of extensions: 24005329
Number of successful extensions: 101160
Number of sequences better than 10.0: 417
Number of HSP's better than 10.0 without gapping: 175
Number of HSP's successfully gapped in prelim test: 261
Number of HSP's that attempted gapping in prelim test: 98188
Number of HSP's gapped (non-prelim): 1500
length of query: 365
length of database: 848,049,833
effective HSP length: 129
effective length of query: 236
effective length of database: 488,017,670
effective search space: 115172170120
effective search space used: 115172170120
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133572.2


