

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC129092.1 - phase: 0
(391 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
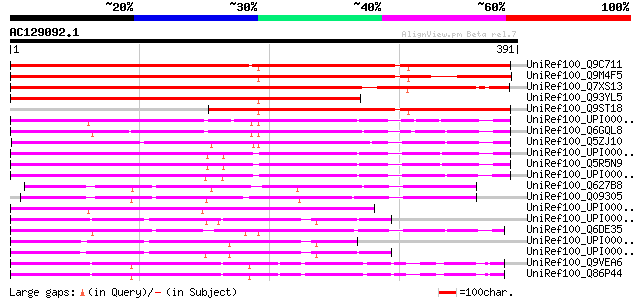
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C711 Hypothetical protein F28G11.5 [Arabidopsis thal... 548 e-155
UniRef100_Q9M4F5 CePP protein [Brassica napus var. napus] 490 e-137
UniRef100_Q7XS13 OSJNBa0095H06.5 protein [Oryza sativa] 430 e-119
UniRef100_Q93YL5 Hypothetical protein cePP [Brassica napus] 377 e-103
UniRef100_Q9ST18 S-locus protein 3 [Brassica campestris] 314 2e-84
UniRef100_UPI0000360221 UPI0000360221 UniRef100 entry 171 3e-41
UniRef100_Q6GQL8 Zgc:92018 [Brachydanio rerio] 166 1e-39
UniRef100_Q5ZJ10 Hypothetical protein [Gallus gallus] 160 6e-38
UniRef100_UPI000036C43D UPI000036C43D UniRef100 entry 151 3e-35
UniRef100_Q5R5N9 Hypothetical protein DKFZp469P0814 [Pongo pygma... 150 5e-35
UniRef100_UPI0000180804 UPI0000180804 UniRef100 entry 150 6e-35
UniRef100_Q627B8 Hypothetical protein CBG00743 [Caenorhabditis b... 150 6e-35
UniRef100_Q09305 Hypothetical protein F10B5.2 in chromosome II [... 149 1e-34
UniRef100_UPI000027CE83 UPI000027CE83 UniRef100 entry 146 1e-33
UniRef100_UPI00004337FB UPI00004337FB UniRef100 entry 145 2e-33
UniRef100_Q6DE35 MGC80226 protein [Xenopus laevis] 141 4e-32
UniRef100_UPI00004337FE UPI00004337FE UniRef100 entry 137 4e-31
UniRef100_UPI00004337FD UPI00004337FD UniRef100 entry 137 4e-31
UniRef100_Q9VEA6 CG12320-PA [Drosophila melanogaster] 129 1e-28
UniRef100_Q86P44 SD27779p [Drosophila melanogaster] 128 2e-28
>UniRef100_Q9C711 Hypothetical protein F28G11.5 [Arabidopsis thaliana]
Length = 399
Score = 548 bits (1413), Expect = e-155
Identities = 267/393 (67%), Positives = 330/393 (83%), Gaps = 11/393 (2%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MDS+ ALELVK+G TLLFLDVPQYTLV IDTQ+F+VGP FKGIKMIPPG HFV+YSSSTR
Sbjct: 1 MDSEKALELVKHGATLLFLDVPQYTLVGIDTQIFAVGPAFKGIKMIPPGIHFVFYSSSTR 60
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
DG+EFSP IGFF+D PS+VIVRKW+QQ+E L KVSEEE+ERY AV+++EFD+ LGPYN
Sbjct: 61 DGREFSPTIGFFVDVAPSQVIVRKWNQQDEWLTKVSEEEEERYSQAVRSLEFDKNLGPYN 120
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNSA 180
L Y +W+ LS++ITK ++E+ EP+GGE++V E+ + + PKT ME ALDTQ+K
Sbjct: 121 LKQYGEWRHLSNYITKDVVEKFEPVGGEITVTYESAILKGGPKTAMEIALDTQMKKSKFT 180
Query: 181 TSVGKLQRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDLLLG 238
TS + Q KG YYTSIPR++K KG+SGQELTS+NLDKTQLLE++L K+Y SEDLLLG
Sbjct: 181 TSSTE-QPKGNRFYYTSIPRIIKHKGMSGQELTSMNLDKTQLLESVLSKEYKDSEDLLLG 239
Query: 239 ELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQK 298
ELQF+F+A +MGQSLE+F+QWKS+VSLLLGCT APF TR++LFTKFIKVIY+QLKYGLQ
Sbjct: 240 ELQFSFVAFLMGQSLESFMQWKSIVSLLLGCTSAPFQTRSQLFTKFIKVIYHQLKYGLQ- 298
Query: 299 DRKDNTGP-----LLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLE 353
K+N+GP LLDDSW ++DSFLH CK+FF+LV + SV+DGDLL WTRKFK+LLE
Sbjct: 299 --KENSGPETGIHALLDDSWLASDSFLHFLCKDFFALVEETSVVDGDLLSWTRKFKELLE 356
Query: 354 SNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
+ LGWEFQ+ +AVDG+YF+E+DE+APVVEMLD+
Sbjct: 357 NRLGWEFQKKSAVDGIYFEEDDEYAPVVEMLDE 389
>UniRef100_Q9M4F5 CePP protein [Brassica napus var. napus]
Length = 441
Score = 490 bits (1261), Expect = e-137
Identities = 256/396 (64%), Positives = 309/396 (77%), Gaps = 31/396 (7%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MDS+ ALELVK+G TLLFLDVPQ+TL+ IDTQ+F+VGP FKGIKMIPPG HFV+YSSSTR
Sbjct: 43 MDSEKALELVKHGATLLFLDVPQHTLIGIDTQMFTVGPAFKGIKMIPPGIHFVFYSSSTR 102
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
DGKEFSP IGFFID PS+VIVRKW+QQ+E L KVSEEE+ERY AVK++EFD+ LGPY+
Sbjct: 103 DGKEFSPTIGFFIDVTPSQVIVRKWNQQDEWLAKVSEEEEERYSQAVKSLEFDKHLGPYD 162
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNS- 179
LS Y WK LS++ITK +IE+ EP+GGE++V E+ + + KT ME+ALD Q+K + S
Sbjct: 163 LSQYGAWKHLSNYITKDVIEKFEPVGGEITVIYESAILKGGHKTEMERALDAQMKKNKSE 222
Query: 180 ATSVGKLQRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDLLL 237
A+S Q KG YYTSIPR++K KGISGQELTSLNLDKTQLLE++L K+Y SEDLLL
Sbjct: 223 ASSSSTEQPKGNRFYYTSIPRIIKHKGISGQELTSLNLDKTQLLESVLSKEYKASEDLLL 282
Query: 238 GELQFAFIA-LMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGL 296
GELQF+F+A L+MGQSLE+F+QWKSLVSLLLGCTEAPF TR+ LFTKFIKVIY+QLKYGL
Sbjct: 283 GELQFSFVAFLVMGQSLESFMQWKSLVSLLLGCTEAPFQTRSELFTKFIKVIYHQLKYGL 342
Query: 297 QKDRKDNTGP-----LLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKL 351
Q K+++GP LLDDSW ++DSFLH CK TRKFK+L
Sbjct: 343 Q---KESSGPEMGVLALLDDSWLASDSFLHLLCK-------------------TRKFKEL 380
Query: 352 LESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDDE 387
LES LGWEFQ+ + VDG+YFDE+DE + +D E
Sbjct: 381 LESRLGWEFQKKSDVDGIYFDEDDETSSYSSYIDVE 416
>UniRef100_Q7XS13 OSJNBa0095H06.5 protein [Oryza sativa]
Length = 390
Score = 430 bits (1105), Expect = e-119
Identities = 221/392 (56%), Positives = 281/392 (71%), Gaps = 21/392 (5%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
+D + A ELV+ G TLL LDVPQ TL+ +DTQVFSVGP FKGIKM+PPG HF+YY S R
Sbjct: 11 VDPEAATELVRKGATLLLLDVPQRTLLGVDTQVFSVGPKFKGIKMVPPGPHFLYYCSPNR 70
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
EF+P +GFF+ PSEVIVRKW QEERL+K+ EEE+ RY AV++ EFD QLGPYN
Sbjct: 71 HANEFAPTVGFFLTTHPSEVIVRKWHAQEERLIKLPEEEEIRYSEAVRHFEFDSQLGPYN 130
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNSA 180
L + DWK+LS ++ +S+IERLEPIGGE+++ E+ AP+T ME+ L QLK A
Sbjct: 131 LDSFGDWKQLSSYLPQSVIERLEPIGGEITIAWESSWMDKAPQTDMERRLMDQLKDGKFA 190
Query: 181 TSVG-KLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDLLLGE 239
+ + +R+GCYYT+IP +K ISG ELT+LNLDKT LLE++L K+Y G EDLLLGE
Sbjct: 191 KNAPVQSERRGCYYTTIPASIKHSNISGDELTALNLDKTCLLESVLAKNYQGQEDLLLGE 250
Query: 240 LQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYGLQKD 299
LQFAFIA MMGQSLEAF+QWK+LVSLLL C+EA FI+ IYYQLK+G Q
Sbjct: 251 LQFAFIAFMMGQSLEAFMQWKALVSLLLSCSEA-----------FIRAIYYQLKHGFQHT 299
Query: 300 RKDNTG-----PLLLDDSWFSTDSFLHHHCK-EFFSLVLDGSVIDGDLLKWTRKFKKLLE 353
+ + +G L LD++WFS D FL+ K +FF+++L+ +V+DGDLL WTRK K LLE
Sbjct: 300 QDNRSGEEMGNSLFLDEAWFSRDIFLYRLSKTDFFAVILEATVVDGDLLSWTRKLKSLLE 359
Query: 354 SNLGWEFQQNNAVDGLYFDENDEFAPVVEMLD 385
+ GW+ NN V+ DE+DEFAPVV +D
Sbjct: 360 TTFGWDL-DNNTVN--LIDEDDEFAPVVVEMD 388
>UniRef100_Q93YL5 Hypothetical protein cePP [Brassica napus]
Length = 315
Score = 377 bits (969), Expect = e-103
Identities = 189/273 (69%), Positives = 228/273 (83%), Gaps = 3/273 (1%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MDS+ ALELVK+G TLLFLDVPQ+TL+ IDTQ+F+VGP FKGIKMIPPG HFV+YSSSTR
Sbjct: 43 MDSEKALELVKHGATLLFLDVPQHTLIGIDTQMFTVGPAFKGIKMIPPGIHFVFYSSSTR 102
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
GKEFSP IGFFID PS+VIVRKW+QQ+E L KVSEEE+ERY AVK++EFD+ LGPY+
Sbjct: 103 GGKEFSPTIGFFIDVTPSQVIVRKWNQQDEWLAKVSEEEEERYSQAVKSLEFDKHLGPYD 162
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDNS- 179
LS Y WK LS++ITK +IE+ EP+GGE++V E+ + + KT ME+ALD Q+K + S
Sbjct: 163 LSQYGAWKHLSNYITKDVIEKFEPVGGEITVIYESAILKGGHKTEMERALDAQMKKNKSE 222
Query: 180 ATSVGKLQRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDLLL 237
A+S Q KG YYTSIPR++K KGISGQELTSLNLDKTQLLE++L K+Y SEDLLL
Sbjct: 223 ASSSSTEQPKGNRFYYTSIPRIIKHKGISGQELTSLNLDKTQLLESVLSKEYKASEDLLL 282
Query: 238 GELQFAFIALMMGQSLEAFLQWKSLVSLLLGCT 270
GELQF+F+A +MGQSLE+F+QWKSLVSLLLGCT
Sbjct: 283 GELQFSFVAFLMGQSLESFMQWKSLVSLLLGCT 315
>UniRef100_Q9ST18 S-locus protein 3 [Brassica campestris]
Length = 249
Score = 314 bits (805), Expect = 2e-84
Identities = 159/241 (65%), Positives = 197/241 (80%), Gaps = 11/241 (4%)
Query: 154 ENDMFRNAPKTPMEKALDTQLK-VDNSATSVGKLQRKG--CYYTSIPRVVKCKGISGQEL 210
E+ + + PK M++ALD Q+K + + A S Q KG YYTSIPR++K KGISGQEL
Sbjct: 4 ESAILKGGPKMEMDRALDEQMKKIKSGAASSTTEQPKGNRFYYTSIPRIIKHKGISGQEL 63
Query: 211 TSLNLDKTQLLETLLVKDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCT 270
TSLNLDKTQLLE++L K+Y SEDLLLGELQF+F+A +MGQSLE+F+QWKSLVSLLLGCT
Sbjct: 64 TSLNLDKTQLLESVLSKEYKDSEDLLLGELQFSFVAFLMGQSLESFMQWKSLVSLLLGCT 123
Query: 271 EAPFHTRTRLFTKFIKVIYYQLKYGLQKDRKDNTGP-----LLLDDSWFSTDSFLHHHCK 325
EAPF TR+ LFTKF+KVIY+QLKYGLQ K+++GP LLDDSW ++DSFLH CK
Sbjct: 124 EAPFQTRSELFTKFVKVIYHQLKYGLQ---KESSGPEMGVLALLDDSWLASDSFLHLLCK 180
Query: 326 EFFSLVLDGSVIDGDLLKWTRKFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLD 385
+FF+LV + SV+DGDLL WTRKFK+LLE+ LGWEFQ+ + VDG+YFDE+DE+APVVEMLD
Sbjct: 181 DFFALVEEASVVDGDLLSWTRKFKELLETRLGWEFQKKSDVDGIYFDEDDEYAPVVEMLD 240
Query: 386 D 386
+
Sbjct: 241 E 241
>UniRef100_UPI0000360221 UPI0000360221 UniRef100 entry
Length = 380
Score = 171 bits (433), Expect = 3e-41
Identities = 128/399 (32%), Positives = 185/399 (46%), Gaps = 38/399 (9%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSST- 59
MD A L + G L+ L VP+ TL+ ID + + VGP FKG+KMIPPG HF++Y S+
Sbjct: 5 MDPDVARRLFEEGAFLVLLGVPKGTLLGIDCKSWQVGPRFKGVKMIPPGLHFLHYCSANP 64
Query: 60 -RDGKEFSPMIGFFIDAGPSEVIVRKWDQQEERL-VKVSEEEDERYRLAVKNMEFDRQLG 117
G E P G F+ P E+++ WD + E L S+ E+E R+ + D LG
Sbjct: 65 PSHGGELGPKTGLFLSLKPREIVLANWDPKIEDLDFSASQNEEELNRIRSNLKDLDPFLG 124
Query: 118 PYNLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVD 177
PY W L+D + + L+P+ G V +D+ T E + Q + D
Sbjct: 125 PYPYEVMRKWVSLTDHLRCELANNLQPLSGRVC--AFSDVIPELQLTHTEDRAE-QPRND 181
Query: 178 NSATSVGK--------LQRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVK 227
+ S+ + QR+G ++ IP G S E+T ++D + LET+L K
Sbjct: 182 QACQSMKEGLDRLPKMKQREGTELRFSVIPEKTYPPGASPAEITKCSMDFSYALETVLEK 241
Query: 228 DYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKV 287
+Y LLGELQFAF+ ++G E F WK L+ LL +E R LF I V
Sbjct: 242 NYSLQPLNLLGELQFAFVCFLLGNVYEGFEHWKHLLGLLCR-SEESIQKRKELFLGLISV 300
Query: 288 IYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRK 347
+Y+QL P D S ++FL ++FF G +DG L K K
Sbjct: 301 LYHQL---------GEIPPDFFVDI-VSQNNFLTSTLQDFFQFA-SGPGVDGTLRKRAEK 349
Query: 348 FKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
FK L W+F+ + D+ APVV L +
Sbjct: 350 FKAHLTKKFNWDFEA----------DLDDCAPVVVELPE 378
>UniRef100_Q6GQL8 Zgc:92018 [Brachydanio rerio]
Length = 384
Score = 166 bits (420), Expect = 1e-39
Identities = 121/399 (30%), Positives = 190/399 (47%), Gaps = 38/399 (9%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A L + G TL+ L VP+ T + +D + +++GP F+G+KMIPPG HF++Y S +
Sbjct: 6 MDPEVARGLFEEGATLVLLGVPEGTELGLDYKTWTLGPRFRGVKMIPPGLHFLHYCSVST 65
Query: 61 DG--KEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGP 118
+G E SP G F+ P +V+V W++ + L ++S+ +E R+ E DR LGP
Sbjct: 66 NGGCGEISPKTGLFLSLKPRQVLVAHWNKSADDL-EISQNSEEVERVRANLKELDRYLGP 124
Query: 119 YNLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDN 178
Y W L+D +++ + L+P+ G V +D+ T + + L ++
Sbjct: 125 YPYDTLRKWVSLTDRLSQEVAVALQPLSGRVC--AFSDVIPELQLTHTKDRVQQNLSRND 182
Query: 179 SATSVGK---------LQRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVK 227
K QR+G ++SIP+ G + ++T ++D++ L +L +
Sbjct: 183 QECQSMKEGLDRLPRMKQREGTELRFSSIPKQAYPPGATPAQITQYSMDRSYTLNAVLER 242
Query: 228 DYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKV 287
Y +LGELQFAF+ ++G EAF WKSL++LL EA R L+ I V
Sbjct: 243 HYKEQPLNVLGELQFAFVCFLVGNVYEAFEHWKSLLALLCRSEEA-MKDRKELYLGLIAV 301
Query: 288 IYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRK 347
+Y+QL + LD S +FL ++FF V D L K K
Sbjct: 302 LYHQL--------GEIPPDFFLD--IVSQSNFLTSTLQDFFQFASSPGV-DSTLRKRAEK 350
Query: 348 FKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
FK L W+F + D+ APVV L D
Sbjct: 351 FKVHLTKKFRWDFDA----------DLDDCAPVVVELPD 379
>UniRef100_Q5ZJ10 Hypothetical protein [Gallus gallus]
Length = 382
Score = 160 bits (405), Expect = 6e-38
Identities = 125/395 (31%), Positives = 183/395 (45%), Gaps = 34/395 (8%)
Query: 2 DSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSS-TR 60
D + A L G ++ L VP+ T ID ++VGP F+G+KM+PPG HFV+ S++ T
Sbjct: 7 DPELAKRLFFEGAAVVVLGVPEGTEFGIDYNSWTVGPRFRGVKMVPPGVHFVHCSAARTA 66
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPYN 120
G E P G F+ P +V +W+ E L + + E+E L E D LGPY
Sbjct: 67 GGTETGPRAGLFLCLRPRQVRALRWEPGSEELREAAPGEEE--ALGRDLREMDAFLGPYP 124
Query: 121 LSHYEDWKRLSDFITKSIIERLEPIGGEVSVECE---NDMFRNAPKTPMEKALDTQLKVD 177
+ W LS F+ ++ +ERL+P G++S E R+ +K
Sbjct: 125 YETLKKWISLSSFVGEAAVERLQPESGQISAFSEVLPVRAGRHGRDRAGQKLPPYDAGCG 184
Query: 178 NSATSVGKL----QRKG--CYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGG 231
+ A V +L R G +T +PR G S +E+T ++D + LE ++ + Y G
Sbjct: 185 SYAEGVARLPEMRPRAGTEIRFTELPRQTYPDGASPEEITRHSMDLSYALEKVIARRYAG 244
Query: 232 SEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQ 291
LL ELQFAFI ++G +AF WK L+++L +A R L+ I V+Y+Q
Sbjct: 245 RPQDLLAELQFAFICFLIGNVYDAFEHWKRLLNILCRSEDAIAKYRD-LYANLIFVLYHQ 303
Query: 292 LKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKL 351
L N P S D+FL + FFS + +V DG L K KFK
Sbjct: 304 L----------NEIPSDFFVDIVSQDNFLTSTLQVFFSYTCNAAV-DGALRKKAEKFKAH 352
Query: 352 LESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
L W+F E D+ APVV L +
Sbjct: 353 LTKKFKWDFDA----------EPDDCAPVVVELPE 377
>UniRef100_UPI000036C43D UPI000036C43D UniRef100 entry
Length = 384
Score = 151 bits (382), Expect = 3e-35
Identities = 113/400 (28%), Positives = 185/400 (46%), Gaps = 40/400 (10%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A L G T++ L++P+ T ID + VGP F+G+KMIPPG HF++YSS +
Sbjct: 6 MDPELAKRLFFEGATVVILNMPKGTEFGIDYNSWEVGPKFRGVKMIPPGIHFLHYSSVDK 65
Query: 61 -DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPY 119
+ KE P +GFF+ + V +W E + E E + E D+ LGPY
Sbjct: 66 ANPKEVGPRMGFFLSLHQRGLTVLRWSTLREEVDLSPAPESEVEAMRANLQELDQFLGPY 125
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEVSV-----------ECENDMFRNAPK--TPM 166
+ + W L++FI+++ +E+L+P ++ ++ + +N P+ T
Sbjct: 126 PYATLKKWISLTNFISEATVEKLQPENRQICAFSDVLPVLSMKHTKDRVGQNLPRCGTEC 185
Query: 167 EKALDTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLV 226
+ + ++ G R ++ +P + +G + E+T ++D + LET+L
Sbjct: 186 KSYQEGLARLPEMKPRAGTEIR----FSELPTQMFPEGATPAEITKHSMDLSYALETVLN 241
Query: 227 KDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIK 286
K + S +LGELQFAF+ ++G EAF WK L++LL +EA L+ I
Sbjct: 242 KQFPSSPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SEAAMMKHHTLYINLIS 300
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
++Y+QL P S D+FL + FFS +D L K
Sbjct: 301 ILYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSAC-SIAVDATLRKKAE 349
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
KF+ L W+F E ++ APVV L +
Sbjct: 350 KFQAHLTKKFRWDFAA----------EPEDCAPVVVELPE 379
>UniRef100_Q5R5N9 Hypothetical protein DKFZp469P0814 [Pongo pygmaeus]
Length = 384
Score = 150 bits (380), Expect = 5e-35
Identities = 113/400 (28%), Positives = 184/400 (45%), Gaps = 40/400 (10%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A L G T++ L++P+ T ID + VGP F+G+K IPPG HF++YSS +
Sbjct: 6 MDPELAKRLFFEGATVVILNMPRGTEFGIDYNSWEVGPKFRGVKTIPPGIHFLHYSSVDK 65
Query: 61 -DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPY 119
+ KE P +GFF+ + V +W E + E E + E D+ LGPY
Sbjct: 66 ANPKEVGPRMGFFLSLHQRGLTVLRWSTLREEVDLSPAPESEVEAMRANLQELDQFLGPY 125
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEVSV-----------ECENDMFRNAPK--TPM 166
+ + W L++FI+++ +E+L+P ++ ++ M +N P+ T
Sbjct: 126 PYATLKKWISLTNFISEATVEKLQPENRQICAFSDVLPVLSMKHTKDRMGQNLPRCGTEC 185
Query: 167 EKALDTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLV 226
+ + ++ G R ++ +P + +G + E+T ++D + LET+L
Sbjct: 186 KSYQEGLARLPEMKPRAGTEIR----FSELPTQMFPEGATPAEITKHSMDLSYALETVLN 241
Query: 227 KDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIK 286
K + S +LGELQFAF+ ++G EAF WK L++LL +EA L+ I
Sbjct: 242 KQFPSSPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SEAAMMKHHTLYINLIS 300
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
++Y+QL P S D+FL + FFS +D L K
Sbjct: 301 ILYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSAC-SIAVDATLRKKAE 349
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
KF+ L W+F E ++ APVV L +
Sbjct: 350 KFQAHLTKKFRWDFAA----------EPEDCAPVVVELPE 379
>UniRef100_UPI0000180804 UPI0000180804 UniRef100 entry
Length = 384
Score = 150 bits (379), Expect = 6e-35
Identities = 115/400 (28%), Positives = 187/400 (46%), Gaps = 40/400 (10%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A +L G T++ L++P+ T ID + VGP F+G+KMIPPG HF+YYSS +
Sbjct: 6 MDPELAKQLFFEGATVVILNMPKGTEFGIDYNSWEVGPKFRGVKMIPPGVHFLYYSSVDK 65
Query: 61 -DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGPY 119
+ +E P +GFF+ E+ V +W+ +E + E E + E D+ LGPY
Sbjct: 66 ANPREVGPRMGFFLSLKQRELTVLRWNAVQEEVDLSPAPEAEVEAMRANLPELDQFLGPY 125
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEV----------SVECEND-MFRNAP--KTPM 166
+ W L++FI+++ +E+L+P ++ S+ D + +N P T
Sbjct: 126 PYGTLKKWISLTNFISEATMEKLQPESRQICAFSDVLPVLSMRYTKDRVEQNLPLCGTEC 185
Query: 167 EKALDTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLV 226
+ ++ G R ++ +P + G + E+T ++D + LET+L
Sbjct: 186 RSYQEGLARLPEMKPRAGTEIR----FSELPTQMFPAGATPAEITRHSMDLSYALETVLS 241
Query: 227 KDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIK 286
K + G+ +LGELQFAF+ ++G EAF WK L++LL +E L+ I
Sbjct: 242 KQFPGNPQDVLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCR-SETAMVKHHALYVSLIS 300
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
++Y+QL P S D+FL + FFS +V + L K
Sbjct: 301 ILYHQL----------GEIPADFFVDIVSQDNFLTSTLQVFFSSACSVAV-EATLRKKAE 349
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVVEMLDD 386
KF+ L W+F E ++ APVV L +
Sbjct: 350 KFQAHLTKKFRWDFT----------SEPEDCAPVVVELPE 379
>UniRef100_Q627B8 Hypothetical protein CBG00743 [Caenorhabditis briggsae]
Length = 357
Score = 150 bits (379), Expect = 6e-35
Identities = 111/366 (30%), Positives = 170/366 (46%), Gaps = 44/366 (12%)
Query: 12 NGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTRDGKEFSPMIGF 71
NG LLFL PQ + ID + + G F G+KMIPPG HFVY S + +P IGF
Sbjct: 16 NGAFLLFLGFPQASEFGIDYKTWKTGERFMGLKMIPPGVHFVYCSIKS------APRIGF 69
Query: 72 FIDAGPSEVIVRKWDQQEERLV--KVSEEEDERYRLAVKNMEFDRQLGPYNLSHYEDWKR 129
F + E++V+KW+++ E +V E+ + +K+M D L PY +Y W
Sbjct: 70 FHNFKAGEIVVKKWNKEAETFEDGEVPAEQIPEKKRQLKSM--DNSLAPYPYENYRSWYG 127
Query: 130 LSDFITKSIIERLEPIGGEVSVECE---------NDMFRNAPKTPMEKALDTQLKVDNSA 180
L+DFIT +ER+ P+ G ++ + E D R +T + +D Q V
Sbjct: 128 LTDFITPETVERVHPVLGRITSQAELVSLETQFMEDAEREHKETHFKNRVDRQNPVRTRF 187
Query: 181 TSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQL------LETLLVKDYGGSED 234
L I ++ + I QE+ +L + K Q+ +L++ GG
Sbjct: 188 VDQHGL--------PIMKIREGYEIRFQEIPALTVTKHQVGIDYSDRLYVLLRGLGGDWK 239
Query: 235 LLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKY 294
LL E+Q AF+ + GQ E F QWK ++ L+ C+ A + LF FI+V+++QLK
Sbjct: 240 QLLAEMQIAFVCFLQGQVFEGFEQWKRIIHLMSCCSNA-LGSEKELFMAFIRVLFFQLK- 297
Query: 295 GLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLES 354
P S D+FL F+ V D + +L K T +FK+ L +
Sbjct: 298 ---------ECPTDFFVDIVSRDNFLTTTLSMLFANVRDSAHGGAELKKKTAQFKQYLTN 348
Query: 355 NLGWEF 360
W+F
Sbjct: 349 QFKWDF 354
>UniRef100_Q09305 Hypothetical protein F10B5.2 in chromosome II [Caenorhabditis
elegans]
Length = 357
Score = 149 bits (377), Expect = 1e-34
Identities = 108/365 (29%), Positives = 171/365 (46%), Gaps = 36/365 (9%)
Query: 9 LVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTRDGKEFSPM 68
+ +NG LLFL PQ + ID + + G F G+KMIPPG HFVY S + +P
Sbjct: 13 MYRNGAFLLFLGFPQASEFGIDYKSWKTGEKFMGLKMIPPGVHFVYCSIKS------APR 66
Query: 69 IGFFIDAGPSEVIVRKWDQQEERL--VKVSEEEDERYRLAVKNMEFDRQLGPYNLSHYED 126
IGFF + E++V+KW+ + E +V ++ + +KNM D L PY +Y
Sbjct: 67 IGFFHNFKAGEILVKKWNTESETFEDEEVPTDQISEKKRQLKNM--DSSLAPYPYENYRS 124
Query: 127 WKRLSDFITKSIIERLEPIGGEVS-----VECENDMFRNAPKTPMEKALDTQLKVDNSAT 181
W L+DFIT +ER+ PI G ++ V E + NA K + ++ +N
Sbjct: 125 WYGLTDFITADTVERIHPILGRITSQAELVSLETEFMENAEKEHKDSHFRNRVDRENPV- 183
Query: 182 SVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQL-LE-----TLLVKDYGGSEDL 235
+ + + I ++ + I Q++ L + + ++ +E L++ GG
Sbjct: 184 ---RTRFTDQHGLPIMKIREGYEIRFQDIPPLTVSQNRVGIEYSDRLYRLLRALGGDWKQ 240
Query: 236 LLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIKVIYYQLKYG 295
LL E+Q AF+ + GQ E F QWK ++ L+ C + LF FI+V+++QLK
Sbjct: 241 LLAEMQIAFVCFLQGQVFEGFEQWKRIIH-LMSCCPNSLGSEKELFMSFIRVLFFQLK-- 297
Query: 296 LQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTRKFKKLLESN 355
P S D+FL F+ V D + DL K T +FK+ L +
Sbjct: 298 --------ECPTDFFVDIVSRDNFLTTTLSMLFANVRDSAHAGDDLKKKTAQFKQYLTNQ 349
Query: 356 LGWEF 360
W+F
Sbjct: 350 FKWDF 354
>UniRef100_UPI000027CE83 UPI000027CE83 UniRef100 entry
Length = 368
Score = 146 bits (368), Expect = 1e-33
Identities = 93/291 (31%), Positives = 144/291 (48%), Gaps = 10/291 (3%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSST- 59
MD A L + G L+ L VP+ T + ID + + VGP FKG+KMIPPG HF++Y S+
Sbjct: 10 MDPDVARRLFEEGAFLVLLGVPKGTELGIDCKSWQVGPRFKGVKMIPPGLHFLHYCSANP 69
Query: 60 -RDGKEFSPMIGFFIDAGPSEVIVRKWDQQEERL-VKVSEEEDERYRLAVKNMEFDRQLG 117
G E P G F+ P E+++ WD + E L S+ E+E R+ + D LG
Sbjct: 70 PSHGGELGPKTGLFLSLKPREILLANWDPKIEDLDFSASQNEEEVQRIRSNLKDLDPFLG 129
Query: 118 PYNLSHYEDWKRLSDFITKSIIERLEPIGG------EVSVECENDMFRNAPKTPM-EKAL 170
PY W L+D + + L+P+ G EV E + ++ + P ++A
Sbjct: 130 PYPYEGMRKWVSLTDHLKYELANNLQPLSGRVCAFSEVIPELQLRHTKDRAEQPRNDQAC 189
Query: 171 DTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYG 230
+ + + + + + +++IP G + E+T ++D + LET+L K+Y
Sbjct: 190 QSMKEGLDRLPKMKEREGTELRFSAIPERKYPPGATPAEITRCSMDFSYALETVLEKNYS 249
Query: 231 GSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLF 281
+LGELQFAF+ ++G E F WKSL+SLL E R LF
Sbjct: 250 LQPLNVLGELQFAFVCFLLGNVYEGFEHWKSLLSLLCRGPEESMQKRKELF 300
>UniRef100_UPI00004337FB UPI00004337FB UniRef100 entry
Length = 307
Score = 145 bits (365), Expect = 2e-33
Identities = 104/308 (33%), Positives = 154/308 (49%), Gaps = 27/308 (8%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD A L G TL+ L+VP T ID + ++ FKGIKMIPPG H+++YS+
Sbjct: 3 MDQTLAQRLFFEGATLVTLNVPSGTEFGIDLKSWNTADKFKGIKMIPPGFHYIHYSAIDE 62
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNM-EFDRQLGPY 119
G E P +GFF SE +V+ WD EE L +E ER + KNM + D+ LG Y
Sbjct: 63 FG-EAKPKVGFFHTFKKSEFLVKHWDTTEEDLSSEKQETVERLK---KNMKDLDKFLGCY 118
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEVS---VECENDMFR-----NAPKTPMEKALD 171
++ WK L++ IT S++ER PI G V+ +E +D R + PK +
Sbjct: 119 PYDIWKQWKDLTNHITPSLVERCSPICGYVNFTKLEYCSDATRPRGGESKPK-KRRSGIT 177
Query: 172 TQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGG 231
+ K D + + IP G S E+T +LD + +L+ +L
Sbjct: 178 VEEKEDELLPDMKPKPGTELRLSKIPNKQYPDGASPTEITKYSLDSSYVLDIIL------ 231
Query: 232 SEDL-----LLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFIK 286
DL ++GE+Q F+ ++GQSL+AF WK LVSL+ G + R ++ +F+K
Sbjct: 232 -NDLTLPIEIIGEIQLTFVCFLVGQSLDAFEHWKKLVSLICG-ADCAISQRRAIYVEFMK 289
Query: 287 VIYYQLKY 294
I QL +
Sbjct: 290 AIEIQLMH 297
>UniRef100_Q6DE35 MGC80226 protein [Xenopus laevis]
Length = 388
Score = 141 bits (355), Expect = 4e-32
Identities = 116/401 (28%), Positives = 182/401 (44%), Gaps = 53/401 (13%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD + A +L G TL+ L VP+ + ID + VGP F+G+KM+PPG HF++Y+ +
Sbjct: 1 MDPELARQLFFEGATLVILGVPEGSEFGIDYNSWQVGPRFRGVKMVPPGVHFMHYNVVGK 60
Query: 61 DG--KEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNMEFDRQLGP 118
G E SP F+ E+ + W+ QEE +V EE E+ R ++++ D LGP
Sbjct: 61 GGGLGEMSPRSSMFLYLQQRELRLFHWNPQEEEMVVAPHEEAEKLREELQSL--DSFLGP 118
Query: 119 YNLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDN 178
Y W LS+ I K + RL+P G + F P+E T +V +
Sbjct: 119 YPYESMRRWVSLSNHIEKETMFRLQPTCGTI--------FSFPEVLPLEAMTHTADRVQH 170
Query: 179 SA---TSVGKLQRKG--------------CYYTSIPRVVKCKGISGQELTSLNLDKTQLL 221
+ SV + ++G ++ IP + + E+T ++D + L
Sbjct: 171 NLPRYDSVCQSYKEGMARLPQMKQKEGTEIRFSKIPAKMYPDDATPTEITQHSMDLSYAL 230
Query: 222 ETLLVKDYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTR-TRL 280
E LL Y G LL ELQF+F+ ++G EAF QWKSL++LL C F + L
Sbjct: 231 EQLLKTHYTGQPLQLLAELQFSFVCFVLGNVYEAFEQWKSLLNLL--CRAETFSLQHPEL 288
Query: 281 FTKFIKVIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGD 340
+ K I V+Y+QL P S ++FL + FS + + +
Sbjct: 289 YIKVISVLYHQLA----------QVPTDFFVDIVSQNNFLTSTLQVLFSFLCSPNA-NPS 337
Query: 341 LLKWTRKFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVV 381
L K +F+ L W+F+ +E D+ AP++
Sbjct: 338 LRKKAIRFRAHLTKKFQWDFE----------EEPDDCAPLI 368
>UniRef100_UPI00004337FE UPI00004337FE UniRef100 entry
Length = 262
Score = 137 bits (346), Expect = 4e-31
Identities = 93/276 (33%), Positives = 137/276 (48%), Gaps = 22/276 (7%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD A L G TL+ L+VP T ID + ++ FKGIKMIPPG H+++Y R
Sbjct: 1 MDQTLAQRLFFEGATLVTLNVPSGTEFGIDLKSWNTADKFKGIKMIPPGFHYIHY----R 56
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNM-EFDRQLGPY 119
+ E P +GFF SE +V+ WD EE L +E ER + KNM + D+ LG Y
Sbjct: 57 EFGEAKPKVGFFHTFKKSEFLVKHWDTTEEDLSSEKQETVERLK---KNMKDLDKFLGCY 113
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEK--ALDTQLKVD 177
++ WK L++ IT S++ER PI E+ + R P ++ + + K D
Sbjct: 114 PYDIWKQWKDLTNHITPSLVERCSPISLELEYCSDATRPRGGESKPKKRRSGITVEEKED 173
Query: 178 NSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYGGSEDL-- 235
+ + IP G S E+T +LD + +L+ +L DL
Sbjct: 174 ELLPDMKPKPGTELRLSKIPNKQYPDGASPTEITKYSLDSSYVLDIIL-------NDLTL 226
Query: 236 ---LLGELQFAFIALMMGQSLEAFLQWKSLVSLLLG 268
++GE+Q F+ ++GQSL+AF WK LVSL+ G
Sbjct: 227 PIEIIGEIQLTFVCFLVGQSLDAFEHWKKLVSLICG 262
>UniRef100_UPI00004337FD UPI00004337FD UniRef100 entry
Length = 302
Score = 137 bits (346), Expect = 4e-31
Identities = 101/309 (32%), Positives = 149/309 (47%), Gaps = 30/309 (9%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD A L G TL+ L+VP T ID + ++ FKGIKMIPPG H++ T
Sbjct: 3 MDQTLAQRLFFEGATLVTLNVPSGTEFGIDLKSWNTADKFKGIKMIPPGFHYII----TD 58
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERLVKVSEEEDERYRLAVKNM-EFDRQLGPY 119
+ E P +GFF SE +V+ WD EE L +E ER + KNM + D+ LG Y
Sbjct: 59 EFGEAKPKVGFFHTFKKSEFLVKHWDTTEEDLSSEKQETVERLK---KNMKDLDKFLGCY 115
Query: 120 NLSHYEDWKRLSDFITKSIIERLEPIGGEV---SVECENDMFRNAPKTPMEK------AL 170
++ WK L++ IT S++ER PI G V ++E E P+ K +
Sbjct: 116 PYDIWKQWKDLTNHITPSLVERCSPICGYVNFTTLELEYCSDATRPRGGESKPKKRRSGI 175
Query: 171 DTQLKVDNSATSVGKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVKDYG 230
+ K D + + IP G S E+T +LD + +L+ +L
Sbjct: 176 TVEEKEDELLPDMKPKPGTELRLSKIPNKQYPDGASPTEITKYSLDSSYVLDIIL----- 230
Query: 231 GSEDL-----LLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRLFTKFI 285
DL ++GE+Q F+ ++GQSL+AF WK LVSL+ G + R ++ +F+
Sbjct: 231 --NDLTLPIEIIGEIQLTFVCFLVGQSLDAFEHWKKLVSLICG-ADCAISQRRAIYVEFM 287
Query: 286 KVIYYQLKY 294
K I QL +
Sbjct: 288 KAIEIQLMH 296
>UniRef100_Q9VEA6 CG12320-PA [Drosophila melanogaster]
Length = 379
Score = 129 bits (325), Expect = 1e-28
Identities = 110/395 (27%), Positives = 187/395 (46%), Gaps = 45/395 (11%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD Q AL L+ +G L+ VP+ T ID +++GP F+G+KMIPPG H+V+ SS
Sbjct: 10 MDPQLALRLLADGGVLVIAGVPEGTEFGIDLCAYTIGPDFRGVKMIPPGVHYVWCSSRGP 69
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERL--VKVSEEEDERYRLAVKNMEFDRQLGP 118
G + +P +GF P+E++VR+WD + E L +++E E ER R+ + +R L P
Sbjct: 70 YG-DAAPRVGFVHFFHPNEIVVREWDHELEELRPRRIAEPEVERDRIRKNLAQLERVLAP 128
Query: 119 YNLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDN 178
Y+ + WK L+ +T+S ++R P G + E +A + P A + N
Sbjct: 129 YDYRYVVQWKELTGSVTESCVDRCRPTEGTIRTNIELQSCPDADR-PRGGAATVSISRRN 187
Query: 179 SATSV-----------GKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVK 227
+A + ++ +T++P V + +++ LD ++ LL
Sbjct: 188 AAARILLDESELLPDLKPVEGTAPRFTTVPLRVPQDALPA-DVSRHALDCIDAVDKLL-- 244
Query: 228 DYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRL-FTKFIK 286
+ + D L+ ELQ A+ ++G S+E+ W+ L+ LL A T+ +L + K+ +
Sbjct: 245 EGFETADGLIEELQLAYAFFLVGYSVESLAHWRKLLGLLAHSQTAV--TKHKLTYMKYSE 302
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
V+ +QL + + GP + ++ +E ++ G L
Sbjct: 303 VLAHQLPH--LPEELMVPGP----------HNTVYKDVRELLV-----NLHAGGLSVSAE 345
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVV 381
+ K LE LGWEF +GL DE+ E PVV
Sbjct: 346 RLTKRLEKKLGWEF------EGL-LDEDPEDQPVV 373
>UniRef100_Q86P44 SD27779p [Drosophila melanogaster]
Length = 388
Score = 128 bits (322), Expect = 2e-28
Identities = 109/395 (27%), Positives = 187/395 (46%), Gaps = 45/395 (11%)
Query: 1 MDSQTALELVKNGVTLLFLDVPQYTLVAIDTQVFSVGPTFKGIKMIPPGTHFVYYSSSTR 60
MD Q AL L+ +G L+ VP+ T ID +++GP F+G+KMIPPG H+V+ SS
Sbjct: 19 MDPQLALRLLADGGVLVIAGVPEGTEFGIDLCAYTIGPDFRGVKMIPPGVHYVWCSSRGP 78
Query: 61 DGKEFSPMIGFFIDAGPSEVIVRKWDQQEERL--VKVSEEEDERYRLAVKNMEFDRQLGP 118
G + +P +GF P+E++VR+WD + E L +++E E ER R+ + +R L P
Sbjct: 79 YG-DAAPRVGFVHFFHPNEIVVREWDHELEELRPRRIAEPEVERDRIRKNLAQLERVLAP 137
Query: 119 YNLSHYEDWKRLSDFITKSIIERLEPIGGEVSVECENDMFRNAPKTPMEKALDTQLKVDN 178
Y+ + WK L+ +T+S ++R P G + E +A + P A + N
Sbjct: 138 YDYRYVVQWKELTGSVTESCVDRCRPTEGTIRTNIELQSCPDADR-PRGGAATVSISRRN 196
Query: 179 SATSV-----------GKLQRKGCYYTSIPRVVKCKGISGQELTSLNLDKTQLLETLLVK 227
+A + ++ +T++P V + +++ LD ++ LL
Sbjct: 197 AAARILLDESELLPDLKPVEGTAPRFTTVPLRVPQDALPA-DVSRHALDCIDAVDKLL-- 253
Query: 228 DYGGSEDLLLGELQFAFIALMMGQSLEAFLQWKSLVSLLLGCTEAPFHTRTRL-FTKFIK 286
+ + D L+ ELQ A+ ++G S+E+ W+ L+ L+ A T+ +L + K+ +
Sbjct: 254 EGFETADGLIEELQLAYAFFLVGYSVESLAHWRKLLGLVAHSQTAV--TKHKLTYMKYSE 311
Query: 287 VIYYQLKYGLQKDRKDNTGPLLLDDSWFSTDSFLHHHCKEFFSLVLDGSVIDGDLLKWTR 346
V+ +QL + + GP + ++ +E ++ G L
Sbjct: 312 VLAHQLPH--LPEELMVPGP----------HNTVYKDVRELLV-----NLHAGGLSVSAE 354
Query: 347 KFKKLLESNLGWEFQQNNAVDGLYFDENDEFAPVV 381
+ K LE LGWEF +GL DE+ E PVV
Sbjct: 355 RLTKRLEKKLGWEF------EGL-LDEDPEDQPVV 382
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 662,845,641
Number of Sequences: 2790947
Number of extensions: 28247626
Number of successful extensions: 66239
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 66100
Number of HSP's gapped (non-prelim): 68
length of query: 391
length of database: 848,049,833
effective HSP length: 129
effective length of query: 262
effective length of database: 488,017,670
effective search space: 127860629540
effective search space used: 127860629540
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC129092.1


