

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127428.4 + phase: 0
(358 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
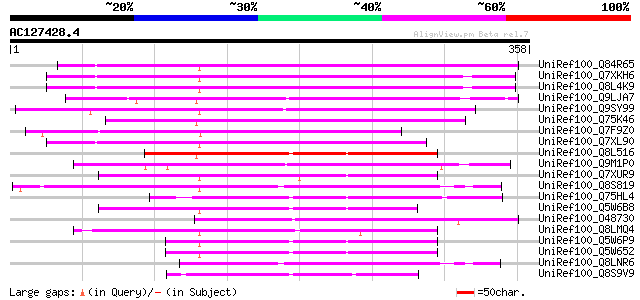
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sa... 224 2e-57
UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa] 220 4e-56
UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sa... 220 5e-56
UniRef100_Q9LJA7 Similarity to En/Spm-like transposon protein [A... 215 2e-54
UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana] 210 6e-53
UniRef100_Q75K46 Hypothetical protein OSJNBb0108E08.7 [Oryza sat... 207 5e-52
UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa] 202 9e-51
UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa] 194 4e-48
UniRef100_Q8L516 Putative transposase [Oryza sativa] 188 2e-46
UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis tha... 181 4e-44
UniRef100_Q7XUR9 OSJNBa0084K11.21 protein [Oryza sativa] 179 8e-44
UniRef100_Q8S819 Putative transposase [Oryza sativa] 173 6e-42
UniRef100_Q75HL4 Expressed protein [Oryza sativa] 173 8e-42
UniRef100_Q5W6B8 Putative polyprotein [Oryza sativa] 172 2e-41
UniRef100_O48730 En/Spm-like transposon protein [Arabidopsis tha... 165 2e-39
UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sat... 163 6e-39
UniRef100_Q5W6P9 Putative polyprotein [Oryza sativa] 155 2e-36
UniRef100_Q5W652 Hypothetical protein OSJNBb0052F16.10 [Oryza sa... 155 2e-36
UniRef100_Q8LNR6 Hypothetical protein OSJNBa0042E19.20 [Oryza sa... 145 2e-33
UniRef100_Q8S9V9 P0519D04.27 protein [Oryza sativa] 141 3e-32
>UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sativa]
Length = 411
Score = 224 bits (572), Expect = 2e-57
Identities = 132/366 (36%), Positives = 181/366 (49%), Gaps = 49/366 (13%)
Query: 34 YATKYLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVE-HDLKSS 92
Y + Y+ K RTS L+G WVQE L P CY M RM + F L LV+ + L +
Sbjct: 41 YHSLYMNKAAPRTSRLSGMGWVQETL-ATPGECYRMLRMNGNTFLALHDLLVDKYKLAPT 99
Query: 93 KHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR----------------------- 129
HM E +A+FL ++G G N+ Q F+HSGET+
Sbjct: 100 VHMHTMEALAIFLYILGDGSSNQRAQNCFKHSGETISRKIEEVLFAVVELGRDIVRPKDP 159
Query: 130 ------------------------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDW 165
+DGTH+ VV ++ R+IGR TQN+MA+CD
Sbjct: 160 NFSNVHERIRKDRRMWPHFKDCIGAVDGTHILAVVPDDDKIRYIGRSKSTTQNVMAICDH 219
Query: 166 NMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCE 225
+M FT+ G G+ HD V AL T FPHPPQGKYYLVD+GYP GY+ PY+ +
Sbjct: 220 DMRFTYASIGQPGSMHDTTVLFNALRTDIDIFPHPPQGKYYLVDAGYPNRPGYLAPYKGQ 279
Query: 226 RYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIV 285
RYH+PEFRR S E FN+ H S+R IER FGVWK ++ +L MP F Q +V
Sbjct: 280 RYHVPEFRRGSAPSGVKEKFNFLHCSVRTIIERCFGVWKMKWRVLLKMPSFPLWKQKMVV 339
Query: 286 VATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARN 345
ATMA+HNFIR D F+ +E + + + + ++ + D + +
Sbjct: 340 AATMALHNFIRDHNAPDRHFHRFERNPDYVPTIPSRFRRYVISQDASDTSTEGQSDVSMD 399
Query: 346 SIRDQI 351
+ RD +
Sbjct: 400 AFRDDL 405
>UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa]
Length = 877
Score = 220 bits (561), Expect = 4e-56
Identities = 142/373 (38%), Positives = 186/373 (49%), Gaps = 55/373 (14%)
Query: 26 ILAALLGEY-ATKYLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHEL 84
+LA ++G Y +Y K P R + +GH WV L N T CY MFRM + +F +L L
Sbjct: 503 MLATMIGTYYLERYSNKAPRRVAPESGHDWVMRTL-ANRTACYNMFRMHRPVFERLHSVL 561
Query: 85 VE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR-------------- 129
VE + LKS+ +M E + +FL +VG R Q+RF S +TV
Sbjct: 562 VESYQLKSTNNMDSMECLGLFLWIVGAPQSVRQAQDRFVRSLQTVHSKFNAVLTALLKLA 621
Query: 130 --------------------------------VIDGTHVSCVVSASEQPRFIGRKGYPTQ 157
IDGTH+ VV S + R +Q
Sbjct: 622 QDIIRPKDPLFTTVHKKLLSPQYTPYLDNCIGAIDGTHIQVVVPNSAAVQHRNRHQEKSQ 681
Query: 158 NIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIG 217
N+M VCD++M FTFVLAGW G+ HD RVF+ A T + FP PP GK+YLVDSGYP G
Sbjct: 682 NVMCVCDFDMRFTFVLAGWPGSVHDMRVFNDAQTRFSAKFPKPPLGKFYLVDSGYPNRPG 741
Query: 218 YIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFK 277
Y+ PY+ YH E+ S+ E FNY HSS R IER+FGV KN++ IL S+P +
Sbjct: 742 YLAPYKGITYHFHEYNESTLPRGRREHFNYCHSSCRNVIERSFGVLKNKWRILFSLPSYS 801
Query: 278 YETQVHIVVATMAIHNFIRRSAEMDVDF-NLYEDENTVIHHDDDHRSTNLNQSQSFNVAS 336
E Q I+ A +A+HNFIR S D +F N DEN D T+ S+ N
Sbjct: 802 QEKQSRIIHACIALHNFIRDSQMADTEFDNCDHDENY-----DPLGGTSSPSSEPTNELD 856
Query: 337 SSEMDHARNSIRD 349
S M+ R+ I D
Sbjct: 857 SRVMNQFRDWIAD 869
>UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sativa]
Length = 916
Score = 220 bits (560), Expect = 5e-56
Identities = 142/373 (38%), Positives = 188/373 (50%), Gaps = 55/373 (14%)
Query: 26 ILAALLGEYATK-YLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHEL 84
+LAA++G Y + Y K P R + +GH WV L N T CY MFRM + +F +L L
Sbjct: 542 MLAAMIGTYYLELYSNKAPRRVAPESGHDWVMRTL-ANRTACYNMFRMHRPVFERLHSVL 600
Query: 85 VE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR-------------- 129
VE ++LKS+ +M E + +FL +VG R Q+RF S +TV
Sbjct: 601 VESYELKSTNNMDSMECLGLFLWIVGAPQSVRQAQDRFVRSLKTVHSKFKAVLTALLKLA 660
Query: 130 --------------------------------VIDGTHVSCVVSASEQPRFIGRKGYPTQ 157
IDGTH+ VV S + R +Q
Sbjct: 661 QDIIRPKDPLFTTVHKKLLSPQYTPYLDNCIGAIDGTHIQVVVPNSAAVQHRNRHKEKSQ 720
Query: 158 NIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIG 217
N+M VCD++M F+FVLAGW G+ HD RVF+ A T + FP PP GK+YLVDSGYP G
Sbjct: 721 NVMCVCDFDMRFSFVLAGWPGSVHDMRVFNDAHTRFSAKFPKPPPGKFYLVDSGYPNRPG 780
Query: 218 YIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFK 277
Y+ PY+ YH E+ S+ E FNY HSS R IER+FGV KN++ IL S+P +
Sbjct: 781 YLAPYKGITYHFQEYNESTLPRGKREHFNYCHSSCRNVIERSFGVLKNKWRILFSLPSYS 840
Query: 278 YETQVHIVVATMAIHNFIRRSAEMDVDF-NLYEDENTVIHHDDDHRSTNLNQSQSFNVAS 336
E Q I+ A +A+HNFIR S D +F N DEN D T+ S+ N
Sbjct: 841 QEKQSRIIHACIALHNFIRDSQMADTEFDNCDHDENY-----DPLGGTSSPSSEPTNELD 895
Query: 337 SSEMDHARNSIRD 349
S M+ R+ I D
Sbjct: 896 SRVMNQFRDWIAD 908
>UniRef100_Q9LJA7 Similarity to En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 329
Score = 215 bits (547), Expect = 2e-54
Identities = 120/329 (36%), Positives = 183/329 (55%), Gaps = 25/329 (7%)
Query: 39 LCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVE--HDLKSSKHMG 96
+ K+P G +V++++ G+P +C + RM +F LC E++E + ++SS+++
Sbjct: 1 MVKQPLCAHPNCGRQFVEDLIHGHPYKCDFLLRMRLKVFFDLC-EIIEKTYKIRSSQNLS 59
Query: 97 VEEMVAMFLVVVGHGVGNRMIQERFQHSGET--------------VRVIDGTHVSCVVSA 142
V+E A+FL + GH R I F HS ET V +DGTHV +VS
Sbjct: 60 VKESGAIFLYICGHNASQRSIMRMFGHSQETICRKIYEVLNLIGIVGAMDGTHVPAMVSG 119
Query: 143 SEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQ 202
+ R+ RK + NI+AVC+++M FT++ G G+AHD +V A+ + NFPHP
Sbjct: 120 RDHQRYWNRKSICSMNILAVCNFDMLFTYIYVGVFGSAHDTKVLSLAME-GDPNFPHPHI 178
Query: 203 GKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGV 262
GKYYLVDSGY GY+GP+R YH +F+ + NH E FN+ HS LRC IERTFGV
Sbjct: 179 GKYYLVDSGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHSLLRCVIERTFGV 238
Query: 263 WKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDDDHR 322
WK ++ I++ + T I+VATMA+HNF+ + D+DF++ + D+DH
Sbjct: 239 WKGKWRIMQDRAWYNIVTTRKIMVATMALHNFVWKLGIPDLDFDIDWMQ------DNDHH 292
Query: 323 STNLNQSQSFNVASSSEMDHARNSIRDQI 351
T ++ ++ + H RD+I
Sbjct: 293 PTLDDEDETVEQDMTGSRQH-MEGFRDEI 320
>UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana]
Length = 404
Score = 210 bits (534), Expect = 6e-53
Identities = 134/369 (36%), Positives = 186/369 (50%), Gaps = 53/369 (14%)
Query: 5 KEQLEHNTQEEEDDDTFEESYILAALLGE-YATKYLCKEPCRTSELTGHAW--VQEILQG 61
KE ++++ D EE + AL+ E Y K + E G W VQ ++
Sbjct: 4 KETFRFLVEDDDMDLVLEEENNMYALVKELYGGKNVNTERQLVRINRGGGWRRVQRLMVE 63
Query: 62 NPTRCYEMFRMEKHIFHKLCHELVE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQER 120
+ C+++ RM + F LC L E + L+ S ++ +EE VAMF+ +V + R+I ER
Sbjct: 64 SKVECFDILRMHQSTFRTLCKILSEQYKLEESCNIYLEESVAMFIEMVAQDLTVRVIAER 123
Query: 121 FQHSGETVR-----------------------------------------------VIDG 133
+QHS ETV+ +DG
Sbjct: 124 YQHSLETVKRKLDEVLSALLKLAADIVKPTRDDVTGISPFLVNDKRYMPYFIDCIGALDG 183
Query: 134 THVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTA 193
THVS + + R+ GRK T NI+AVC+++M F G G AHD +V T
Sbjct: 184 THVSVRPPSGDVERYRGRKSEATMNILAVCNFSMKFNIAYVGVPGRAHDTKVLTYC-ATH 242
Query: 194 NLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLR 253
+FPHPP GKYYLVDSGYPT GY+GP+R RYHL F R N E+FN HSSLR
Sbjct: 243 EASFPHPPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLR 302
Query: 254 CTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDEN 312
IERTFGVWK ++ IL R PK++ + + IV +TMA+HN+IR S + D DF +E
Sbjct: 303 SVIERTFGVWKAKWRILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVE 362
Query: 313 TVIHHDDDH 321
+ H D++
Sbjct: 363 SYEQHGDEN 371
>UniRef100_Q75K46 Hypothetical protein OSJNBb0108E08.7 [Oryza sativa]
Length = 767
Score = 207 bits (526), Expect = 5e-52
Identities = 119/275 (43%), Positives = 158/275 (57%), Gaps = 27/275 (9%)
Query: 67 YEMFRMEKHIFHKLCHELVE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSG 125
Y M+RM +F +L LV+ ++LKSS E +AMFL +VG R ++RF+ S
Sbjct: 457 YGMYRMSPTMFLRLHDLLVQSYELKSSSKSTSVEALAMFLWMVGAPQSIRQAEDRFERSM 516
Query: 126 ET-------------------------VRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIM 160
T + IDGTHV CVV +++ + + KG TQN+M
Sbjct: 517 GTLVDPQFTTMHTRLRNRRFFPYFKDCIGAIDGTHVPCVVPSNKLVQHLCCKGMTTQNVM 576
Query: 161 AVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIG 220
VCD++M FTFVLAGW G+ HD RVFD A TT FPH P GKYYLVDSGYP GY+
Sbjct: 577 VVCDFDMRFTFVLAGWPGSVHDMRVFDDAHTTYTHVFPHSPTGKYYLVDSGYPNRPGYLA 636
Query: 221 PYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYET 280
PY+ +YHL E+R + + E FNY HSSLR IER+FGV K ++ +L +P +
Sbjct: 637 PYKGTKYHLQEYRDAPEPQGKEENFNYAHSSLRNVIERSFGVLKMKWRMLEKIPSYDPRK 696
Query: 281 QVHIVVATMAIHNFIRRSAEMDVDFNLYE-DENTV 314
Q I++A A+HNFIR+S D F+ + DEN V
Sbjct: 697 QTQIIIACCALHNFIRKSGIRDKHFDRCDRDENYV 731
>UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa]
Length = 728
Score = 202 bits (515), Expect = 9e-51
Identities = 121/317 (38%), Positives = 165/317 (51%), Gaps = 59/317 (18%)
Query: 12 TQEEEDDDTF-----------EESYILAALLGEYATKYLCKEPCRTSELTGHAWVQEILQ 60
+ +E++D+TF + +L A YA YL K R + L G+ WV L
Sbjct: 400 SDDEDEDETFVVASRQLDMMRKVGPVLTAFGMLYAETYLNKSKYREATLAGYDWVMRNLN 459
Query: 61 GNPTRCYEMFRMEKHIFHKLCHELV-EHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQE 119
P CY+MFRM + +F +L + LV + LKS+ M E + MFL +G ++
Sbjct: 460 F-PQDCYDMFRMSRPLFERLHNLLVTSYGLKSTSKMDSIEALGMFLWTIGAPQSFVQVKN 518
Query: 120 RFQHSGETVRV----------------------------------------------IDG 133
RF+ S T+ V IDG
Sbjct: 519 RFERSKGTISVKFEEVLQSVYLLSKDLVKPRDPHFTTIHPRLHGDRFQPHFNNCIGAIDG 578
Query: 134 THVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTA 193
TH+ VV AS+ + +GR YPTQN++AVCD++M FTF++AGW G+AHD RVF+ AL
Sbjct: 579 THILVVVPASKVVQHVGRNKYPTQNVLAVCDFDMRFTFIVAGWPGSAHDMRVFNDALCKY 638
Query: 194 NLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLR 253
FPHPP GK+YLVDSGYP +G++ PY+ +YHLPEFR EVFN+ HSSLR
Sbjct: 639 ATIFPHPPPGKFYLVDSGYPNRMGFLAPYKGTKYHLPEFRAGPRPSGKKEVFNHLHSSLR 698
Query: 254 CTIERTFGVWKNRFAIL 270
IER+FGV K ++ IL
Sbjct: 699 NVIERSFGVLKMKWRIL 715
>UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa]
Length = 804
Score = 194 bits (492), Expect = 4e-48
Identities = 120/310 (38%), Positives = 158/310 (50%), Gaps = 49/310 (15%)
Query: 26 ILAALLGEYATK-YLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHEL 84
+LAA++G Y + Y K P R + +GH WV L N T CY MFRM + +F +L L
Sbjct: 496 MLAAMIGTYYLELYSNKAPRRVAPESGHDWVMRTL-ANRTVCYNMFRMHRPVFERLHSVL 554
Query: 85 VE-HDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR-------------- 129
VE ++LKS+ +M E + +FL +VG R Q+RF S +TV
Sbjct: 555 VESYELKSTNNMDSMECLGLFLWIVGAPQSVRQAQDRFVRSLKTVHSKFKAVLTALLKLA 614
Query: 130 --------------------------------VIDGTHVSCVVSASEQPRFIGRKGYPTQ 157
IDGTH+ VV S + R +Q
Sbjct: 615 QDIIRPKDPLFTTVHKKLLSPQYTPYLDNCIGAIDGTHIQVVVPNSAAVQHRNRHEEKSQ 674
Query: 158 NIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIG 217
N+M VCD++M FTFVLAGW G+ HD RVF+ A T + FP P GK+YLVDSGYP G
Sbjct: 675 NVMCVCDFDMRFTFVLAGWPGSVHDMRVFNDAQTRFSAKFPKLPLGKFYLVDSGYPNRPG 734
Query: 218 YIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFK 277
Y+ PY+ YH E+ S+ E FNY HSS R IER+FGV KN++ IL S+P +
Sbjct: 735 YLAPYKGITYHFQEYNESTLPRGRREHFNYCHSSCRNVIERSFGVLKNKWRILFSLPSYS 794
Query: 278 YETQVHIVVA 287
E Q I+ A
Sbjct: 795 QEKQSRIIHA 804
>UniRef100_Q8L516 Putative transposase [Oryza sativa]
Length = 535
Score = 188 bits (477), Expect = 2e-46
Identities = 101/205 (49%), Positives = 132/205 (64%), Gaps = 6/205 (2%)
Query: 94 HMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGET--VRVIDGTHVSCVVSASEQPRFIGR 151
HM +EE VAMFL VGH + NR+++ + SGET + IDGTH+ V + + F GR
Sbjct: 21 HMCIEEQVAMFLHTVGHNLRNRLVRTNYGRSGETDCIGAIDGTHIRASVRKNVESSFRGR 80
Query: 152 KGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSG 211
K + TQN+MA D+++ FT+VLAGWEGTAHDA V AL N H PQGK+YLVD G
Sbjct: 81 KSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERE--NGLHVPQGKFYLVDGG 138
Query: 212 YPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAIL- 270
Y G++ P+R RYHL E+ ++ +N E+FN HSSLR T+ER FG K RF +L
Sbjct: 139 YGAKQGFLPPFRAVRYHLKEW-GNNPVQNEKELFNLRHSSLRITVERAFGSLKRRFKVLD 197
Query: 271 RSMPKFKYETQVHIVVATMAIHNFI 295
+ P F + TQV IVVA IHN++
Sbjct: 198 DATPFFPFRTQVDIVVACCIIHNWV 222
>UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis thaliana]
Length = 377
Score = 181 bits (458), Expect = 4e-44
Identities = 102/319 (31%), Positives = 173/319 (53%), Gaps = 25/319 (7%)
Query: 45 RTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDLKSS------KHMGVE 98
R+ G+ ++Q L NP +++RME +FHKLC ++ ++ S KH +
Sbjct: 50 RSVSRVGYNYIQTALTVNPAHFRQLYRMEPEVFHKLCDVILLYNGHISEIKLFYKHKFHK 109
Query: 99 EMVAMFLVV--------VGHGVGNRMIQERFQHSGETVRVIDGTHVSCVVSASEQPRFIG 150
+ A+ V V R + + + + IDGTH+ +V + F
Sbjct: 110 ALKALAFVAPNWMAKPEVRVPAKIRESTSFYPYFKDCIGAIDGTHIFAMVPTCDAASFRN 169
Query: 151 RKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDS 210
RKG+ +QN++A C++++ F ++L+GW+G+AHD++V + AL T N N P+GK+YLVD
Sbjct: 170 RKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDAL-TRNTNRLQVPEGKFYLVDC 228
Query: 211 GYPTPIGYIGPYRCERYHLPEFR-RSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAI 269
GY ++ P+R RYHL +FR + + NE+FN H+SLR IER FG++K+RF I
Sbjct: 229 GYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIFGIFKSRFLI 288
Query: 270 LRSMPKFKYETQVHIVVATMAIHNFIR---RSAEMDVDFNLYEDENTVIHHDDDHRSTNL 326
+S P + ++TQ +V+A +HNF+ RS E + ++ E+ + DD+ N
Sbjct: 289 FKSAPPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEE------NSDDNEENNY 342
Query: 327 NQSQSFNVASSSEMDHARN 345
++ + S + ++A N
Sbjct: 343 EENGDVGILESQQREYANN 361
>UniRef100_Q7XUR9 OSJNBa0084K11.21 protein [Oryza sativa]
Length = 377
Score = 179 bits (455), Expect = 8e-44
Identities = 111/283 (39%), Positives = 148/283 (52%), Gaps = 50/283 (17%)
Query: 62 NPTRCYEMFRMEKHIFHKLCHELVEHDL-KSSKHMGVEEMVAMFLVVVGHGVGNRMIQER 120
N T C M R+ + F + C + L + + HM +EE VAMFL VGH + NR+++
Sbjct: 42 NDTTCINMLRLRRASFFRFCKLFRDRGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTN 101
Query: 121 FQHSGETVR--------------------------------------------VIDGTHV 136
F S ETV IDGTH+
Sbjct: 102 FDRSRETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPYFKDCIGAIDGTHI 161
Query: 137 SCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTAN-L 195
V + + F GRK + TQN+MA D+++ FT+VLAGWEGTAHDA V AL N L
Sbjct: 162 RASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERENGL 221
Query: 196 NFP--HPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLR 253
P + QGKYYLVD+GY G++ P+R RYHL E+ ++ +N E+FN HSSLR
Sbjct: 222 RVPQGNRLQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKELFNLRHSSLR 280
Query: 254 CTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFI 295
T+ER FG K RF +L + P F ++TQV IVVA IHN++
Sbjct: 281 VTVERAFGSLKRRFKVLDDATPFFPFQTQVDIVVACCIIHNWV 323
>UniRef100_Q8S819 Putative transposase [Oryza sativa]
Length = 1003
Score = 173 bits (439), Expect = 6e-42
Identities = 124/387 (32%), Positives = 179/387 (46%), Gaps = 70/387 (18%)
Query: 3 FWKE---QLEHNTQEEEDDDTFEESYILAALLGEYATKYLCKEPCRTSELTGHAWVQEIL 59
+W+ L +++EED++ + L AL + + K TS ++ IL
Sbjct: 260 YWRRGMNSLIRKSKDEEDEEII--MFWLPALYLLTSNGGIEKRVRHTSSQYSEEKLRNIL 317
Query: 60 QGNPTRCYEMFRMEKHIFHKLCHEL-VEHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQ 118
+G+ C FRME +IF + L EH L+ ++ + VEE + FL ++ H +Q
Sbjct: 318 EGHEKNCLVAFRMEPNIFRAIVTYLRTEHLLRDTRGITVEEKLGHFLYMISHNASYEDLQ 377
Query: 119 ERFQHSGETVR---------------------------------------------VIDG 133
F HSGET+ IDG
Sbjct: 378 HEFHHSGETIHRHIKAVFKVIPSLTYRFIKQTTRVETHWKISTDQLFFPYFQNCLGAIDG 437
Query: 134 THVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTA 193
THV +S Q + RKG +QN+M VCD+++ F F+ +GWEG+A DARV L +A
Sbjct: 438 THVPITISQDLQAPYRNRKGTLSQNVMLVCDFDLNFLFIPSGWEGSATDARV----LRSA 493
Query: 194 NLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGF-ENHNEVFNYYHSSL 252
L + PQGKYYLVD GY ++ PYR RYHL EF R N+ E+FN+ H+ L
Sbjct: 494 MLKGFNVPQGKYYLVDGGYANTPSFLAPYRGVRYHLKEFGRGQQRPRNYKELFNHRHAIL 553
Query: 253 RCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDEN 312
R IER GV K RF IL+ + + QV I VAT+ HN IR L DE
Sbjct: 554 RNHIERAIGVLKKRFPILKVGTHHRIKNQVKIPVATVVFHNLIRM---------LNGDEG 604
Query: 313 TVIHHDDDHRSTNLNQSQSFNVASSSE 339
+ +H+ +N++ Q +V +
Sbjct: 605 WL-----NHQGSNISPEQFIDVPEGDD 626
>UniRef100_Q75HL4 Expressed protein [Oryza sativa]
Length = 608
Score = 173 bits (438), Expect = 8e-42
Identities = 102/245 (41%), Positives = 139/245 (56%), Gaps = 17/245 (6%)
Query: 97 VEEMVAMFLVVVGHGVGNRMIQERFQHSGETVRVIDGTHVSCVVSASEQPRFIGRKGYPT 156
VEE VAMFL +GH + NR + + IDGTHV V + + F GRK + T
Sbjct: 120 VEEQVAMFLHTIGHNLRNR----------DCIGAIDGTHVRASVPKNMESSFRGRKNHAT 169
Query: 157 QNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPI 216
QN+MA D+++ FT+VLAGWEG AHD V AL N H PQGK+YLVD GY
Sbjct: 170 QNVMAAVDFDLRFTYVLAGWEGAAHDVVVLRDALERE--NGLHVPQGKFYLVDVGYGAKP 227
Query: 217 GYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAIL-RSMPK 275
G++ P+R RYHL E+ ++ +N E+FN HSSLR T+ER FG K RF IL + P
Sbjct: 228 GFLPPFRSTRYHLNEW-GNNPVQNEKELFNLRHSSLRITVERAFGSLKRRFKILDDATPF 286
Query: 276 FKYETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVA 335
F ++TQV+IVVA IHN++ D+D + ++I +D + N + +
Sbjct: 287 FPFQTQVNIVVACCIIHNWVIND---DIDELVMGPNESIIQDNDTSTDMDTNNGKGVVAS 343
Query: 336 SSSEM 340
+S M
Sbjct: 344 WTSSM 348
>UniRef100_Q5W6B8 Putative polyprotein [Oryza sativa]
Length = 561
Score = 172 bits (435), Expect = 2e-41
Identities = 103/266 (38%), Positives = 138/266 (51%), Gaps = 49/266 (18%)
Query: 62 NPTRCYEMFRMEKHIFHKLCHELVEHDL-KSSKHMGVEEMVAMFLVVVGHGVGNRMIQER 120
N T C +M R+ K F + C + L + + HM +EE VAMFL VGH + NR+++
Sbjct: 19 NDTTCIDMLRLRKGSFFRFCKLFRDRGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTN 78
Query: 121 FQHSGETVR--------------------------------------------VIDGTHV 136
+ SGETV IDGTH+
Sbjct: 79 YSRSGETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPYFKDCIGAIDGTHI 138
Query: 137 SCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLN 196
V + + F GRK + TQN+MA D+++ FT+VLAGWEGTAHDA V AL N
Sbjct: 139 RASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERE--N 196
Query: 197 FPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNYYHSSLRCTI 256
H PQGK+YLVD+GY G++ P+R RYHL E+ ++ +N E+FN HSSLR T+
Sbjct: 197 GLHVPQGKFYLVDAGYGAKQGFLPPFRAVRYHLKEW-GNNPVQNEKELFNLRHSSLRITV 255
Query: 257 ERTFGVWKNRFAIL-RSMPKFKYETQ 281
ER FG K RF +L + P F + TQ
Sbjct: 256 ERAFGSLKRRFKVLDDATPFFPFRTQ 281
>UniRef100_O48730 En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 292
Score = 165 bits (417), Expect = 2e-39
Identities = 94/238 (39%), Positives = 126/238 (52%), Gaps = 15/238 (6%)
Query: 128 VRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFD 187
V +DGTHV + ++ GRK PT N++A+C+++M F + G G AHD +V +
Sbjct: 45 VGALDGTHVPVRPPSQTAKKYKGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKVLN 104
Query: 188 KALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNEVFNY 247
T F HPP GKYYLVDSGYPT GY+GP+R RYHL +F R E+FN
Sbjct: 105 YCATNEPY-FSHPPNGKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTARELFNR 163
Query: 248 YHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFIRRSAEMDVDFN 306
HS LR IERTFGVWK ++ I+ R PK+ + IV ATMA+HNFIR S D DF
Sbjct: 164 KHSGLRSVIERTFGVWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHREDHDFL 223
Query: 307 LY-------------EDENTVIHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQI 351
+ E+E ++ + + +VA D A ++RD I
Sbjct: 224 QWQSIEEYHVDDEEEEEEEEGEEEEEGEEEEGEGEEEEEHVAYEPTGDRAMEALRDNI 281
>UniRef100_Q8LMQ4 Hypothetical protein OSJNBa0011L14.7 [Oryza sativa]
Length = 1202
Score = 163 bits (413), Expect = 6e-39
Identities = 110/304 (36%), Positives = 145/304 (47%), Gaps = 62/304 (20%)
Query: 45 RTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDLK-SSKHMGVEEMVAM 103
RTSEL ++ + C + RM + +F+KLC L L + H+ VEE VAM
Sbjct: 577 RTSELI------RLINISDRTCTQQLRMSRAVFYKLCARLRNKGLLVDTFHVSVEEQVAM 630
Query: 104 FLVVVGHGVGNRMIQERFQHSGETVR---------------------------------- 129
FL VG + F SGETV
Sbjct: 631 FLKKVGQHHSVSCVGFSFWRSGETVSRYFRIVLRAMCEIARELIYIRSTNTHSKITSKKN 690
Query: 130 -----------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEG 178
+DGTH+ V A + RF GRK YPTQN++A D+++ F ++LAGWEG
Sbjct: 691 KFYPYFKDCIGALDGTHIRASVPAKKVDRFRGRKPYPTQNVLAAVDFDLRFIYILAGWEG 750
Query: 179 TAHDARVFDKALTTAN-LNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSG 237
+AHD+ V AL+ N L +GK++L D+GY G + YR RYHL EFR G
Sbjct: 751 SAHDSLVLQDALSRPNGLKI---LEGKFFLADAGYAARPGILPSYRRVRYHLKEFRGPQG 807
Query: 238 FEN------HNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAI 291
E+FNY HSSLR TIER FG KNRF IL + P + Q +V+A A+
Sbjct: 808 PHGPQGPKCPKELFNYRHSSLRTTIERAFGALKNRFKILMNKPFIPLKAQSMVVIACCAL 867
Query: 292 HNFI 295
HN+I
Sbjct: 868 HNWI 871
>UniRef100_Q5W6P9 Putative polyprotein [Oryza sativa]
Length = 1067
Score = 155 bits (392), Expect = 2e-36
Identities = 93/233 (39%), Positives = 121/233 (51%), Gaps = 48/233 (20%)
Query: 108 VGHGVGNRMIQERFQHSGETVR-------------------------------------- 129
VGH + NR+++ F SGETV
Sbjct: 18 VGHNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPY 77
Query: 130 ------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDA 183
IDGTH+ V + + F GRK + TQN+MA D+++ FT+VLAGWEGT HDA
Sbjct: 78 FKDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDA 137
Query: 184 RVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNE 243
V AL N PQGKYYLVD+GY G++ P+R RYHL E+ ++ +N E
Sbjct: 138 VVLRDALERE--NSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKE 194
Query: 244 VFNYYHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFI 295
+FN HSSLR T+ER FG K RF +L + P F + TQV IVVA IHN++
Sbjct: 195 LFNLRHSSLRVTVERAFGSLKRRFKVLDDATPFFPFRTQVDIVVACCIIHNWV 247
>UniRef100_Q5W652 Hypothetical protein OSJNBb0052F16.10 [Oryza sativa]
Length = 1220
Score = 155 bits (392), Expect = 2e-36
Identities = 93/233 (39%), Positives = 121/233 (51%), Gaps = 48/233 (20%)
Query: 108 VGHGVGNRMIQERFQHSGETVR-------------------------------------- 129
VGH + NR+++ F SGETV
Sbjct: 171 VGHNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPY 230
Query: 130 ------VIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDA 183
IDGTH+ V + + F GRK + TQN+MA D+++ FT+VLAGWEGT HDA
Sbjct: 231 FKDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDA 290
Query: 184 RVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFRRSSGFENHNE 243
V AL N PQGKYYLVD+GY G++ P+R RYHL E+ ++ +N E
Sbjct: 291 VVLRDALERE--NSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEW-GNNPVQNEKE 347
Query: 244 VFNYYHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQVHIVVATMAIHNFI 295
+FN HSSLR T+ER FG K RF +L + P F + TQV IVVA IHN++
Sbjct: 348 LFNLRHSSLRVTVERAFGSLKRRFKVLDDATPFFPFRTQVDIVVACCIIHNWV 400
>UniRef100_Q8LNR6 Hypothetical protein OSJNBa0042E19.20 [Oryza sativa]
Length = 481
Score = 145 bits (366), Expect = 2e-33
Identities = 89/222 (40%), Positives = 122/222 (54%), Gaps = 19/222 (8%)
Query: 118 QERFQHSGETVRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWE 177
Q+ F + + IDGTHV +S Q + RKG +QN+M VCD+++ F+F+ +GWE
Sbjct: 31 QQFFPYFQNCLGAIDGTHVPITISQDLQAPYRNRKGTLSQNVMLVCDFDLNFSFISSGWE 90
Query: 178 GTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYHLPEFR-RSS 236
G+A DARV L +A L + PQGKYYLVD GY ++ PYR RYHL EF
Sbjct: 91 GSATDARV----LRSAMLKGFNVPQGKYYLVDGGYANTPSFLAPYRGVRYHLKEFGCGQQ 146
Query: 237 GFENHNEVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIR 296
N+ E+FN+ H+ LR IERT GV K RF IL+ E QV I VAT+ HN IR
Sbjct: 147 RPRNYKELFNHRHAILRNHIERTIGVLKKRFPILKVGTHHSIENQVKIPVATVVFHNLIR 206
Query: 297 RSAEMDVDFNLYEDENTVIHHDDDHRSTNLNQSQSFNVASSS 338
L DE + +H+ +N++ Q +V ++
Sbjct: 207 M---------LNGDEGWL-----NHQGSNISPEQFIDVPEAT 234
>UniRef100_Q8S9V9 P0519D04.27 protein [Oryza sativa]
Length = 538
Score = 141 bits (355), Expect = 3e-32
Identities = 80/175 (45%), Positives = 105/175 (59%), Gaps = 8/175 (4%)
Query: 109 GHGVGNRMIQERFQHSGETVRVIDGTHVSCVVSASEQPRFIGRKGYPTQNIMAVCDWNMC 168
G G G R + Q + + IDGTH+ V + + F GRK + TQN+MA D+++
Sbjct: 95 GRGGGARRAR---QLQEDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLR 151
Query: 169 FTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQGKYYLVDSGYPTPIGYIGPYRCERYH 228
FT+VLAGWEGTAHDA V AL N H PQGK+YLVD+GY G++ P+R RYH
Sbjct: 152 FTYVLAGWEGTAHDAVVLRDALERE--NGIHVPQGKFYLVDAGYGAKQGFLPPFRAVRYH 209
Query: 229 LPEFRRSSGFENHNEVFNYYHSSLRCTIERTFGVWKNRFAIL-RSMPKFKYETQV 282
L E+ + + E+FN HSSLR T+ER FG K RF +L + P F + TQV
Sbjct: 210 LKEWGNNP--VQNEELFNLRHSSLRITVERAFGSLKRRFKVLDDATPFFPFRTQV 262
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 622,731,140
Number of Sequences: 2790947
Number of extensions: 26199667
Number of successful extensions: 62121
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 131
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 61778
Number of HSP's gapped (non-prelim): 244
length of query: 358
length of database: 848,049,833
effective HSP length: 128
effective length of query: 230
effective length of database: 490,808,617
effective search space: 112885981910
effective search space used: 112885981910
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC127428.4


