

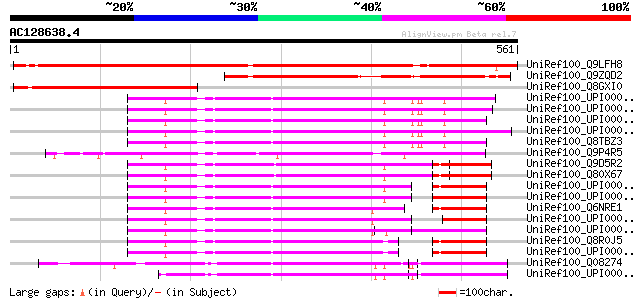
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
(561 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LFH8 Hypothetical protein F4P12_90 [Arabidopsis thal... 811 0.0
UniRef100_Q9ZQD2 Hypothetical protein At2g37160 [Arabidopsis tha... 436 e-121
UniRef100_Q8GXI0 Hypothetical protein At2g37160 [Arabidopsis tha... 285 2e-75
UniRef100_UPI000019C56F UPI000019C56F UniRef100 entry 239 2e-61
UniRef100_UPI0000369EAE UPI0000369EAE UniRef100 entry 238 4e-61
UniRef100_UPI000019C572 UPI000019C572 UniRef100 entry 237 6e-61
UniRef100_UPI00001CFAC8 UPI00001CFAC8 UniRef100 entry 235 2e-60
UniRef100_Q8TBZ3 WD-repeat protein 20 [Homo sapiens] 234 6e-60
UniRef100_Q9P4R5 Catabolite repression protein creC [Emericella ... 221 4e-56
UniRef100_Q9D5R2 WD-repeat protein 20 [Mus musculus] 214 5e-54
UniRef100_Q80X67 WD repeat domain 20 [Mus musculus] 213 1e-53
UniRef100_UPI000025D922 UPI000025D922 UniRef100 entry 211 3e-53
UniRef100_UPI000036088A UPI000036088A UniRef100 entry 209 2e-52
UniRef100_Q6NRE1 Hypothetical protein [Xenopus laevis] 203 9e-51
UniRef100_UPI0000361C87 UPI0000361C87 UniRef100 entry 202 2e-50
UniRef100_UPI0000361C85 UPI0000361C85 UniRef100 entry 202 2e-50
UniRef100_Q8R0J5 2310040A13Rik protein [Mus musculus] 196 2e-48
UniRef100_UPI000017F7C1 UPI000017F7C1 UniRef100 entry 194 4e-48
UniRef100_Q08274 Dystrophia myotonica-containing WD repeat motif... 191 6e-47
UniRef100_UPI00001CE7D9 UPI00001CE7D9 UniRef100 entry 190 8e-47
>UniRef100_Q9LFH8 Hypothetical protein F4P12_90 [Arabidopsis thaliana]
Length = 556
Score = 811 bits (2095), Expect = 0.0
Identities = 411/568 (72%), Positives = 469/568 (82%), Gaps = 26/568 (4%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++A SSSSS P QS G+KTYFKTPEG+YKL ++KTH SGLL + HGKTV
Sbjct: 4 TSNGMISAPSSSSS--PAANPQSP--GIKTYFKTPEGKYKLHYEKTHSSGLLHYAHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VT+A LKE+ AP TPT +SS +SASSG RSA ARLLG NG+RALSFVGGNG KS
Sbjct: 60 TQVTIAQLKERAAPSTPTGTSSGYSASSGFRSATARLLGTGNGSRALSFVGGNGGGKSVS 119
Query: 125 GASRI-GSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRI GS +S+ ++S+ N NFDGKG+YLVFN DAI I DLNSQ+KDP+KSIHFSNSN
Sbjct: 120 TSSRISGSFAASNSNTSMTNTNFDGKGTYLVFNVSDAIFICDLNSQEKDPVKSIHFSNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNN--SRC 241
P+CHAFD DAKDGHDLLIGL SGDVY+VSLRQQLQDV KK+VGA HYNKDG +NN RC
Sbjct: 180 PMCHAFDPDAKDGHDLLIGLNSGDVYTVSLRQQLQDVSKKLVGALHYNKDGSVNNRQDRC 239
Query: 242 TCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
T ISWVPGGDGAFVVAHADGNLY NKDGA ES+FP ++D T FSV A+YSKSNP+A
Sbjct: 240 TSISWVPGGDGAFVVAHADGNLY----NKDGATESTFPAIRDPTQFSVDKAKYSKSNPVA 295
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
RWHICQGSINSI+FS DGA+LATVGRDGYLR+FD+ + LVCGGKSYYG LLCC+WSMDG
Sbjct: 296 RWHICQGSINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGGKSYYGALLCCSWSMDG 355
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGS 421
KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS WSSPNS+ +GE + YRFGS
Sbjct: 356 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSNWSSPNSDGSGEHVMYRFGS 415
Query: 422 VGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPS 481
VGQDTQLLLW+LEMDEIVVPLRR P GSPT+S GS S+HWDN +P+GTLQPAP
Sbjct: 416 VGQDTQLLLWDLEMDEIVVPLRRAPG------GSPTYSTGS-SAHWDNVIPMGTLQPAPC 468
Query: 482 MRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE---- 537
RDVPK+SP++AHRVHTEPLS L+FTQESV+TACREGHIKIWTRP +E+Q ++SE
Sbjct: 469 KRDVPKLSPIIAHRVHTEPLSGLMFTQESVVTACREGHIKIWTRPSESETQTNSSEANPT 528
Query: 538 -TLLATSLKE--KPSLSSK-ISNSIYKQ 561
LL+TS + K SLSSK IS S +KQ
Sbjct: 529 TALLSTSFTKDNKASLSSKIISGSTFKQ 556
>UniRef100_Q9ZQD2 Hypothetical protein At2g37160 [Arabidopsis thaliana]
Length = 444
Score = 436 bits (1121), Expect = e-121
Identities = 220/317 (69%), Positives = 251/317 (78%), Gaps = 36/317 (11%)
Query: 238 NSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKS 297
NSRCT I+WVPGGDG+FV AHADGNLY NK+GA +SSF ++D T FSV A+YSKS
Sbjct: 150 NSRCTSIAWVPGGDGSFVAAHADGNLY----NKEGATDSSFSAIRDPTQFSVDKAKYSKS 205
Query: 298 NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAW 357
NP+ARWHI QG+INSI+FS DGAYLATVGRDGYLR+FD++ + LVCG KSYYG LLCCAW
Sbjct: 206 NPVARWHIGQGAINSIAFSNDGAYLATVGRDGYLRIFDFSTQKLVCGVKSYYGALLCCAW 265
Query: 358 SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITY 417
SMDGKY+LTGGEDDLVQVWSMEDRKVVAW EG +GE + Y
Sbjct: 266 SMDGKYLLTGGEDDLVQVWSMEDRKVVAW-EG---------------------SGENVMY 303
Query: 418 RFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQ 477
RFGSVGQDTQLLLW+LEMDEIVVPLRR PPG GSPT+S GSQS+HWDN VP+GTLQ
Sbjct: 304 RFGSVGQDTQLLLWDLEMDEIVVPLRR-PPG-----GSPTYSTGSQSAHWDNIVPMGTLQ 357
Query: 478 PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
PAP MRDVPK+SP+VAHRVHTEPLS LIFTQES++TACREGH+KIWTRP ++QPS+S
Sbjct: 358 PAPCMRDVPKLSPVVAHRVHTEPLSGLIFTQESLITACREGHLKIWTRP---DTQPSSSS 414
Query: 538 TLLATSLKEKPSLSSKI 554
KPSL+SK+
Sbjct: 415 E-ATNPTTSKPSLTSKV 430
>UniRef100_Q8GXI0 Hypothetical protein At2g37160 [Arabidopsis thaliana]
Length = 206
Score = 285 bits (730), Expect = 2e-75
Identities = 143/204 (70%), Positives = 165/204 (80%), Gaps = 5/204 (2%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++ SSSS+S Q G+KTYFKTPEG+YKL ++KTHPSGLL + HGKTV
Sbjct: 4 TSNGMISGPSSSSASA----ANPQSPGIKTYFKTPEGKYKLHYEKTHPSGLLHYTHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VTLAHLK+KPAP TPT +SSS +ASSG RSA ARLLGG NGNRALSFVGGNG +K+ G
Sbjct: 60 TQVTLAHLKDKPAPSTPTGTSSSSTASSGFRSATARLLGGGNGNRALSFVGGNGGAKNVG 119
Query: 125 GASRIG-SIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRIG S +SS ++S N NFDGKG+YLVFN GDAI ISDLNSQDKDP+KSIH SNSN
Sbjct: 120 ASSRIGASFAASSSNTSATNTNFDGKGTYLVFNVGDAIFISDLNSQDKDPVKSIHISNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGD 207
P+CHAF DAKDGHDLLIGL SGD
Sbjct: 180 PMCHAFHPDAKDGHDLLIGLNSGD 203
>UniRef100_UPI000019C56F UPI000019C56F UniRef100 entry
Length = 581
Score = 239 bits (610), Expect = 2e-61
Identities = 179/540 (33%), Positives = 242/540 (44%), Gaps = 142/540 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG S S S+
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPGKVGSLSSPSQ 572
>UniRef100_UPI0000369EAE UPI0000369EAE UniRef100 entry
Length = 581
Score = 238 bits (606), Expect = 4e-61
Identities = 178/537 (33%), Positives = 240/537 (44%), Gaps = 142/537 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPS 534
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG S S
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPGKVGSLSS 569
>UniRef100_UPI000019C572 UPI000019C572 UniRef100 entry
Length = 569
Score = 237 bits (605), Expect = 6e-61
Identities = 176/530 (33%), Positives = 238/530 (44%), Gaps = 142/530 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPG 562
>UniRef100_UPI00001CFAC8 UPI00001CFAC8 UniRef100 entry
Length = 595
Score = 235 bits (600), Expect = 2e-60
Identities = 179/558 (32%), Positives = 250/558 (44%), Gaps = 143/558 (25%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ G++Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGSMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGDSGPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKAKTDPAKTLGTS 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG +PS
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCLVTACQEGFICTWARPGKV-CRPSLGL 571
Query: 538 TLLATSLKEKPSLSSKIS 555
L S +++K++
Sbjct: 572 CPLTNSAVSAARVAAKVA 589
>UniRef100_Q8TBZ3 WD-repeat protein 20 [Homo sapiens]
Length = 569
Score = 234 bits (596), Expect = 6e-60
Identities = 175/530 (33%), Positives = 237/530 (44%), Gaps = 142/530 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WV G + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVHGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPG 562
>UniRef100_Q9P4R5 Catabolite repression protein creC [Emericella nidulans]
Length = 630
Score = 221 bits (563), Expect = 4e-56
Identities = 165/540 (30%), Positives = 255/540 (46%), Gaps = 82/540 (15%)
Query: 40 EGRYKLQFD---KTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRS 96
EG Y+LQ D T P H ++ L P P PT+ S G+R+
Sbjct: 87 EGTYQLQDDLHLATPPP------HPSEAPIINPNPLATVPNP--PTSGVKLSLISLGLRN 138
Query: 97 ---------AAARLLGGSNGNRALSFVGGNGSSKSNGGASRIGSIGSSSLSSSVANP--- 144
AR G +GN AL+ SK + I SS +S + +
Sbjct: 139 KTAFPSKVQVTARPFG--DGNSALAAAPVKDPSKKRKPKNNIIKSSSSFVSRVIIHEATT 196
Query: 145 ----NFDGKGSYLVFNAGDAILISDLNSQDKD-PIKSIHFSNSNPVCHAFDQDAKDGH-- 197
+ D +G + N A DL+S+ KD P+ I F+ ++ + H ++ K
Sbjct: 197 KRLNDRDPEGLFAFANINRAFQWLDLSSKHKDEPLSKILFTKAHMISHDVNEITKSSAHI 256
Query: 198 DLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVA 257
D+++G +GD++ + + +K NK+GI+N+S T I W+PG + F+ A
Sbjct: 257 DVIMGSSAGDIF------WYEPISQKYA---RINKNGIINSSPVTHIKWIPGSENFFIAA 307
Query: 258 HADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYS------------KSNPIARWHI 305
H +G L VY+K K+ A P L +Q+ SV +R+S K+NP+A W +
Sbjct: 308 HENGQLVVYDKEKEDA--LFIPELPEQSAESVKPSRWSLQVLKSVNSKNQKANPVAVWRL 365
Query: 306 CQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
I +FS D +LA V DG LR+ DY +E ++ +SYYGG C WS DGKYI+
Sbjct: 366 ANQKITQFAFSPDHRHLAVVLEDGTLRLMDYLQEEVLDVFRSYYGGFTCVCWSPDGKYIV 425
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
TGG+DDLV +WS+ +RK++A +GH+SWVS VAFD + + TYR GSVG D
Sbjct: 426 TGGQDDLVTIWSLPERKIIARCQGHDSWVSAVAFDPW--------RCDERTYRIGSVGDD 477
Query: 426 TQLLLWELEM-----------------DEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWD 468
LLLW+ + ++ P + P + SQ + D
Sbjct: 478 CNLLLWDFSVGMLHRPKVHHQTSARHRTSLIAPSSQQPNRHRADSSGNRMRSDSQRTAAD 537
Query: 469 N-AVPLGTLQ-PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRP 526
+ + P +Q P S + P+++ V +P+ L F +++++T+ EGHI+ W RP
Sbjct: 538 SESAPDQPVQHPVESRARTALLPPIMSKAVGEDPICWLGFQEDTIMTSSLEGHIRTWDRP 597
>UniRef100_Q9D5R2 WD-repeat protein 20 [Mus musculus]
Length = 567
Score = 214 bits (545), Expect = 5e-54
Identities = 146/406 (35%), Positives = 201/406 (48%), Gaps = 63/406 (15%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G+ L F+ G + + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQTGNGNRLCFSVGRELYVYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
C+ WVPG D F+VAH+ GN+Y Y ++ G + +LK F+V + + SKS
Sbjct: 154 CVKWVPGSDSLFLVAHSSGNMYSYNVEHTYGTTAPHYQLLKQGESFAVHNCK-SKSTRDL 212
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
+W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS DG
Sbjct: 213 KWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSAELHGTMKSYFGGLLCLCWSPDG 272
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE-------- 413
KYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 KYIVTGGEDDLVTVWSFLDCRVIARGRGHKSWVSVVAFDPYTTSVEESDPMEFSDSDKNF 332
Query: 414 -----------------------------TITYRFGSVGQDTQLLLWELEMDEIV--VPL 442
++TYRFGSVGQDTQL LW+L D + PL
Sbjct: 333 QDLLHFGRDRANSTQSRLSKQNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQPL 392
Query: 443 RRGPPGGS--PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
R + G P S GS + N+VP P P +P
Sbjct: 393 SRARAHANVMNATGLPAGSNGSAVTTPGNSVP----PPLPRSNSLP 434
Score = 56.2 bits (134), Expect = 2e-06
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 502 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 560
Query: 528 -VAESQP 533
V++ QP
Sbjct: 561 KVSKFQP 567
>UniRef100_Q80X67 WD repeat domain 20 [Mus musculus]
Length = 567
Score = 213 bits (542), Expect = 1e-53
Identities = 146/406 (35%), Positives = 200/406 (48%), Gaps = 63/406 (15%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G+ L F+ G + + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQTGNGNRLCFSVGRELYVYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSVGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
C+ WVPG D F+VAH+ GN+Y Y ++ G + +LK F+V + + SKS
Sbjct: 154 CVKWVPGSDSLFLVAHSSGNMYSYNVEHTYGTTAPHYQLLKQGESFAVHNCK-SKSTRDL 212
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
+W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS DG
Sbjct: 213 KWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSAELHGTMKSYFGGLLCLCWSPDG 272
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE-------- 413
KYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 KYIVTGGEDDLVTVWSFLDCRVIARGRGHKSWVSVVAFDPYTTSVEESDPMEFSDSDKNF 332
Query: 414 -----------------------------TITYRFGSVGQDTQLLLWELEMDEIV--VPL 442
++TYRFGSVGQDTQL LW+L D + PL
Sbjct: 333 QDLLHFGRDRANSTQSRLSKQNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQPL 392
Query: 443 RRGPPGGS--PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
R + G P S GS + N+VP P P +P
Sbjct: 393 SRARAHANVMNATGLPAGSNGSAVTTPGNSVP----PPLPRSNSLP 434
Score = 56.2 bits (134), Expect = 2e-06
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 502 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 560
Query: 528 -VAESQP 533
V++ QP
Sbjct: 561 KVSKFQP 567
>UniRef100_UPI000025D922 UPI000025D922 UniRef100 entry
Length = 626
Score = 211 bits (538), Expect = 3e-53
Identities = 136/365 (37%), Positives = 187/365 (50%), Gaps = 60/365 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGDRICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+V+HA GN+Y+Y ++ G + +LK +SV H SKS NP
Sbjct: 154 CVKWVPGSESLFLVSHASGNMYLYNVEHTCGTTAPHYQLLKQGESYSV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDAVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+ GGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 DGKYIVAGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDHYTTSVEESDPMEFSGSDE 332
Query: 414 --------------------------------TITYRFGSVGQDTQLLLWELEMDEIV-- 439
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDHVIHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPH 392
Query: 440 VPLRR 444
+PL R
Sbjct: 393 LPLSR 397
Score = 56.2 bits (134), Expect = 2e-06
Identities = 28/60 (46%), Positives = 38/60 (62%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT Q P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 549 DPAKTLGT-QLCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCLVTACQEGFICTWARPG 607
>UniRef100_UPI000036088A UPI000036088A UniRef100 entry
Length = 589
Score = 209 bits (532), Expect = 2e-52
Identities = 135/365 (36%), Positives = 187/365 (50%), Gaps = 60/365 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGDRICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+V+HA G++Y+Y ++ G + +LK +SV H SKS NP
Sbjct: 154 CVKWVPGSESLFLVSHASGSMYLYNVEHTCGTTAPHYQLLKQGENYSV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDAVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+ GGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 DGKYIVAGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDHYTTSVEESDPMEFSGSDE 332
Query: 414 --------------------------------TITYRFGSVGQDTQLLLWELEMDEIV-- 439
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDQMIHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPH 392
Query: 440 VPLRR 444
+PL R
Sbjct: 393 LPLSR 397
Score = 56.2 bits (134), Expect = 2e-06
Identities = 28/60 (46%), Positives = 38/60 (62%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGTL P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 527 DPAKTLGTLL-CPRMEDVPLLEPLICKKIAHERLTVLIFLEDCLVTACQEGFICTWARPG 585
>UniRef100_Q6NRE1 Hypothetical protein [Xenopus laevis]
Length = 574
Score = 203 bits (517), Expect = 9e-51
Identities = 132/354 (37%), Positives = 181/354 (50%), Gaps = 57/354 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGDRICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ D LL+G +G V QL D KK + +N++ +++ +R T
Sbjct: 102 TCHDFNHLTATADSVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKARVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYEKNKD-GAGESSFPILKDQTLFSVAHARYSKS--NP 299
+ WVPG + F+VAH+ G++++Y G + +LK F+V H SKS NP
Sbjct: 154 SVKWVPGSESLFLVAHSSGSMFLYNVEYTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD------------------- 400
DGKYI+TGGEDDLV VWS D +V+A G+GH SWVS VAFD
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGQGHKSWVSVVAFDPNTTSVEETDPMEFSGSDE 332
Query: 401 -----------------SYWSSPNSNDNGE-TITYRFGSVGQDTQLLLWELEMD 436
S S NS D+ ++TYRFGSVGQDTQL LW+L D
Sbjct: 333 DFQELMSFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTED 386
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/60 (45%), Positives = 38/60 (63%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D+A LGT P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 509 DSAKTLGT-PLCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWARPG 567
>UniRef100_UPI0000361C87 UPI0000361C87 UniRef100 entry
Length = 572
Score = 202 bits (515), Expect = 2e-50
Identities = 134/367 (36%), Positives = 185/367 (49%), Gaps = 62/367 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 33 SQGSNPVKVSFVNVNDQSGNGERICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 92
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 93 TCHDFNPLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 144
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ G++Y+Y +N G + +LK ++V H SKS NP
Sbjct: 145 CVRWVPGSESLFLVAHSSGSMYLYNVENTCGTTAPHYQLLKQGENYAV-HTCKSKSARNP 203
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ RW + +G++N +FS DG +LA +DG+LRVF + L KSY+GGLLC WS
Sbjct: 204 LLRWTVGEGALNEFAFSPDGKFLACASQDGFLRVFGFDAAELHGTMKSYFGGLLCVCWSP 263
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD------------------- 400
DG+YI+ GGEDDLV VWS D +V+A G GH SWVS VAFD
Sbjct: 264 DGRYIVAGGEDDLVTVWSFSDCRVIARGHGHKSWVSVVAFDHCTTSVEDGDVPAEFSGSD 323
Query: 401 --------------------SYWSSPNSNDNGE-TITYRFGSVGQDTQLLLWELEMDEIV 439
S S NS ++ ++TYRFGSVGQDTQL LW+L D +
Sbjct: 324 EDFHEHINFGAGRERANSAHSRLSKRNSTESRPVSVTYRFGSVGQDTQLCLWDLTEDILF 383
Query: 440 --VPLRR 444
+PL R
Sbjct: 384 PHLPLSR 390
Score = 50.8 bits (120), Expect = 1e-04
Identities = 21/48 (43%), Positives = 32/48 (65%)
Query: 480 PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M +VP + PLV ++ E L+ LIF ++ ++TAC+EG + W RPG
Sbjct: 523 PRMEEVPLLEPLVCKKIAHERLTVLIFLEDCLVTACQEGFVCTWARPG 570
>UniRef100_UPI0000361C85 UPI0000361C85 UniRef100 entry
Length = 612
Score = 202 bits (515), Expect = 2e-50
Identities = 134/367 (36%), Positives = 185/367 (49%), Gaps = 62/367 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGERICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNPLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ G++Y+Y +N G + +LK ++V H SKS NP
Sbjct: 154 CVRWVPGSESLFLVAHSSGSMYLYNVENTCGTTAPHYQLLKQGENYAV-HTCKSKSARNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ RW + +G++N +FS DG +LA +DG+LRVF + L KSY+GGLLC WS
Sbjct: 213 LLRWTVGEGALNEFAFSPDGKFLACASQDGFLRVFGFDAAELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD------------------- 400
DG+YI+ GGEDDLV VWS D +V+A G GH SWVS VAFD
Sbjct: 273 DGRYIVAGGEDDLVTVWSFSDCRVIARGHGHKSWVSVVAFDHCTTSVEDGDVPAEFSGSD 332
Query: 401 --------------------SYWSSPNSNDNGE-TITYRFGSVGQDTQLLLWELEMDEIV 439
S S NS ++ ++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 EDFHEHINFGAGRERANSAHSRLSKRNSTESRPVSVTYRFGSVGQDTQLCLWDLTEDILF 392
Query: 440 --VPLRR 444
+PL R
Sbjct: 393 PHLPLSR 399
Score = 51.6 bits (122), Expect = 6e-05
Identities = 41/129 (31%), Positives = 57/129 (43%), Gaps = 5/129 (3%)
Query: 404 SSPNSNDNGETI---TYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSA 460
SS NSN G +I + V + L L E D +R G S
Sbjct: 480 SSNNSNAKGNSIIDSAFIATGVSKFATLSLHESRKDRHEKDHKRNHSMGHINSKSSDKLN 539
Query: 461 GSQSSHWDNAVPLGTLQPA--PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREG 518
SS A + TL P M +VP + PLV ++ E L+ LIF ++ ++TAC+EG
Sbjct: 540 QLNSSRTAKAEAVKTLGTTLCPRMEEVPLLEPLVCKKIAHERLTVLIFLEDCLVTACQEG 599
Query: 519 HIKIWTRPG 527
+ W RPG
Sbjct: 600 FVCTWARPG 608
>UniRef100_Q8R0J5 2310040A13Rik protein [Mus musculus]
Length = 539
Score = 196 bits (498), Expect = 2e-48
Identities = 123/311 (39%), Positives = 169/311 (53%), Gaps = 25/311 (8%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFD--QDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNLLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRF 419
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E F
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPME-----F 327
Query: 420 GSVGQDTQLLL 430
+D Q LL
Sbjct: 328 SGSDEDFQDLL 338
Score = 54.7 bits (130), Expect = 7e-06
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 474 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 532
>UniRef100_UPI000017F7C1 UPI000017F7C1 UniRef100 entry
Length = 539
Score = 194 bits (494), Expect = 4e-48
Identities = 122/311 (39%), Positives = 169/311 (54%), Gaps = 25/311 (8%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ G++Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGSMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRF 419
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E F
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPME-----F 327
Query: 420 GSVGQDTQLLL 430
+D Q LL
Sbjct: 328 SGSDEDFQDLL 338
Score = 53.9 bits (128), Expect = 1e-05
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 474 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCLVTACQEGFICTWARPG 532
>UniRef100_Q08274 Dystrophia myotonica-containing WD repeat motif protein [Mus
musculus]
Length = 650
Score = 191 bits (484), Expect = 6e-47
Identities = 149/472 (31%), Positives = 220/472 (46%), Gaps = 83/472 (17%)
Query: 32 LKTYFKTPEGRYKL-QFDKTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTASSSSFSA 90
+K+ F+T EG YKL D T SG T A P P P ++ S
Sbjct: 7 IKSQFRTREGFYKLLPGDATRRSG------------PTSAQTPAPPQPTQPPPGPAAASG 54
Query: 91 SSGVRSAAARLLGGSNGNRALSFV--------GGNGSSKSNGGASRIGSIGSSSLSSSVA 142
A++ G AL V +G+ + S +G+ G +
Sbjct: 55 PGAAGPASSPPPAGPGPGPALPAVRLSLVRLGDPDGAGEPPSTPSGLGAGGDRVCFNL-- 112
Query: 143 NPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSNPVCHAFDQ--DAKDGHDLL 200
G L F G S + PI + + P CH F+Q A + LL
Sbjct: 113 -------GRELYFYPGCCRSGSQRSIDLNKPIDKRIYKGTQPTCHDFNQFTAATETISLL 165
Query: 201 IGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVAHAD 260
+G +G V + L ++ D K +N++ +++ ++ T + W+P + F+ +HA
Sbjct: 166 VGFSAGQVQYLDLIKK--DTSKL------FNEERLIDKTKVTYLKWLPESESLFLASHAS 217
Query: 261 GNLYVYEKNKDGAGES-SFPILKDQTLFSVAHARYSKS--NPIARWHICQGSINSISFSA 317
G+LY+Y + + +LK F+V +A SK+ NP+A+W + +G +N +FS
Sbjct: 218 GHLYLYNVSHPCTSTPPQYSLLKQGEGFAV-YAAKSKAPRNPLAKWAVGEGPLNEFAFSP 276
Query: 318 DGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWS 377
DG +LA V +DG LRVF + L KSY+GGLLC WS DG+Y++TGGEDDLV VWS
Sbjct: 277 DGRHLACVSQDGCLRVFHFDSMLLRGLMKSYFGGLLCVCWSPDGRYVVTGGEDDLVTVWS 336
Query: 378 MEDRKVVAWGEGHNSWVSGVAFDSYW--------SSPNSNDNGE---------------- 413
+ +VVA G GH SWV+ VAFD Y +S + + +GE
Sbjct: 337 FTEGRVVARGHGHKSWVNAVAFDPYTTRAEEAASASADGDPSGEEEEPEVTSSDTGAPVS 396
Query: 414 ------TITYRFGSVGQDTQLLLWELEMDEIVVP---LRR-----GPPGGSP 451
+ITYRFGS GQDTQ LW+L ++++ P L R G PG +P
Sbjct: 397 PLPKAGSITYRFGSAGQDTQFCLWDL-TEDVLSPHPSLARTRTLPGTPGATP 447
Score = 57.4 bits (137), Expect = 1e-06
Identities = 36/110 (32%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 442 LRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPL 501
+ RG GG+ + + S + S D A LGT P + +VP + PLV ++ E L
Sbjct: 526 ISRGGSGGNSS--NDKLSGPAPRSRLDPAKVLGTAL-CPRIHEVPLLEPLVCKKIAQERL 582
Query: 502 SSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLS 551
+ L+F ++ ++TAC+EG I W RPG A + S PS S
Sbjct: 583 TVLLFLEDCIITACQEGLICTWARPGKAFTDEETEAQAGQASWPRSPSKS 632
>UniRef100_UPI00001CE7D9 UPI00001CE7D9 UniRef100 entry
Length = 1177
Score = 190 bits (483), Expect = 8e-47
Identities = 121/329 (36%), Positives = 176/329 (52%), Gaps = 55/329 (16%)
Query: 165 DLNSQDKDPIKSIHFSNSNPVCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGK 222
DLN PI + + P CH F+Q A + LL+G +G V + L ++ D K
Sbjct: 659 DLNK----PIDKRIYKGTQPTCHDFNQFTAATETISLLVGFSAGQVQYLDLIKK--DTSK 712
Query: 223 KIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGES-SFPIL 281
+N++ +++ ++ T + W+P + F+ +HA G+LY+Y + A + +L
Sbjct: 713 L------FNEERLIDKTKVTYLKWLPESESLFLASHASGHLYLYNVSHPCASTPPQYSLL 766
Query: 282 KDQTLFSVAHARYSKS--NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKE 339
K F+V +A SK+ NP+A+W + +G +N +FS DG +LA V +DG LRVF +
Sbjct: 767 KQGEGFAV-YAAKSKAPRNPLAKWAVGEGPLNEFAFSPDGRHLACVSQDGCLRVFHFDSM 825
Query: 340 HLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
L KSY+GGLLC WS DG+Y++TGGEDDLV VWS + +VVA G GH SWV+ VAF
Sbjct: 826 LLRGLMKSYFGGLLCVCWSPDGRYVVTGGEDDLVTVWSFTEGRVVARGHGHKSWVNAVAF 885
Query: 400 DSYW--------SSPNSNDNGE----------------------TITYRFGSVGQDTQLL 429
D Y +S + + +GE ++TYRFGS GQDTQ
Sbjct: 886 DPYTTRAEETASASGDGDPSGEEEEPEATSSDTGAPVSPLPKAGSVTYRFGSAGQDTQFC 945
Query: 430 LWELEMDEIV--VPLRR-----GPPGGSP 451
LW+L D + PL R G PG +P
Sbjct: 946 LWDLTEDVLSPHPPLARTRTLPGTPGATP 974
Score = 55.8 bits (133), Expect = 3e-06
Identities = 34/110 (30%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 442 LRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPL 501
+ RG GG+ + + + + S D A LGT P + +VP + PLV ++ E L
Sbjct: 1053 ISRGGSGGNSS--NDKLNGPAPRSRLDPAKVLGTAL-CPRIHEVPLLEPLVCKKIAQERL 1109
Query: 502 SSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLS 551
+ L+F ++ ++TAC+EG + W RPG A + S PS S
Sbjct: 1110 TVLLFLEDCIITACQEGLVCTWARPGKAFTDEETETQAGEASWPRSPSKS 1159
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,056,209,204
Number of Sequences: 2790947
Number of extensions: 50647261
Number of successful extensions: 173355
Number of sequences better than 10.0: 3079
Number of HSP's better than 10.0 without gapping: 1387
Number of HSP's successfully gapped in prelim test: 1754
Number of HSP's that attempted gapping in prelim test: 150335
Number of HSP's gapped (non-prelim): 14453
length of query: 561
length of database: 848,049,833
effective HSP length: 133
effective length of query: 428
effective length of database: 476,853,882
effective search space: 204093461496
effective search space used: 204093461496
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC128638.4


