

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.4 + phase: 0
(301 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
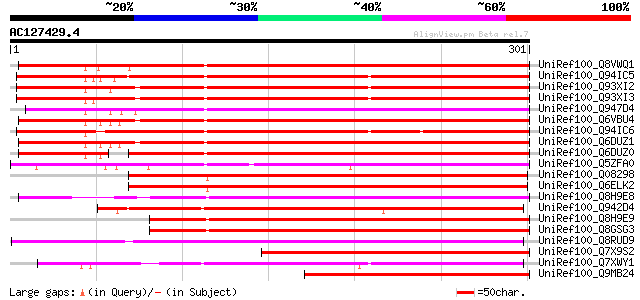
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VWQ1 Dehydration-induced protein RD22-like protein [... 301 1e-80
UniRef100_Q94IC5 BURP domain-containing protein [Bruguiera gymno... 301 2e-80
UniRef100_Q93XI2 BURP domain-containing protein [Bruguiera gymno... 300 4e-80
UniRef100_Q93XI3 BURP domain-containing protein [Bruguiera gymno... 299 5e-80
UniRef100_Q947D4 Dehydration-responsive protein RD22 [Prunus per... 293 3e-78
UniRef100_Q6VBU4 BURP domain-containing protein [Gossypium hirsu... 291 1e-77
UniRef100_Q94IC6 BURP domain-containing protein [Bruguiera gymno... 289 5e-77
UniRef100_Q6DUZ1 Dehydration-induced protein RD22-like protein 1... 288 9e-77
UniRef100_Q6DUZ0 Dehydration-induced protein RD22-like protein 2... 268 2e-70
UniRef100_Q5ZFA0 BURP-domain containing protein [Plantago major] 254 2e-66
UniRef100_Q08298 Dehydration-responsive protein RD22 precursor [... 241 1e-62
UniRef100_Q6ELK2 BURP domain-containing protein [Brassica napus] 237 3e-61
UniRef100_Q8H9E8 Resistant specific protein-3 [Vigna radiata] 231 1e-59
UniRef100_Q942D4 Putative dehydration-responsive protein RD22 [O... 219 5e-56
UniRef100_Q8H9E9 Resistant specific protein-2 [Vigna radiata] 214 2e-54
UniRef100_Q8GSG3 Resistant specific protein-1(4) (Resistant spec... 213 4e-54
UniRef100_Q8RUD9 Seed coat BURP domain protein 1 [Glycine max] 205 1e-51
UniRef100_Q7X9S2 Putative dehydration-induced protein [Gossypium... 201 2e-50
UniRef100_Q7XWY1 OSJNBa0036B17.11 protein [Oryza sativa] 198 1e-49
UniRef100_Q9MB24 S1-3 protein [Vigna unguiculata] 192 7e-48
>UniRef100_Q8VWQ1 Dehydration-induced protein RD22-like protein [Gossypium hirsutum]
Length = 335
Score = 301 bits (772), Expect = 1e-80
Identities = 157/326 (48%), Positives = 208/326 (63%), Gaps = 31/326 (9%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISV------------- 50
L LA+V +HA L PE YW KLP T +PKA+ +ILHP M++ SV
Sbjct: 11 LALAVVVSHAALSPEQYWSYKLPNTPMPKAVKEILHPELMEEKSTSVNVGGGGVNVNTGK 70
Query: 51 -----DGKISVNGKTRIVYDAAP----------IYFYENDANEAELHDNRNLAMFLLEKD 95
D ++V GK V P + Y A+E ++H++ N+A+F LEKD
Sbjct: 71 GKPGGDTHVNVGGKGVGVNTGKPGGGTHVNVGDPFNYLYAASETQIHEDPNVALFFLEKD 130
Query: 96 LHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTI 155
+H G ++ FT+ ++ FLP A IPFSS+KL I N FS+K GS ++E++KNTI
Sbjct: 131 MHPGATMSLHFTENTEKS-AFLPYQTAQKIPFSSDKLPEIFNKFSVKPGSLKAEMMKNTI 189
Query: 156 SECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMG 215
ECE I+GEEK C TSLESMID++ KLG VSTEV ++ +Q+Y IA GV+KM
Sbjct: 190 KECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQAVSTEVEKQTPMQKYTIAAGVQKMT 249
Query: 216 ENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAF 275
++ VVCHK+NY YAVFYCHK++ T+ Y VPLEGADG++ KAVA+CH+DTS W+ KHLAF
Sbjct: 250 DDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEGADGTKAKAVAVCHTDTSAWNPKHLAF 309
Query: 276 QVLKVQQGTFPVCHILQQGQVVWFSK 301
QVLKV+ GT PVCH L + +VW K
Sbjct: 310 QVLKVEPGTIPVCHFLPRDHIVWVPK 335
>UniRef100_Q94IC5 BURP domain-containing protein [Bruguiera gymnorrhiza]
Length = 346
Score = 301 bits (770), Expect = 2e-80
Identities = 166/348 (47%), Positives = 217/348 (61%), Gaps = 54/348 (15%)
Query: 5 VLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP------------------MDDG 46
+L LALVA+ A L PE YW S LP T +PKAI D+LHP +D G
Sbjct: 2 LLMLALVASDAALAPEAYWNSVLPNTPMPKAIKDLLHPDMLEDKGTSVAVGKGGVNVDAG 61
Query: 47 K---------------ISVD---------------GKISVNGK---TRIVYDAAPIYFYE 73
K + VD G +SVN T + + +P + Y
Sbjct: 62 KGKPGGGTHVGVGGKGVGVDAGTPGGGGTNVGVGKGGVSVNTGHKGTPVNVNVSP-FVYR 120
Query: 74 NDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLE 133
A+E ++HDN N+A+F LEKD+ G N+ F+++S+ TFLP VA+SIPFSSNKL+
Sbjct: 121 YAASEDQIHDNPNVALFFLEKDMKPGKSMNLDFSESSNTA-TFLPRHVADSIPFSSNKLQ 179
Query: 134 NILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVS 193
+ FS+K GS E+EI++NT+ ECE GI+GEEK C TSLESM+DF+T KLG ++ +S
Sbjct: 180 DAFREFSVKPGSMEAEIMENTVKECENPGIEGEEKYCATSLESMVDFSTSKLGKDIQAIS 239
Query: 194 TEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGS 253
TEV ++G QQY IA GVKK + VVCHK+NYPYAVFYCH T +T+ Y V L+GAD +
Sbjct: 240 TEVEKQTGRQQYTIA-GVKKTAGDKSVVCHKQNYPYAVFYCHSTQSTRAYLVQLKGADRT 298
Query: 254 RVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
R++AVAICH++TS W+ KHLAFQVL V+ GT P+CH L Q VVW K
Sbjct: 299 RLQAVAICHTNTSAWNPKHLAFQVLAVKPGTVPICHFLPQSHVVWVPK 346
>UniRef100_Q93XI2 BURP domain-containing protein [Bruguiera gymnorrhiza]
Length = 307
Score = 300 bits (767), Expect = 4e-80
Identities = 161/310 (51%), Positives = 203/310 (64%), Gaps = 17/310 (5%)
Query: 5 VLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP---MDDGKISVDGKISVN---- 57
+L LALVA+ A L PE YW S LP T +PKAI D+LHP D G GK N
Sbjct: 2 LLMLALVASGAALAPEAYWNSVLPNTPMPKAIKDLLHPDMLEDKGTSVAVGKGGENVDAG 61
Query: 58 ------GKTRIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSD 111
G T D A Y A+E ++HDN N+A+F L+KDL G N+ F +S+
Sbjct: 62 KGKPGGGGTNFGIDRAVSNIYA--ASEDQIHDNPNVALFFLQKDLKPGKSMNLDFPASSN 119
Query: 112 HGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCV 171
TFLP VA+SIPFSSNKL++ FS+K GS E+EI++NT+ ECE GI+GEEK C
Sbjct: 120 TA-TFLPRHVADSIPFSSNKLQDAFREFSVKPGSLEAEIMENTVKECENPGIEGEEKYCA 178
Query: 172 TSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAV 231
TSLESM+DF+T KLG +V +STEV ++G Q+Y IA GVKK+ + VVCHK+NYPYAV
Sbjct: 179 TSLESMVDFSTSKLGKDVQAISTEVEKQTGRQKYTIA-GVKKIAGDKCVVCHKQNYPYAV 237
Query: 232 FYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHIL 291
FYCH +T+ Y V L+G D +R++AVAICH++TS W+ KH AFQVL V+ GT P CH L
Sbjct: 238 FYCHSIQSTRAYLVQLKGVDRTRLQAVAICHTNTSAWNPKHPAFQVLAVKPGTVPTCHFL 297
Query: 292 QQGQVVWFSK 301
Q VVW K
Sbjct: 298 PQSHVVWVPK 307
>UniRef100_Q93XI3 BURP domain-containing protein [Bruguiera gymnorrhiza]
Length = 336
Score = 299 bits (766), Expect = 5e-80
Identities = 161/330 (48%), Positives = 208/330 (62%), Gaps = 37/330 (11%)
Query: 5 VLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP------------------MDDG 46
+L LALVA+ A L PE YW S LP T +PKAI D+LHP +D G
Sbjct: 11 LLMLALVASGAALAPEAYWNSVLPNTPMPKAIKDLLHPDMLEDKGTSVAVGKGGVNVDAG 70
Query: 47 K---------------ISVDGKISVNGKTRIVYDAAPIYFYENDANEAELHDNRNLAMFL 91
K + VD G T +D A Y A+E ++HDN NLA+F
Sbjct: 71 KGKPGGGTHVGVGGKGVGVDAGKPGGGGTNFGFDRAVSNIYA--ASEDQIHDNPNLALFF 128
Query: 92 LEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIV 151
L+KDL G N+ F ++S+ TFLP VA+SIPFSSNKL++ FS+K GS E+EI+
Sbjct: 129 LQKDLKPGKSMNLDFPESSNTA-TFLPRHVADSIPFSSNKLQDAFREFSVKPGSLEAEIM 187
Query: 152 KNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGV 211
+NT+ ECE GI+GEEK C TSLESM+DF+T KLG ++ +STEV ++G Q+Y IA GV
Sbjct: 188 ENTVKECENPGIEGEEKYCATSLESMVDFSTSKLGKDIQAISTEVEKQTGRQKYTIA-GV 246
Query: 212 KKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLK 271
KK+ + VVCHK+NYPYAVFYCH +T+ Y V L+G D +R++AVAICH++TS W+ K
Sbjct: 247 KKIAGDKCVVCHKQNYPYAVFYCHSIQSTRAYLVQLKGVDRTRLQAVAICHTNTSAWNPK 306
Query: 272 HLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
H AFQVL V+ GT P+CH L Q VVW K
Sbjct: 307 HPAFQVLAVKPGTVPICHFLPQSHVVWVPK 336
>UniRef100_Q947D4 Dehydration-responsive protein RD22 [Prunus persica]
Length = 349
Score = 293 bits (751), Expect = 3e-78
Identities = 158/347 (45%), Positives = 202/347 (57%), Gaps = 56/347 (16%)
Query: 10 LVATHATLPPELYWKSKLPTTQIPKAITDILHP------------------MDDGKISVD 51
LVA+HA LPP+LYW S LP TQ+PK+I+++L P + GK
Sbjct: 1 LVASHAALPPQLYWNSVLPNTQMPKSISELLQPDFAEEKGTTVGVGKGGVNVQAGKGKPG 60
Query: 52 GKISVN----------------GKTRIV-----------YDAAPIYF----------YEN 74
G +VN G T + + P+Y Y
Sbjct: 61 GGTNVNVGGKGGGVGVTTGKPGGGTNVGVGKGGVSVNTGHKGKPVYVGVKPGPNPFNYVY 120
Query: 75 DANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLEN 134
A E +LH NRN A+F LEKD+ GT + F+ S+ F+P A+SIPFSSNKL
Sbjct: 121 AATETQLHGNRNAAIFFLEKDIRPGTSMTLTFSGNSNTA-AFVPRKTADSIPFSSNKLPE 179
Query: 135 ILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVST 194
I + FS+K S E++I+K TI ECE+ GI+GEEK C TSLESM+DF+T KLG N+ +ST
Sbjct: 180 IFSQFSVKPESVEADIIKGTIEECESSGIRGEEKYCATSLESMVDFSTSKLGGNIQAIST 239
Query: 195 EVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSR 254
E + LQ+Y I GVKK+ VVCHK+ YPYAVFYCH T T+ Y VPL+GADG
Sbjct: 240 EAEKGATLQKYTITPGVKKLAAGKSVVCHKQTYPYAVFYCHATKTTRAYVVPLKGADGLE 299
Query: 255 VKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
KAVA+CH+DTS+W+ KHLAFQVLKV+ GT PVCH L + +VW K
Sbjct: 300 AKAVAVCHTDTSEWNPKHLAFQVLKVKPGTVPVCHFLPKDHIVWVPK 346
>UniRef100_Q6VBU4 BURP domain-containing protein [Gossypium hirsutum]
Length = 335
Score = 291 bits (745), Expect = 1e-77
Identities = 154/328 (46%), Positives = 208/328 (62%), Gaps = 35/328 (10%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISVD---GKISVN--- 57
L LA+V +HA L PE YW KLP +PKA+ +ILHP M++ SV+ G ++VN
Sbjct: 11 LALAVVVSHAVLSPEQYWSYKLPNIPMPKAVKEILHPELMEEKSTSVNVGGGGVNVNTGK 70
Query: 58 ----GKTRI-------------------VYDAAPI-YFYENDANEAELHDNRNLAMFLLE 93
G T + V D P Y Y A+E ++H++ N+A+F LE
Sbjct: 71 GKPGGDTHVNVGGKGVGVNTGKPGGGTHVNDPDPFNYLYA--ASETQIHEDPNVALFFLE 128
Query: 94 KDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKN 153
KD+H G ++ F + ++ FLP A IPFSS+KL I N FS+K GS ++E++KN
Sbjct: 129 KDMHPGATMSLHFIENTEKS-AFLPYQTAQKIPFSSDKLPEIFNKFSVKPGSVKAEMMKN 187
Query: 154 TISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKK 213
TI ECE I+GEEK C TSLESMID++ KLG VSTEV ++ +Q+Y IA GV+K
Sbjct: 188 TIKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQAVSTEVEKQTPMQKYTIAAGVQK 247
Query: 214 MGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHL 273
M ++ VVCHK+NY YAVFYCHK++ T+ Y VPLEGA G++ +A+A+CH+DTS W+ KHL
Sbjct: 248 MTDDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEGAGGTKAQALAVCHTDTSAWNPKHL 307
Query: 274 AFQVLKVQQGTFPVCHILQQGQVVWFSK 301
AFQ LKV+ GT PVCH L + +VW K
Sbjct: 308 AFQFLKVEPGTIPVCHFLPRDHIVWVPK 335
>UniRef100_Q94IC6 BURP domain-containing protein [Bruguiera gymnorrhiza]
Length = 292
Score = 289 bits (740), Expect = 5e-77
Identities = 155/299 (51%), Positives = 201/299 (66%), Gaps = 10/299 (3%)
Query: 5 VLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISVDGKISVNGKTRI 62
+L LALVA+ A L PE YW S LP T +PKAI +LHP ++D SV GK +
Sbjct: 2 LLMLALVASDAALAPEAYWNSVLPNTPMPKAIKVLLHPDMLEDKGTSV-----AVGKGAV 56
Query: 63 VYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVA 122
DA Y N A+E ++ DN N+ +F LEKDL G N+ F ++S+ TFLP VA
Sbjct: 57 NVDAGSFINYYNAASEDQIRDNSNITIFFLEKDLKPGKSMNLYFFESSNTA-TFLPRHVA 115
Query: 123 NSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTT 182
+S+PFSSNKL+ FS+K GS E+E +++TI ECE I+GEEK CVTSLES++DF++
Sbjct: 116 DSMPFSSNKLQEAFREFSVKPGSLEAETMEDTIKECENPRIEGEEKYCVTSLESIVDFSS 175
Query: 183 LKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKV 242
KLG ++ +STEV ++G Q+Y IA VKKM + VVCHK+NYPYAVFYCH T ++
Sbjct: 176 SKLGKDIQAISTEVEKQTGRQKYTIA-VVKKMAGDKSVVCHKQNYPYAVFYCHTTQ-SRA 233
Query: 243 YSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
Y V L+GAD S +KAVA CH++TS W+ KH+AFQVL V+ GT PVCH L Q VVW K
Sbjct: 234 YLVQLKGADRSELKAVATCHTNTSAWNPKHMAFQVLAVKPGTVPVCHFLTQSHVVWAPK 292
>UniRef100_Q6DUZ1 Dehydration-induced protein RD22-like protein 1 [Gossypium
arboreum]
Length = 335
Score = 288 bits (738), Expect = 9e-77
Identities = 154/328 (46%), Positives = 207/328 (62%), Gaps = 35/328 (10%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISVD---GKISVN--- 57
L LA+V +HA L PE YW KLP T +PKA+ +ILHP M++ SV+ G ++VN
Sbjct: 11 LALAVVVSHAALSPEQYWSYKLPNTPMPKAVKEILHPELMEEKSTSVNVGGGGVNVNTGK 70
Query: 58 ----GKTRI-------------------VYDAAPI-YFYENDANEAELHDNRNLAMFLLE 93
G T + V D P Y Y A+E ++H++ N+A+F LE
Sbjct: 71 GKPGGDTHVNVGGKGVGVNTGKPGGGTHVNDPDPFNYLYA--ASETQIHEDPNVALFFLE 128
Query: 94 KDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKN 153
KD+H G ++ F + ++ FLP A FSS+KL I N FS+K GS ++E++KN
Sbjct: 129 KDMHPGATMSLHFIENTEKS-AFLPYQTAPKNTFSSDKLPEIFNKFSVKPGSVKAEMMKN 187
Query: 154 TISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKK 213
TI ECE I+GEEK C TSLESMID++ KLG VSTEV ++ +Q+Y IA GV+K
Sbjct: 188 TIKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQAVSTEVEKQTPMQKYTIAAGVQK 247
Query: 214 MGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHL 273
M ++ VVCHK+NY YAVFYCHK++ T+ Y VPLEGA G++ KA+A+CH+DTS W+ KHL
Sbjct: 248 MTDDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEGAGGTKAKALAVCHTDTSAWNPKHL 307
Query: 274 AFQVLKVQQGTFPVCHILQQGQVVWFSK 301
AFQ LKV+ GT PVCH L + +VW K
Sbjct: 308 AFQFLKVEPGTIPVCHFLPRDHIVWVPK 335
>UniRef100_Q6DUZ0 Dehydration-induced protein RD22-like protein 2 [Gossypium
arboreum]
Length = 376
Score = 268 bits (684), Expect = 2e-70
Identities = 126/232 (54%), Positives = 165/232 (70%), Gaps = 1/232 (0%)
Query: 70 YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSS 129
+ Y+ A+E ++HD+ N+A+F LEKDLH G ++ FT+ ++ FLP A IPFSS
Sbjct: 146 FLYQYAASETQIHDDPNVALFFLEKDLHPGATMSLHFTENTEKS-AFLPYQTAQKIPFSS 204
Query: 130 NKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNV 189
N+L I N FS+K GS ++E++KNTI ECE I+GEEK C TSLESMID++ KLG
Sbjct: 205 NELPEIFNKFSVKPGSVKAEMMKNTIKECEQPAIEGEEKYCATSLESMIDYSISKLGKVD 264
Query: 190 DTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEG 249
VSTEV ++ +Y I GV+KM + VVCHK+NY YAVFYCHK++ T+ Y VPLEG
Sbjct: 265 QAVSTEVEKQTPTHKYTITAGVQKMTNDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEG 324
Query: 250 ADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
ADG++ KAVA+CH+DTS W+ KHLAFQVLKV+ GT PVCH L + +VW K
Sbjct: 325 ADGTKAKAVAVCHTDTSAWNPKHLAFQVLKVEPGTIPVCHFLPRDHIVWVPK 376
Score = 55.5 bits (132), Expect = 2e-06
Identities = 29/57 (50%), Positives = 39/57 (67%), Gaps = 5/57 (8%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP--MDDGKISVD---GKISVN 57
L LA+VA+HA L PE YW KLP T +PKA+ +ILHP M++ SV+ G ++VN
Sbjct: 11 LALAVVASHAALSPEQYWSYKLPNTPMPKAVKEILHPELMEEKSTSVNVGGGGVNVN 67
>UniRef100_Q5ZFA0 BURP-domain containing protein [Plantago major]
Length = 348
Score = 254 bits (649), Expect = 2e-66
Identities = 157/348 (45%), Positives = 200/348 (57%), Gaps = 50/348 (14%)
Query: 1 MINVVLQLALVATH---ATLPPELYWKSKLPTTQIPKAITDILH-PMDDGKISVDGKI-- 54
++ +VLQ+A+V A P E YWK+ LP + +P+A+ +L P G + +G+I
Sbjct: 3 LVYLVLQVAIVVIRCDGALSPSERYWKAVLPNSPMPQAVKVLLPTPTGVGVDAANGRIER 62
Query: 55 ----------SVNGKT----------------RIVYDAAPIYFYENDANEA--------- 79
+ NGK RIV AAPI A
Sbjct: 63 HAAGRTIYAAAANGKIERHAAAYTIYAAAANGRIVRHAAPIILIYAAATNGRIERANVTG 122
Query: 80 -ELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNY 138
+LHD+ ++F LEKD+H GTK +QF KTS+ TFLP VA+SIPFSS + IL
Sbjct: 123 TQLHDDPTASLFFLEKDMHAGTKMKLQFVKTSNQA-TFLPRQVADSIPFSSKRFPEILRK 181
Query: 139 FSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN-NVDTVSTEV- 196
F K S E++I+K+TI +CE GIKGEEK C TSLESMIDF T KLG+ NV+ +ST
Sbjct: 182 F--KPTSDEADIMKDTIKDCEDEGIKGEEKFCATSLESMIDFCTSKLGSRNVEAISTNAQ 239
Query: 197 -NGE-SGLQQYVIANGVKKMGENNLVV-CHKRNYPYAVFYCHKTDATKVYSVPLEGADGS 253
N E S ++Y + +KM N VV CHK +Y YAVFYCHK T Y V L ADG
Sbjct: 240 ENAENSSPKEYTLVGAPRKMPGNKAVVACHKMDYAYAVFYCHKIAKTVAYEVSLAAADGC 299
Query: 254 RVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
+V+AVA+CH DTSQW+ KH AFQVLKVQ G+ PVCH L Q Q+VW K
Sbjct: 300 KVEAVAVCHHDTSQWNPKHFAFQVLKVQPGSVPVCHFLPQQQIVWVPK 347
>UniRef100_Q08298 Dehydration-responsive protein RD22 precursor [Arabidopsis
thaliana]
Length = 392
Score = 241 bits (616), Expect = 1e-62
Identities = 123/236 (52%), Positives = 159/236 (67%), Gaps = 5/236 (2%)
Query: 70 YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHG--PTFLPGDVANSIPF 127
+ Y A E +LHD+ N A+F LEKDL G + N++F +G FLP A ++PF
Sbjct: 156 FVYNYAAKETQLHDDPNAALFFLEKDLVRGKEMNVRFNAEDGYGGKTAFLPRGEAETVPF 215
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN 187
S K L FS++ GS E+E++K TI ECEA + GEEK C TSLESM+DF+ KLG
Sbjct: 216 GSEKFSETLKRFSVEAGSEEAEMMKKTIEECEARKVSGEEKYCATSLESMVDFSVSKLGK 275
Query: 188 -NVDTVSTEVNGESG-LQQYVIAN-GVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYS 244
+V VSTEV ++ +Q+Y IA GVKK+ ++ VVCHK+ YP+AVFYCHK T VY+
Sbjct: 276 YHVRAVSTEVAKKNAPMQKYKIAAAGVKKLSDDKSVVCHKQKYPFAVFYCHKAMMTTVYA 335
Query: 245 VPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFS 300
VPLEG +G R KAVA+CH +TS W+ HLAF+VLKV+ GT PVCH L + VVWFS
Sbjct: 336 VPLEGENGMRAKAVAVCHKNTSAWNPNHLAFKVLKVKPGTVPVCHFLPETHVVWFS 391
Score = 33.1 bits (74), Expect = 9.2
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 10 LVATHATLPPELYWKSKLPTTQIPKAITDIL 40
+VA A L PE YW + LP T IP ++ ++L
Sbjct: 16 VVAIAADLTPERYWSTALPNTPIPNSLHNLL 46
>UniRef100_Q6ELK2 BURP domain-containing protein [Brassica napus]
Length = 387
Score = 237 bits (604), Expect = 3e-61
Identities = 121/236 (51%), Positives = 160/236 (67%), Gaps = 5/236 (2%)
Query: 70 YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHG--PTFLPGDVANSIPF 127
+ Y A+E +LHD+ A+F LEKD+ G N++F + FLP A ++PF
Sbjct: 151 FAYNYAASETQLHDDPKAALFFLEKDMVPGKAMNLRFNAEDGYNGKTAFLPRGEAETVPF 210
Query: 128 SSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLG- 186
S K ILN FS+K GS E+E++K TI ECEA + GEEK C TSLESM+DF+ KLG
Sbjct: 211 GSEKSSEILNTFSVKPGSGEAEMMKKTIEECEAKRVGGEEKYCATSLESMVDFSVSKLGK 270
Query: 187 NNVDTVSTEVNGESG-LQQY-VIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYS 244
++V VSTEV ++ +Q+Y + A GVKK+ ++ VVCHK+ YP+AVFYCHK T VY+
Sbjct: 271 DHVRAVSTEVAEKNAPMQKYRIAAAGVKKLSDDKSVVCHKQKYPFAVFYCHKAMMTSVYA 330
Query: 245 VPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFS 300
VPLEG +G R KAVA+CH +TS W+ HLAF+VLKV+ G+ PVCH L + VVWFS
Sbjct: 331 VPLEGENGLRAKAVAVCHKNTSAWNPNHLAFKVLKVKPGSVPVCHFLPETHVVWFS 386
Score = 33.1 bits (74), Expect = 9.2
Identities = 14/33 (42%), Positives = 20/33 (60%)
Query: 4 VVLQLALVATHATLPPELYWKSKLPTTQIPKAI 36
+ L +++ A A L PE YW S LP T IP ++
Sbjct: 8 ICLLVSVTAIAADLTPERYWNSALPNTPIPNSL 40
>UniRef100_Q8H9E8 Resistant specific protein-3 [Vigna radiata]
Length = 275
Score = 231 bits (590), Expect = 1e-59
Identities = 120/296 (40%), Positives = 168/296 (56%), Gaps = 32/296 (10%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTRIVYD 65
L + L+A A LPPE+YW+ LP T IPK I R +
Sbjct: 12 LIVILMAAQAALPPEVYWERMLPNTPIPKVI------------------------RQFSE 47
Query: 66 AAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSI 125
PI + + HD+ + F E+ L G K ++ F K + P L ++A +
Sbjct: 48 LGPIVNHHH-------HDHLKPSNFFSEERLRRGAKLDVLFRKRNFSTP-LLTREIAEHL 99
Query: 126 PFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKL 185
PFSS K+ IL ++K S +++ V+ T++ CE +KGEEK C TS+ESM+DF T KL
Sbjct: 100 PFSSEKINEILEILAVKPDSKDAKNVEETLNHCEKPALKGEEKQCATSVESMVDFVTSKL 159
Query: 186 GNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSV 245
GNN STE+ S Q++++ +GVK + E ++ CH +YPY VFYCHK + + +
Sbjct: 160 GNNARVTSTELEIGSKFQKFIVKDGVKILAEEKIIACHPMSYPYVVFYCHKMANSTAHFL 219
Query: 246 PLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
PLEG DG+RVKAVAICH DTSQW H+AFQVLKV+ GT CH +G +VW++K
Sbjct: 220 PLEGEDGTRVKAVAICHKDTSQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVWYAK 275
>UniRef100_Q942D4 Putative dehydration-responsive protein RD22 [Oryza sativa]
Length = 429
Score = 219 bits (559), Expect = 5e-56
Identities = 119/255 (46%), Positives = 158/255 (61%), Gaps = 10/255 (3%)
Query: 52 GKISVNGKTR---IVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTK 108
G + VN K R + + AP + Y A E +LHD+ N+A+F LEKDLH G + FT
Sbjct: 173 GGVGVNVKPRGKPVHVNVAP-FIYNYAATETQLHDDPNVALFFLEKDLHPGKTMAVHFTA 231
Query: 109 TSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEK 168
T+ G FLP A+++PFSS K+ IL+ FS+K GS E+ + T+ +CEA +GE K
Sbjct: 232 TTA-GEKFLPRSEADAMPFSSEKVPEILSRFSVKPGSVEAAEMAQTLRDCEAPPAQGERK 290
Query: 169 LCVTSLESMIDFTTLKLG-NNVDTVSTEVNGESGLQQYVIANGVKKMG----ENNLVVCH 223
C TSLESM+DF T LG ++V ST V E +Q VK+ ++ LV CH
Sbjct: 291 ACATSLESMVDFATSSLGTSHVRAASTVVGKEGSPEQEYTVTAVKRAAAGGDQDQLVACH 350
Query: 224 KRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQG 283
Y YAVF CH T AT+ Y+V + G DG+ V+AVA+CH+DT+ W+ KH+AFQVLKV+ G
Sbjct: 351 AEPYAYAVFACHLTRATRAYAVSMAGRDGTGVEAVAVCHADTAGWNPKHVAFQVLKVKPG 410
Query: 284 TFPVCHILQQGQVVW 298
T PVCH L Q VVW
Sbjct: 411 TVPVCHFLPQDHVVW 425
Score = 39.3 bits (90), Expect = 0.13
Identities = 19/48 (39%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDG-KISVDG 52
L +A V +HA PE YWKS LP + IP +++ +L G ++V G
Sbjct: 12 LLIASVGSHAARTPEQYWKSALPNSPIPSSLSQLLSTAGGGTSVNVGG 59
>UniRef100_Q8H9E9 Resistant specific protein-2 [Vigna radiata]
Length = 440
Score = 214 bits (545), Expect = 2e-54
Identities = 100/220 (45%), Positives = 139/220 (62%), Gaps = 1/220 (0%)
Query: 82 HDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSI 141
HD+ + + E+ L G K + F K P L ++A +PFSS K+ IL ++
Sbjct: 222 HDHLKPSSYFSEEGLRRGAKLVMLFHKRKFSTP-LLTREIAEHLPFSSEKINEILEILAV 280
Query: 142 KQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESG 201
K S ++ V+ T++ CE +KGEEK C TS+ESM+DF T KLGNN STE+ ES
Sbjct: 281 KPDSKNAKNVEKTLNNCEEPALKGEEKHCATSVESMVDFVTSKLGNNARVTSTELEIESK 340
Query: 202 LQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAIC 261
Q++++ +GVK + E ++ CH +YPY VFYCHK + + VPLEG DG+RVKA+ IC
Sbjct: 341 FQKFIVKDGVKILAEEEIIACHPMSYPYVVFYCHKMSNSTAHVVPLEGEDGTRVKAIVIC 400
Query: 262 HSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
H DTSQW H+AFQVLKV+ GT PVCH G ++W++K
Sbjct: 401 HKDTSQWDPDHVAFQVLKVKPGTSPVCHFFPNGHLLWYAK 440
Score = 39.3 bits (90), Expect = 0.13
Identities = 20/42 (47%), Positives = 25/42 (58%), Gaps = 1/42 (2%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGK 47
L + L+A A+LP E+YW+ KLP T IPK I D GK
Sbjct: 12 LIVILMAAQASLPSEVYWERKLPNTPIPKVIRQ-FSKQDGGK 52
>UniRef100_Q8GSG3 Resistant specific protein-1(4) (Resistant specific protein- 1(8))
[Vigna radiata]
Length = 402
Score = 213 bits (543), Expect = 4e-54
Identities = 100/220 (45%), Positives = 141/220 (63%), Gaps = 1/220 (0%)
Query: 82 HDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSI 141
HD+ + F E+ L G K ++ F K + P L ++A +PFSS K+ IL ++
Sbjct: 184 HDHLKPSNFFSEERLRRGAKLDVLFRKRNFSTP-LLTREIAEHLPFSSEKINEILEILAV 242
Query: 142 KQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESG 201
K S +++ V+ T++ CE +KGEEK C TS+ESM+DF T KLGNN STE+ S
Sbjct: 243 KPDSKDAKNVEETLNHCEKPALKGEEKQCATSVESMVDFVTSKLGNNARVTSTELEIGSK 302
Query: 202 LQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAIC 261
Q++++ +GVK + E ++ CH +YPY VFYCHK + + +PLEG DG+RVKAVAIC
Sbjct: 303 FQKFIVKDGVKILAEEKIIACHPMSYPYVVFYCHKMANSTAHFLPLEGEDGTRVKAVAIC 362
Query: 262 HSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
H DTSQW H+AFQVLKV+ GT CH +G +VW++K
Sbjct: 363 HKDTSQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVWYAK 402
Score = 39.3 bits (90), Expect = 0.13
Identities = 17/31 (54%), Positives = 21/31 (66%)
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAI 36
L + L+A A LPPE+YW+ LP T IPK I
Sbjct: 12 LIVILMAAQAALPPEVYWERMLPNTPIPKVI 42
>UniRef100_Q8RUD9 Seed coat BURP domain protein 1 [Glycine max]
Length = 305
Score = 205 bits (521), Expect = 1e-51
Identities = 109/299 (36%), Positives = 163/299 (54%), Gaps = 6/299 (2%)
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTR 61
I + L L L+ +A L P YW++ LP T +PKAIT++L G +
Sbjct: 8 IFLYLNLMLMTANAALTPRHYWETMLPRTPLPKAITELLSLESRSIFEYAGNDDQSESRS 67
Query: 62 IVYDAAPIYFYENDANEAELHDNRNL-AMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGD 120
I+ A Y D ++ H+ + +F LE+DL G FN++F + LP
Sbjct: 68 ILGYAG----YNQDEDDVSKHNIQIFNRLFFLEEDLRAGKIFNMKFVNNTKATVPLLPRQ 123
Query: 121 VANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDF 180
++ IPFS +K + +L ++ S+ ++I+ TI C+ +GE K C TSLESM+DF
Sbjct: 124 ISKQIPFSEDKKKQVLAMLGVEANSSNAKIIAETIGLCQEPATEGERKHCATSLESMVDF 183
Query: 181 TTLKLGNNVDTVSTEVNGESGLQQYVIA-NGVKKMGENNLVVCHKRNYPYAVFYCHKTDA 239
LG NV STE E+ ++V+ NGV+K+G++ ++ CH +YPY VF CH
Sbjct: 184 VVSALGKNVGAFSTEKERETESGKFVVVKNGVRKLGDDKVIACHPMSYPYVVFGCHLVPR 243
Query: 240 TKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ Y V L+G DG RVKAV CH DTS+W H AF+VL ++ G VCH+ +G ++W
Sbjct: 244 SSGYLVRLKGEDGVRVKAVVACHRDTSKWDHNHGAFKVLNLKPGNGTVCHVFTEGNLLW 302
>UniRef100_Q7X9S2 Putative dehydration-induced protein [Gossypium barbadense]
Length = 156
Score = 201 bits (511), Expect = 2e-50
Identities = 92/155 (59%), Positives = 118/155 (75%)
Query: 147 ESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYV 206
++E++KNTI ECE I+GEEK C TSLESMID++ KLG VSTEV ++ +Q+Y
Sbjct: 2 KAEMMKNTIKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQAVSTEVEKQTPMQKYT 61
Query: 207 IANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTS 266
IA GV+KM ++ VVCHK+NY YAVFYCHK++ T+ Y VPLEGADG++ KAVA+CH+DTS
Sbjct: 62 IAAGVQKMTDDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEGADGTKAKAVAVCHTDTS 121
Query: 267 QWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
W+ KHLAFQVLK + GT PVCH L + +VW K
Sbjct: 122 AWNPKHLAFQVLKAEPGTVPVCHFLPRDHIVWVPK 156
>UniRef100_Q7XWY1 OSJNBa0036B17.11 protein [Oryza sativa]
Length = 285
Score = 198 bits (504), Expect = 1e-49
Identities = 114/293 (38%), Positives = 165/293 (55%), Gaps = 23/293 (7%)
Query: 17 LPPELYWKSKLP-TTQIPKAITDIL---HPMDD--GKISVDGKISVNGKTRIVYDAAPIY 70
+ PE YW+S LP +T +P +I+ +L +P G ++ + I AA
Sbjct: 1 MSPEQYWRSILPDSTPMPISISQLLGDGYPYSPAVGLPKRGDRVQIRYGPNIYGLAASQQ 60
Query: 71 FYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSN 130
F+++ + +F LE +L + F G FLP A+++PFSS
Sbjct: 61 FFKDPT----------MGLFFLETNLQSSKSIKLHFANMMA-GTKFLPRGEADAVPFSSK 109
Query: 131 KLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGN-NV 189
L+ IL F ++ GS ++ +VKNT+ ECE KGE+K C TSLESM+DF LG ++
Sbjct: 110 DLQEILARFGVRPGSVDASVVKNTLLECELPANKGEKKACATSLESMVDFVASSLGTRDI 169
Query: 190 DTVSTEVNGESG---LQQYVIANGVKKMGENN-LVVCHKRNYPYAVFYCHKTDATKVYSV 245
ST + G+ G Q+Y + G ++M E L+ CH +YPYAVF CH T+AT+ Y
Sbjct: 170 KAASTFLVGKDGDTPAQEYTVT-GARRMAETGQLIACHPESYPYAVFMCHLTEATRAYKA 228
Query: 246 PLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
L G DG+ V+AVA+CH+DT++W+ KH AFQVL V+ GT PVCH +Q VVW
Sbjct: 229 SLVGKDGAAVEAVAVCHTDTAEWNPKHAAFQVLGVKPGTVPVCHFVQPDVVVW 281
>UniRef100_Q9MB24 S1-3 protein [Vigna unguiculata]
Length = 132
Score = 192 bits (489), Expect = 7e-48
Identities = 89/130 (68%), Positives = 107/130 (81%)
Query: 172 TSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAV 231
TSLESM+DF+T KLG NV +STEV+ E+GLQQY IA GVKK+ +N VVCHK++YPYAV
Sbjct: 3 TSLESMVDFSTSKLGKNVAVLSTEVDQETGLQQYTIAPGVKKVSGDNAVVCHKQSYPYAV 62
Query: 232 FYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHIL 291
FYCHKT+ T+ Y VPLEGA+G RVKAVA+CH+DTSQW+ KHLAF+VLKV+ GT PVCH L
Sbjct: 63 FYCHKTETTRTYPVPLEGANGIRVKAVAVCHTDTSQWNPKHLAFEVLKVKPGTVPVCHFL 122
Query: 292 QQGQVVWFSK 301
+ VVW K
Sbjct: 123 PEDHVVWVQK 132
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 514,208,851
Number of Sequences: 2790947
Number of extensions: 21500436
Number of successful extensions: 50465
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 50231
Number of HSP's gapped (non-prelim): 114
length of query: 301
length of database: 848,049,833
effective HSP length: 127
effective length of query: 174
effective length of database: 493,599,564
effective search space: 85886324136
effective search space used: 85886324136
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC127429.4


