

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC129091.6 - phase: 0 /pseudo
(1077 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
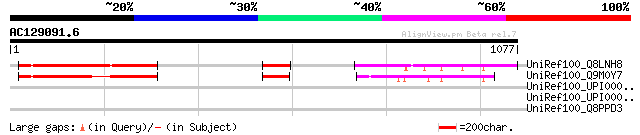
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LNH8 Putative glycine-rich protein [Oryza sativa] 386 e-105
UniRef100_Q9M0Y7 Hypothetical protein AT4g04920 [Arabidopsis tha... 340 1e-91
UniRef100_UPI0000260231 UPI0000260231 UniRef100 entry 38 1.5
UniRef100_UPI000035F5E6 UPI000035F5E6 UniRef100 entry 36 7.2
UniRef100_Q8PPD3 Hypothetical protein XAC0753 [Xanthomonas axono... 36 7.2
>UniRef100_Q8LNH8 Putative glycine-rich protein [Oryza sativa]
Length = 1291
Score = 386 bits (992), Expect = e-105
Identities = 193/305 (63%), Positives = 233/305 (76%), Gaps = 13/305 (4%)
Query: 19 EEVEAGTVFHIRLKQSPSTLLHKMNLHESMRRNFSAVAWCAKLNAIACATETCARIPRSS 78
++ TVF IRLKQ PS+L HKM + E + RNFSAVAWC KLNAIACA+ETCARIP S+
Sbjct: 109 QQASPATVFRIRLKQPPSSLRHKMRVPE-LCRNFSAVAWCGKLNAIACASETCARIPSSN 167
Query: 79 VNPPFWIPIHIVIPERPTENAVFNVVADSPLDSVQFIQWSPVCCPRALLIANFHGRVTIW 138
+PPFWIPIHI+ PERPTE +VFNV ADSP D VQFI+WSP CPRALL+ANFHGR+TIW
Sbjct: 168 SSPPFWIPIHILNPERPTECSVFNVKADSPRDFVQFIEWSPRSCPRALLVANFHGRITIW 227
Query: 139 TQPSHGPANLVIDTNCWLREHEWRQDTAVVTKWLSG---YRWLSSKSSAPANLKSIFEEK 195
TQP+ GP NLV D + W EHEWRQD +VVTKWLSG YRWL + SS +NLK+ FEEK
Sbjct: 228 TQPTKGPTNLVRDASSWQCEHEWRQDLSVVTKWLSGISPYRWLPANSSTSSNLKT-FEEK 286
Query: 196 FISQQSQ----TSGSVQLHWSQWPPTQNATLHKWFCTSKGLLACGPSGIITGDAIITDNG 251
F++QQ Q +SGSVQLHWSQW P+QN+ +WF TSKGLL GPSGI+ DAIIT+ G
Sbjct: 287 FLTQQPQSSVFSSGSVQLHWSQW-PSQNSAQPRWFSTSKGLLGAGPSGIMAADAIITETG 345
Query: 252 TLLVAGVPAVNPAAIIVWEVMPGRGNSLQFSPKKSINNGVPP-LSAPNWSGFAPL--YHF 308
L VAGVP VNP+ ++VWEVMPG GN +Q + K + + +PP L+ P W+GFAPL Y F
Sbjct: 346 ALHVAGVPLVNPSTVVVWEVMPGLGNGIQATAKINATSSLPPSLNPPLWAGFAPLASYLF 405
Query: 309 TARQF 313
+ + +
Sbjct: 406 SLQDY 410
Score = 325 bits (833), Expect = 5e-87
Identities = 190/409 (46%), Positives = 236/409 (57%), Gaps = 67/409 (16%)
Query: 733 GGENGTTISGGSTQMNAWVQEAIAKISSTSDGVSNLTPTSPIGGPSSLMPISINTETFPG 792
G + G + GS+QM WVQ AIAKIS+ +DG +N P +PI G SS MPISINT TFPG
Sbjct: 885 GNQGGVASTTGSSQMQEWVQGAIAKISNNTDGAANAAP-NPISGRSSFMPISINTGTFPG 943
Query: 793 TPAVRLIGDCHFLQRLCQLLFFCFFFKRSQLVLYMNGLRRTAETSL-------------- 838
TPAVRLIGDCHFL RLCQLL FC F+R Q ++++++S+
Sbjct: 944 TPAVRLIGDCHFLHRLCQLLLFCLLFRRRQSPRIPANAQKSSDSSMQKQHLMNSKTEDNT 1003
Query: 839 --VRS-------DDGQTGRAGQIVHGSKGGEEPSLGH--IRLGTGNSGQGYSF------- 880
VRS +DG T R GQ+V G+KG EE +G+ R+G+GN+GQGY+
Sbjct: 1004 LAVRSGLGAAKLEDGTTSR-GQMV-GAKGAEENPVGNKSARIGSGNAGQGYTSDEVKVLF 1061
Query: 881 -------KEAALY*GELHCTS*GFGSIPCHTSACNFHYIKMA----------MQNIPRPR 923
K A L + G +I + +Y + MQN+PRPR
Sbjct: 1062 LILVDLCKRTATLQHPLPSSQVGSSNIIIRLHYIDGNYTVLPEVVEASLGPHMQNMPRPR 1121
Query: 924 GVDDTRLLLRKLELLPPAEVWHRLNMFGGPCTDPDD-----SPRPVRSNPLDSRSL---- 974
G D LLLR+LEL PPAE WHR NMFGGP ++PDD + R ++ N +R L
Sbjct: 1122 GADAAGLLLRELELQPPAEEWHRRNMFGGPWSEPDDLGPLDNTRQLKINGSTNRHLSDME 1181
Query: 975 ESNNIDYGTNGLWPRKHRMIERDAAFALNTS------LDIMGSRRDVITTSWKTGLEGVW 1028
E + +G LWPRK R+ ERDAAF L TS L +MGSRRDVIT WKTGLEG W
Sbjct: 1182 EDGDSSFGIQNLWPRKRRLSERDAAFGLKTSVGLGSFLGVMGSRRDVITAVWKTGLEGEW 1241
Query: 1029 YKCIRCMRQTSAFTSPASANIPNQNERESWWVSRWATNCPMCGGRWVRV 1077
YKCIRC+RQT AF P + NE E+WW+SRW CPMCGG WV+V
Sbjct: 1242 YKCIRCLRQTCAFAQPGALAPNTSNELEAWWISRWTHACPMCGGTWVKV 1290
Score = 68.9 bits (167), Expect = 8e-10
Identities = 35/60 (58%), Positives = 46/60 (76%), Gaps = 2/60 (3%)
Query: 538 YVDSVLDLALSFFTRLRCYAKFCRTLASQA--ATAGNVSNQNMVASPAQNSTTPETSQGS 595
YVD+VLDLA F TRLR YA FCRTLAS A A++G+ +++NMV SP +S +P T+QG+
Sbjct: 827 YVDAVLDLASHFITRLRRYASFCRTLASHAVGASSGSGNSRNMVTSPTNSSPSPSTNQGN 886
>UniRef100_Q9M0Y7 Hypothetical protein AT4g04920 [Arabidopsis thaliana]
Length = 1196
Score = 340 bits (873), Expect = 1e-91
Identities = 176/299 (58%), Positives = 203/299 (67%), Gaps = 40/299 (13%)
Query: 18 IEEVEAGTVFHIRLKQSPSTLLHKMNLHESMRRNFSAVAWCAKLNAIACATETCARIPRS 77
I+ V TVF ++LKQ S LLHKM++ E + RNFSAVAWC KLNAIACA+ETCARIP S
Sbjct: 79 IDPVSPATVFCVKLKQPNSNLLHKMSVPE-LCRNFSAVAWCGKLNAIACASETCARIPSS 137
Query: 78 SVNPPFWIPIHIVIPERPTENAVFNVVADSPLDSVQFIQWSPVCCPRALLIANFHGRVTI 137
N PFWIPIHI+IPERPTE AVFNVVADSP DSVQFI+WSP CPRALLIANFHGR+TI
Sbjct: 138 KANTPFWIPIHILIPERPTECAVFNVVADSPRDSVQFIEWSPTSCPRALLIANFHGRITI 197
Query: 138 WTQPSHGPANLVIDTNCWLREHEWRQDTAVVTKWLSGYRWLSSKSSAPANLKSIFEEKFI 197
WTQP+ G ANLV D W EHEWRQD AVVTKWL+G
Sbjct: 198 WTQPTQGSANLVHDATSWQCEHEWRQDIAVVTKWLTGA---------------------- 235
Query: 198 SQQSQTSGSVQLHWSQWPPTQNATLHKWFCTSKGLLACGPSGIITGDAIITDNGTLLVAG 257
S WP Q +T KWF T KGLL GPSGI+ DAIITD+G + VAG
Sbjct: 236 --------------SPWPSNQGSTAPKWFSTKKGLLGAGPSGIMAADAIITDSGAMHVAG 281
Query: 258 VPAVNPAAIIVWEVMPGRGNSLQFSPKKSINNGVPP-LSAPNWSGFAPL--YHFTARQF 313
VP VNP+ I+VWEV PG GN LQ +PK S + VPP LS+ +W+GFAPL Y F+ +++
Sbjct: 282 VPIVNPSTIVVWEVTPGPGNGLQATPKISTGSRVPPSLSSSSWTGFAPLAAYLFSWQEY 340
Score = 244 bits (622), Expect = 1e-62
Identities = 157/347 (45%), Positives = 193/347 (55%), Gaps = 56/347 (16%)
Query: 738 TTISGGSTQMNAWVQEAIAKISSTSDGVSNLTPTSPIGGPSSLMPISINTETFPGTPAVR 797
TT S GS+ + AW+Q AIAKISS++DG SN T SPI G + MPISINT TFPGTPAVR
Sbjct: 832 TTNSSGSSHVQAWMQGAIAKISSSNDG-SNST-ASPISGSPTFMPISINTGTFPGTPAVR 889
Query: 798 LIGDCHFLQRLCQLLFFCFFFKRSQLV----------LYMNGLRRTAETS-------LVR 840
LIGDCHFL RLCQLL FCF + S+ L + E + L R
Sbjct: 890 LIGDCHFLHRLCQLLLFCFLQRSSRFPQRNADVSSQKLQTGATSKLEEVNSAKPTPALNR 949
Query: 841 SDDGQTGRAGQIVHGSKGGEEPSLGHIRLGTGNSGQGYSFKEAALY*G------------ 888
+D Q R Q+ G KG +E S ++G+GN+GQGY+++E +
Sbjct: 950 IEDAQGFRGAQLGTGVKGIDENSARTTKMGSGNAGQGYTYEEVRVLFHILMDLCKRTSGL 1009
Query: 889 --ELHCTS*GFGSIPCHTSACNFHYIKMA----------MQNIPRPRGVDDTRLLLRKLE 936
L + G G+I + +Y + MQN+PRPRG D LLLR+LE
Sbjct: 1010 AHPLPGSQVGSGNIQVRLHYIDGNYTVLPEVVEAALGPHMQNMPRPRGADAAGLLLRELE 1069
Query: 937 LLPPAEVWHRLNMFGGPCTDPDD----SPRPVRSNPLDSRSLESNNIDYGTN---GLWPR 989
L PP+E WHR N+FGGP ++P+D SN LD + I G N LWPR
Sbjct: 1070 LHPPSEEWHRRNLFGGPGSEPEDMILTDDVSKLSNSLDLPDTNFSGICDGYNRVHSLWPR 1129
Query: 990 KHRMIERDAAFALNTS------LDIMGSRRDVITTSWKTGLEGVWYK 1030
K RM ERDAAF NTS L IMGSRRDV+T +WKTGLEGVWYK
Sbjct: 1130 KRRMSERDAAFGSNTSVGLGAYLGIMGSRRDVVTATWKTGLEGVWYK 1176
Score = 76.3 bits (186), Expect = 5e-12
Identities = 38/57 (66%), Positives = 44/57 (76%)
Query: 537 AYVDSVLDLALSFFTRLRCYAKFCRTLASQAATAGNVSNQNMVASPAQNSTTPETSQ 593
AYVD+VLDLA F TRLR YA FCRTLAS AA+AG SN+N V SP QN+++P T Q
Sbjct: 753 AYVDAVLDLASHFITRLRRYASFCRTLASHAASAGTGSNRNNVTSPTQNASSPATPQ 809
>UniRef100_UPI0000260231 UPI0000260231 UniRef100 entry
Length = 333
Score = 38.1 bits (87), Expect = 1.5
Identities = 21/54 (38%), Positives = 32/54 (58%), Gaps = 1/54 (1%)
Query: 838 LVRSDDGQTGRAGQI-VHGSKGGEEPSLGHIRLGTGNSGQGYSFKEAALY*GEL 890
++R +D T A QI + + GG+ P LGH+ L G+ G+G S + +L GEL
Sbjct: 88 IIRGEDHVTNSAAQIQLFKALGGKVPELGHLSLLAGSEGEGLSKRFGSLNLGEL 141
>UniRef100_UPI000035F5E6 UPI000035F5E6 UniRef100 entry
Length = 127
Score = 35.8 bits (81), Expect = 7.2
Identities = 22/84 (26%), Positives = 46/84 (54%), Gaps = 3/84 (3%)
Query: 416 CIKFMYSGLFVTV-VNFNDMLHILFDGEFLKQFSATTFLNTVLSAWFGVFNFCYTYLYLY 474
CI ++Y +++ + + ++IL D K+ +++ +L+ LS + ++ Y +YL
Sbjct: 13 CIIYIYIYIYIYIYIYIYIYIYILVDLFHCKRDTSSIYLSIYLSIYLSIYLSIYLSIYLS 72
Query: 475 SHW*NNLYCYMYNTPYR*IYLPTY 498
+ ++Y +Y + Y IYLPTY
Sbjct: 73 IYL--SIYLSIYLSIYLSIYLPTY 94
>UniRef100_Q8PPD3 Hypothetical protein XAC0753 [Xanthomonas axonopodis]
Length = 526
Score = 35.8 bits (81), Expect = 7.2
Identities = 36/123 (29%), Positives = 50/123 (40%), Gaps = 29/123 (23%)
Query: 209 LHWSQWPPTQNATLHKWFCTSKGLLACGPSGIITGDAI-------------ITDNGTLLV 255
L + W Q AT GLL+ P+G +TG + D+G +LV
Sbjct: 324 LDYPDWASVQQATTQLGLSAPAGLLS--PTGGLTGGTASGWQWRIATDVPSVADSGVVLV 381
Query: 256 AG------------VPAVNPAAIIVWEVMPGRGNSLQFSPKKSINNGVPPLSAPNWSGFA 303
+G +P VN AA VW P G + + + + S+ V P S N S FA
Sbjct: 382 SGLLQGIGLPVSTSIPLVNRAAAAVWVGSPSNG-TRRLAGRPSLRVTVVP-SQSNVSLFA 439
Query: 304 PLY 306
LY
Sbjct: 440 YLY 442
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.340 0.148 0.510
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,743,491,631
Number of Sequences: 2790947
Number of extensions: 72376955
Number of successful extensions: 223351
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 223316
Number of HSP's gapped (non-prelim): 19
length of query: 1077
length of database: 848,049,833
effective HSP length: 138
effective length of query: 939
effective length of database: 462,899,147
effective search space: 434662299033
effective search space used: 434662299033
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC129091.6


