

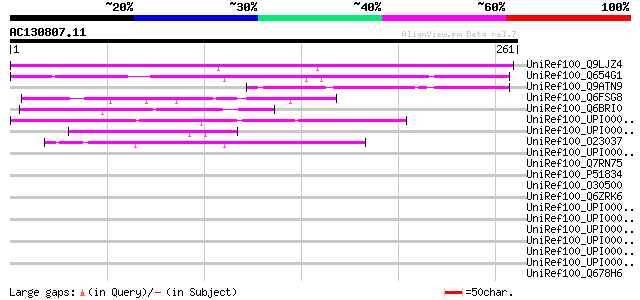
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.11 - phase: 0
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LJZ4 Arabidopsis thaliana genomic DNA, chromosome 3,... 199 8e-50
UniRef100_Q654G1 Hypothetical protein P0412C04.25 [Oryza sativa] 135 1e-30
UniRef100_Q9ATN9 Hypothetical protein [Pennisetum ciliare] 84 3e-15
UniRef100_Q6FSG8 Similar to tr|Q12045 Saccharomyces cerevisiae Y... 51 3e-05
UniRef100_Q6BRI0 Similar to CA4716|IPF8666 Candida albicans IPF8... 50 6e-05
UniRef100_UPI000027AB5E UPI000027AB5E UniRef100 entry 50 8e-05
UniRef100_UPI000042CDFA UPI000042CDFA UniRef100 entry 49 1e-04
UniRef100_O23037 YUP8H12.6 protein [Arabidopsis thaliana] 47 4e-04
UniRef100_UPI00002C5C15 UPI00002C5C15 UniRef100 entry 46 0.001
UniRef100_Q7RN75 Hypothetical protein [Plasmodium yoelii yoelii] 46 0.001
UniRef100_P51834 Chromosome partition protein smc [Bacillus subt... 46 0.001
UniRef100_O30500 YttA [Bacillus subtilis] 45 0.001
UniRef100_Q6ZRK6 Hypothetical protein FLJ46282 [Homo sapiens] 45 0.001
UniRef100_UPI00004983CC UPI00004983CC UniRef100 entry 45 0.002
UniRef100_UPI000025E246 UPI000025E246 UniRef100 entry 45 0.002
UniRef100_UPI000045363A UPI000045363A UniRef100 entry 45 0.002
UniRef100_UPI000034C4FB UPI000034C4FB UniRef100 entry 44 0.003
UniRef100_UPI000033DA4D UPI000033DA4D UniRef100 entry 44 0.003
UniRef100_UPI0000221447 UPI0000221447 UniRef100 entry 44 0.003
UniRef100_Q678H6 Membrane-bound metallopeptidase [Lymphocystis d... 44 0.003
>UniRef100_Q9LJZ4 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MAL21
[Arabidopsis thaliana]
Length = 282
Score = 199 bits (505), Expect = 8e-50
Identities = 114/280 (40%), Positives = 169/280 (59%), Gaps = 21/280 (7%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEALY KLY KYT L+ K SE ++++KEQE KFL FVSA+E++++HLR EN L +
Sbjct: 1 MEALYAKLYDKYTKLQKKKYSEYDEINKEQEEKFLTFVSASEELMEHLRGENQSSLEMVE 60
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK-------EGTSGDL 113
L NE+ S+R +D++ +E ++LL+EE+ KN++L EEV KL++L++ E SG
Sbjct: 61 KLRNEIISIRSGRDDKFLECQKLLMEEELKNKSLSEEVVKLKELVQEEHPRNYEDQSGKK 120
Query: 114 SNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVD--------------TQ 159
RK + + + R R+ ++ D+ + I ++ +
Sbjct: 121 QKRKTPESARVTTRSMIKRSRLSEDLVETDMVSPDISKHHKAKEPLLVSQPQCCRTTYDG 180
Query: 160 ESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLY 219
S + AL ++ L MKLST+ + R C+ A H ++G SFS+++I+ GEE+ELLY
Sbjct: 181 SSSSASCTFQALGKHLLGMKLSTNNKGKRACIVASHPTTGLSFSLTFINNPNGEESELLY 240
Query: 220 HVLSLGTLERLAPEWMREDIMFSPTMCPIFFERVTHVINL 259
SLGT +R+APEWMRE I FS +MCPIFFERV+ VI L
Sbjct: 241 KPASLGTFQRVAPEWMREVIKFSTSMCPIFFERVSRVIKL 280
>UniRef100_Q654G1 Hypothetical protein P0412C04.25 [Oryza sativa]
Length = 270
Score = 135 bits (340), Expect = 1e-30
Identities = 89/281 (31%), Positives = 142/281 (49%), Gaps = 38/281 (13%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
ME L KLY +YT LK KL + E + +++ + A +D + L++EN++L+ ++
Sbjct: 1 MEQLNAKLYHRYTALKKRKLLD-EGLDQKRAADINELRQAMKDWVAELQSENERLVAKLT 59
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT--------SGD 112
K+ +LVE + LLL+E +K + L E+ KLQ LL E S D
Sbjct: 60 Q-----------KEQQLVEAQTLLLDETRKTKELNSEILKLQCLLAEKNDANHIATGSPD 108
Query: 113 LSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPS---ENEGVDTQ---------- 159
+ + N + S + T K +E++ IE +P + EG D
Sbjct: 109 TTAEMTIENQTPISPAKKTPKSNSRERNIRSIEKAIVPRNGFQEEGRDLDSCRRHMSISG 168
Query: 160 ---ESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAE 216
E + H L E + MK S +T LS H++SGY+F+++W+ + G E
Sbjct: 169 SATEESSSTCMFHLLAESMVGMKFSVKNETEGFSLSVSHEASGYNFTLTWVDQPGGSEWS 228
Query: 217 LLYHVLSLGTLERLAPEWMREDIMFSPTMCPIFFERVTHVI 257
Y SLGTL+R+A WM+EDI FS TMCP+FF++++H++
Sbjct: 229 --YQYSSLGTLDRIAMGWMKEDIKFSSTMCPVFFKQISHIL 267
>UniRef100_Q9ATN9 Hypothetical protein [Pennisetum ciliare]
Length = 130
Score = 84.3 bits (207), Expect = 3e-15
Identities = 48/135 (35%), Positives = 76/135 (55%), Gaps = 9/135 (6%)
Query: 123 SNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLST 182
S N+SI + + +E L+ R S D S + H + + + MK++
Sbjct: 2 SKNASI--PHRSLEEESQELECSRRYKCSSENETDEWPSAR---MFHLMLQSLVRMKVTV 56
Query: 183 DCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLYHVLSLGTLERLAPEWMREDIMFS 242
D T +S H++SGYSF ++W+ PGE + Y + SLGTLER+A +WM++DI FS
Sbjct: 57 DDGTESFSVSVCHEASGYSFKLTWLEN-PGEWS---YKLSSLGTLERIAVDWMKQDITFS 112
Query: 243 PTMCPIFFERVTHVI 257
MC +FFER++++I
Sbjct: 113 MNMCRVFFERISNII 127
>UniRef100_Q6FSG8 Similar to tr|Q12045 Saccharomyces cerevisiae YPL253c VIK1 [Candida
glabrata]
Length = 584
Score = 50.8 bits (120), Expect = 3e-05
Identities = 50/178 (28%), Positives = 90/178 (50%), Gaps = 28/178 (15%)
Query: 7 KLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRT--ENDKLLGQINDLGN 64
++ +KY + K +E + ++Q+ +A + I+ LR D+L+ QIN+L +
Sbjct: 111 EIENKYKEVIEQKAAEFSQIFEKQQ-------NAWREKIEELRNMPPEDELINQINNLKH 163
Query: 65 ELTSV------RVAK-DNELVEHKRLL----LEEKKKNEALFEEVEKLQKLLKEGTSGDL 113
L+S ++AK DN+L+ +K+ L E KKKN EE++ K L E T +L
Sbjct: 164 TLSSTEQNLEDKIAKNDNDLILYKQKLQVSFSEYKKKNSTSLEELQSQSKNL-ESTMAEL 222
Query: 114 SNRKVVNNTSNNSSIR-MTRKRMRQEQDALD--IEARCIPSENEGVDTQESDHQNWLV 168
N K T NS +R + ++ E+ +D ++ + +E E V+ + ++QN V
Sbjct: 223 QNIK----TEKNSQLRTLAEEQSELERIVMDKQLQVDKLNAEIEPVENKIKEYQNSFV 276
>UniRef100_Q6BRI0 Similar to CA4716|IPF8666 Candida albicans IPF8666 [Debaryomyces
hansenii]
Length = 734
Score = 50.1 bits (118), Expect = 6e-05
Identities = 42/138 (30%), Positives = 69/138 (49%), Gaps = 15/138 (10%)
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVID------HLRTENDKLLGQI 59
+KL + N +E E + +E +K+ +++ D + LR N KL G++
Sbjct: 600 EKLIEELDKEWANTSTEYEKLCEENNTLSVKYQNSSTDAVQLSSKLIALRDANSKLEGKL 659
Query: 60 NDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEV-EKLQKLLKEGTSGDLSNRKV 118
N G +L + KD EL+ H+ L + K +N+ + EKL KL+KE TS +
Sbjct: 660 NGAGIQLLPSLINKD-ELINHEVELNKLKSENQFMLRFFDEKLDKLIKERTS-------L 711
Query: 119 VNNTSNNSSIRMTRKRMR 136
+++TSN SS R T + R
Sbjct: 712 MDSTSNGSSSRPTNRVSR 729
>UniRef100_UPI000027AB5E UPI000027AB5E UniRef100 entry
Length = 774
Score = 49.7 bits (117), Expect = 8e-05
Identities = 53/210 (25%), Positives = 94/210 (44%), Gaps = 10/210 (4%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHK-EQELKFLK-FVSAAEDVIDHLRTENDKLLGQ 58
++ Y YSK ++K + E + E E++ K +S +E +++ L +KL Q
Sbjct: 465 LQEKYADAYSKNQSIKNESVKRNERIKTIETEIESWKNLLSNSEKMVNELTERKNKLSEQ 524
Query: 59 INDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEE----VEKLQKLLKEGTSGDLS 114
+N+L N+ V+ + ++ E+ R+ EEK +NE + EE +E L+ L E +
Sbjct: 525 LNNLENQ-PRVQAERKGQISENLRISDEEKIENEKIIEETDKKIETLRTQLNEIQEQSIQ 583
Query: 115 NRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEY 174
R+ S+ ++I +KR D + E + EN ++ + H E
Sbjct: 584 IRE--RKASSGATIEGLQKRKNDLLDRISTELN-LTEENILENSNLFGVEELPNHVDQED 640
Query: 175 TLDMKLSTDCQTGRLCLSAMHQSSGYSFSI 204
LD K Q G + L A ++S Y I
Sbjct: 641 ALDKKKQQRDQLGSVNLKADEETSKYEEEI 670
>UniRef100_UPI000042CDFA UPI000042CDFA UniRef100 entry
Length = 681
Score = 48.9 bits (115), Expect = 1e-04
Identities = 36/95 (37%), Positives = 53/95 (54%), Gaps = 8/95 (8%)
Query: 31 ELKFLKF-VSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKK 89
E K LK ++ ++D+I +L K L +I+ L N+L R+ K+N L E+ +L E K
Sbjct: 488 ENKLLKAQIAKSQDLIKNLNDLEKKYLNKIDLLSNQLVDFRIIKENSLAENSKLHDELKT 547
Query: 90 KN---EALFEEVE----KLQKLLKEGTSGDLSNRK 117
N +AL +EVE KL+ LKE T SN+K
Sbjct: 548 LNIAQDALHQEVERVNAKLESTLKEHTDLQESNKK 582
Score = 35.4 bits (80), Expect = 1.5
Identities = 34/126 (26%), Positives = 56/126 (43%), Gaps = 13/126 (10%)
Query: 45 IDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKN--EALFEEVEKLQ 102
+D L+T N L+ Q+ L K+N+ + K L LE K N E+ E+ +
Sbjct: 240 LDKLKTANQLLVQQVEQL---------TKENQSLIQKSLNLENKLHNLEESDLEDNTYYK 290
Query: 103 KLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESD 162
K++K S K+ N+S++ + RQ+ D + I ENE + Q +
Sbjct: 291 KIVKNNQSLQEQIGKLTK--LNSSNVSRLNELERQQNDLKSVIEGEIIEENEKLKQQLQE 348
Query: 163 HQNWLV 168
+N LV
Sbjct: 349 SENNLV 354
>UniRef100_O23037 YUP8H12.6 protein [Arabidopsis thaliana]
Length = 841
Score = 47.4 bits (111), Expect = 4e-04
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 13/175 (7%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLG---NELTSVRVAKDN 75
KLS LE K Q+ K L+ ED+ L +E ++L QI+ L N++ + + N
Sbjct: 524 KLSVLE-AEKYQQAKELQIT--IEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKN 580
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT-------SGDLSNRKVVNNTSNNSSI 128
ELV+ + L +K K++ + ++EKL L+ E + ++ ++ V + +S
Sbjct: 581 ELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTSK 640
Query: 129 RMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTD 183
K ++D L+ +A + E + T S+ + L H E +K S +
Sbjct: 641 LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQE 695
>UniRef100_UPI00002C5C15 UPI00002C5C15 UniRef100 entry
Length = 555
Score = 45.8 bits (107), Expect = 0.001
Identities = 52/213 (24%), Positives = 95/213 (44%), Gaps = 16/213 (7%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHK-EQEL-KFLKFVSAAEDVIDHLRTENDKLLGQ 58
++ +Y +YSK LK + E V ++E+ + + +E ++ LR +KL +
Sbjct: 206 LQEVYADIYSKNQNLKKESIKRSERVKTIDKEIDSWRNLLKNSEKMVQELRERKNKLNFK 265
Query: 59 INDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE------GTSGD 112
+L + S + K ++ E+ RL EK +NE L EE++K L+E +S +
Sbjct: 266 FAELEKQPQS-QAEKKGKISENLRLSESEKNENETLIEEIDKKINDLREELNKVKESSIE 324
Query: 113 LSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALF 172
+ RK S+ ++I KR + +D E +EN ++ D + A+
Sbjct: 325 IRERK----ASSGATIEGLNKRRNDLLERIDTELNL--NENNILEYSNLDKEEEFPDAVS 378
Query: 173 -EYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSI 204
E LD K + G + L A +++ Y I
Sbjct: 379 QEEMLDEKKREREKLGSVNLRADEETTKYETEI 411
>UniRef100_Q7RN75 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 2095
Score = 45.8 bits (107), Expect = 0.001
Identities = 37/153 (24%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query: 16 KTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDN 75
+T ++ E+E V K +E+K +K V E+V ++E K + ++ ++ +V +
Sbjct: 1046 ETEEVKEVEKVEKSEEIKEVKKVEKVEEVEKVEKSEEAKEVDEVKEVKK---VEKVEEVK 1102
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRM 135
E+VE K + EK +N +E+EK++++ K ++ K + N I+ K
Sbjct: 1103 EIVEIKEV---EKVENSEEVKEIEKVKEVEKGEEIKEMEKVKEIEEVEKNEEIKEVEKVE 1159
Query: 136 RQE---QDALDIEARCIPSENEGVDTQESDHQN 165
+ E +D L ENE + +E +H +
Sbjct: 1160 KGEEVKEDKLTEPQIKQAGENETKEIKEGNHND 1192
Score = 37.7 bits (86), Expect = 0.30
Identities = 24/109 (22%), Positives = 42/109 (38%), Gaps = 12/109 (11%)
Query: 24 EDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRL 83
E + +E+E + +D ++H+ + DK +IN+L ++ + +N
Sbjct: 1998 EKMSEEKEFTLSSVIDNIKDPLEHIEFKEDKEKSEINELSGDIFVDNIESEN-------- 2049
Query: 84 LLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTR 132
K EK + L+ EG S SN K N N R +
Sbjct: 2050 ----KNNENPQDTNFEKTETLISEGRSSYASNTKNYKNNKNKKKKRKNK 2094
Score = 35.8 bits (81), Expect = 1.1
Identities = 26/95 (27%), Positives = 50/95 (52%), Gaps = 13/95 (13%)
Query: 16 KTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDL-------GNELTS 68
K ++ E+E + K +E+K ++ V E+V + T+ + + + ++ G E+
Sbjct: 970 KNEEIKEVEKIEKGEEIKEMEKVKEVEEVETNEETKEIERVKETEEVKEVEVEKGGEVKE 1029
Query: 69 VRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQK 103
V V K EL E +E+ K+ E + +EVEK++K
Sbjct: 1030 VEVEKGGELKE-----IEKVKETEEV-KEVEKVEK 1058
>UniRef100_P51834 Chromosome partition protein smc [Bacillus subtilis]
Length = 1186
Score = 45.8 bits (107), Expect = 0.001
Identities = 45/166 (27%), Positives = 87/166 (52%), Gaps = 23/166 (13%)
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNE 65
+KL+ K++TLK E + KE+EL +SA E I+ R + L +N+L
Sbjct: 235 EKLHGKWSTLK-----EKVQMAKEEELAESSAISAKEAKIEDTRDKIQALDESVNELQQV 289
Query: 66 LTSVRVAKDNELVEHKRLLLEEKKKN-----EALFEEVEKLQK---LLKEGTSGDLSNRK 117
L + +++ E +E ++ +L+E+KKN E L E + + Q+ +LKE +LS ++
Sbjct: 290 L--LVTSEELEKLEGRKEVLKERKKNAVQNQEQLEEAIVQFQQKETVLKE----ELSKQE 343
Query: 118 VVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDH 163
V T + ++ R +++++Q AL + + E ++ +SD+
Sbjct: 344 AVFETL-QAEVKQLRAQVKEKQQALSLHNENV---EEKIEQLKSDY 385
>UniRef100_O30500 YttA [Bacillus subtilis]
Length = 248
Score = 45.4 bits (106), Expect = 0.001
Identities = 42/132 (31%), Positives = 67/132 (49%), Gaps = 15/132 (11%)
Query: 2 EALYKKLYSKYTTLKTNKLSELEDVHKE--QELKFLKFVSAAEDVIDHLRTENDKLLGQI 59
E Y L ++Y L T + ELE +K E K LK +D ++ L+ EN
Sbjct: 68 EKKYSALNAEYKNL-TKEHEELEKEYKSVSSEAKKLKDNKEDQDKLEKLKNEN------- 119
Query: 60 NDLGNELTSVRVAKDNELVEHKRLLLEEKK----KNEALFEEVEKLQKLLKEGTSGDLSN 115
+DL S++ A+ EL E+++ L E+ K +NE L ++ KL+ LKE S S+
Sbjct: 120 SDLKKTQKSLK-AEIKELQENQKQLKEDAKTAKAENETLRQDKTKLENQLKETESQTASS 178
Query: 116 RKVVNNTSNNSS 127
+ ++SNN+S
Sbjct: 179 HEDTGSSSNNTS 190
>UniRef100_Q6ZRK6 Hypothetical protein FLJ46282 [Homo sapiens]
Length = 822
Score = 45.4 bits (106), Expect = 0.001
Identities = 53/196 (27%), Positives = 82/196 (41%), Gaps = 13/196 (6%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEA K L TL+ + + E V KE E KFL + E + + ++L G+IN
Sbjct: 44 MEAELKVLKENNQTLERDNELQREKV-KENEEKFLNLQNEHEKALGTWKRHAEELNGEIN 102
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK-EGTSGDLSNRKVV 119
+ NEL+S++ +L EH L N+ FEE +K Q + + + ++S K
Sbjct: 103 KIKNELSSLK-ETHIKLQEHYNKLC-----NQKTFEEDKKFQNVPEVNNENSEMSTEKSE 156
Query: 120 NNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQE---SDHQNWLVHALFEYTL 176
N + + E D E R + EG +E D Q L F+ +
Sbjct: 157 NTIIQKYNTEQEIREENMENFCSDTEYREKEEKKEGSFIEEIIIDDLQ--LFEKSFKNEI 214
Query: 177 DMKLSTDCQTGRLCLS 192
D +S D + LS
Sbjct: 215 DTVVSQDENQSEISLS 230
>UniRef100_UPI00004983CC UPI00004983CC UniRef100 entry
Length = 605
Score = 45.1 bits (105), Expect = 0.002
Identities = 40/160 (25%), Positives = 68/160 (42%), Gaps = 13/160 (8%)
Query: 9 YSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTS 68
Y T NK+ ELE+V+ K K V+ E I+ + EN++ N+L L
Sbjct: 233 YQGKITEYRNKIGELEEVNG----KLTKKVNGMERKIEKMEKENEQNQANTNELIYNLKE 288
Query: 69 VRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRK---VVNNTSNN 125
KDN ++ K L +K E L E+ K++ KE + D+ K +
Sbjct: 289 DIKTKDNIIIGLKTDLNNTDEKIEGLKSEINKMKSAKKENKTDDIFEIKKEHQIQVEEMK 348
Query: 126 SSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQN 165
I K M+++ D +CI E + +E++ ++
Sbjct: 349 KQIEERDKGMKEKND------KCIELMKENIQMKENNEKH 382
>UniRef100_UPI000025E246 UPI000025E246 UniRef100 entry
Length = 229
Score = 45.1 bits (105), Expect = 0.002
Identities = 37/126 (29%), Positives = 67/126 (52%), Gaps = 9/126 (7%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
+E+++ K+ + L ++ED + ELK +K + + D I+ L+ + +KLL +
Sbjct: 56 IESVHLKIKNLEERLNEKLELKIEDFQEMTELKNIKVLDLSSDHIEALQIKLNKLLNERE 115
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQK----LLKEGTSG-DLSN 115
++G +V + + E+ E + LLE K + + L + +EKL+K L KEG + S
Sbjct: 116 NIG----AVNLRAEIEIDESNKKLLEMKSECDDLNKAIEKLKKAITELNKEGRERLETSF 171
Query: 116 RKVVNN 121
KV NN
Sbjct: 172 NKVNNN 177
>UniRef100_UPI000045363A UPI000045363A UniRef100 entry
Length = 893
Score = 44.7 bits (104), Expect = 0.002
Identities = 49/167 (29%), Positives = 81/167 (48%), Gaps = 37/167 (22%)
Query: 5 YKKLYSKYTTL-------KTNK------LSELEDVHKEQEL------KFLKFVSAAEDVI 45
Y KLY KYT L K N+ L EL+D++K+ ++ + K +SA E V+
Sbjct: 511 YNKLYQKYTALLSEYTASKKNEENLNASLVELQDINKKLDIEGRDMKEKYKTLSATERVL 570
Query: 46 DHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKK---KNEAL--FEEVEK 100
+ L+ N +L+G+I+ L E ++ D +++E K EE KN+ L E +K
Sbjct: 571 EQLQESNAELIGEISALKEERDI--LSADYKILEKKYKTSEESMHEIKNQLLDFTLENQK 628
Query: 101 LQKLLKEGTSGDLSNRK---------VVNNTSNNSSIRMTRKRMRQE 138
+KL+K+ + N + +V N NS RM+ K + +E
Sbjct: 629 FKKLIKQTIIPLIPNAEYNSTIGHILMVMNAKENS--RMSTKVLEEE 673
>UniRef100_UPI000034C4FB UPI000034C4FB UniRef100 entry
Length = 189
Score = 44.3 bits (103), Expect = 0.003
Identities = 38/143 (26%), Positives = 78/143 (53%), Gaps = 12/143 (8%)
Query: 10 SKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSV 69
SK +L +N+L +L D KE+E + +S E+ + ++ E L + DL E T+
Sbjct: 33 SKVNSL-SNELDQLNDQLKEKESDYRNNISYLENELRIIKKE----LEETEDLYEEATNE 87
Query: 70 RVAKDNELV---EHKRLLLEE-KKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN-NTSN 124
+++ +N+L+ + R+ L E K++NE+L +E+ L + + T + N+++ N + S
Sbjct: 88 QISSNNQLINSNDELRISLNELKEENESLVDEITVLTEKYDDET--ESLNQQIYNISKSL 145
Query: 125 NSSIRMTRKRMRQEQDALDIEAR 147
N+ + + +RQ + A I+ +
Sbjct: 146 NAQLAIKNNEIRQLKSARVIQKK 168
>UniRef100_UPI000033DA4D UPI000033DA4D UniRef100 entry
Length = 308
Score = 44.3 bits (103), Expect = 0.003
Identities = 49/211 (23%), Positives = 96/211 (45%), Gaps = 12/211 (5%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHK-EQELKFLK-FVSAAEDVIDHLRTENDKLLGQ 58
++ Y YSK ++K + E + E E++ K ++ +E +++ L +KL Q
Sbjct: 98 LQEKYADAYSKNQSIKNESIKRNERIKTIETEIESWKNLLTNSEKMVNELTQRKEKLSKQ 157
Query: 59 INDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEE----VEKLQKLLKEGTSGDLS 114
++DL N+ + + ++ E+ R+ +EK NEA+ +E +E L+ L E +
Sbjct: 158 LDDLENQ-PRAQAERKGQISENLRISDKEKVDNEAIIDETDQKIEMLRTQLNEIQEQSIQ 216
Query: 115 NRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHAL-FE 173
R+ S+ ++I +KR D ++ E +EN ++ + L + + E
Sbjct: 217 IRE--RKASSGATIEGLQKRKNDLLDRINSELNL--NENNILENSNLNGVEELPNPIDQE 272
Query: 174 YTLDMKLSTDCQTGRLCLSAMHQSSGYSFSI 204
LD K + G + L A ++S Y I
Sbjct: 273 DALDKKKQEREKLGSVNLKADEETSKYEEEI 303
>UniRef100_UPI0000221447 UPI0000221447 UniRef100 entry
Length = 306
Score = 44.3 bits (103), Expect = 0.003
Identities = 40/181 (22%), Positives = 82/181 (45%), Gaps = 28/181 (15%)
Query: 4 LYKKLYSKYTTLKTNKLSELED---VHKEQELKFLKFVSAAEDVIDHLR----TENDKLL 56
+ KKL+ Y + L ELED + KEQE K + ++ + D +R EN+K+
Sbjct: 52 IQKKLFGNYVAVHERTLLELEDSRKIQKEQEEK-INIITEENEKFDEIRGKFNEENEKIR 110
Query: 57 GQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKL-------LKEGT 109
++ D LTS + ++ E++ +L+ + + A E+ ++QK + EGT
Sbjct: 111 EELEDFEENLTS----QKSQEQENREILIAKLTEEFAECEKEREIQKKQFEDYVDVHEGT 166
Query: 110 SGDLSNRKVVNN---------TSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQE 160
+L + + + T N R+++ +E + L E + + + E ++ +
Sbjct: 167 LLELEDSRKIQKEQKEKINILTEENEKFSDIRQKLNEENEKLREELQELKLKLESIEENK 226
Query: 161 S 161
+
Sbjct: 227 T 227
>UniRef100_Q678H6 Membrane-bound metallopeptidase [Lymphocystis disease virus -
isolate China]
Length = 454
Score = 44.3 bits (103), Expect = 0.003
Identities = 48/185 (25%), Positives = 78/185 (41%), Gaps = 11/185 (5%)
Query: 15 LKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKD 74
L+ + S LE+ KEQ F+K + + VI+ L+ +N++ L Q D E T + K
Sbjct: 152 LRKSLTSRLEEERKEQNNNFVKVLQEKDAVIESLKKQNEEELKQDVDRLKE-TEDTLLKQ 210
Query: 75 NELV---EHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMT 131
EL+ E R+ EE + + +EVE + +KE + +L K TS +
Sbjct: 211 AELIKDAEDSRVRREEALRKQVQRKEVE--AEKIKEKYTSNLERLKEKEKTSKIVLLDQL 268
Query: 132 RKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCL 191
+K R+ D E + EN + D +L+ T D KL +
Sbjct: 269 KKDRRKPGKLTDDEVKVKVYENPYFERMFKDAVAYLLE-----TEDPKLHRSITIKEIIQ 323
Query: 192 SAMHQ 196
+ HQ
Sbjct: 324 TVAHQ 328
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 420,997,595
Number of Sequences: 2790947
Number of extensions: 17228700
Number of successful extensions: 71534
Number of sequences better than 10.0: 1533
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 1474
Number of HSP's that attempted gapping in prelim test: 69496
Number of HSP's gapped (non-prelim): 3300
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC130807.11


