

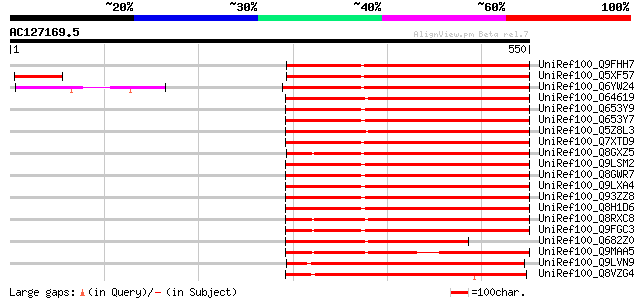
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.5 + phase: 0 /pseudo
(550 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FHH7 Similarity to unknown protein [Arabidopsis thal... 394 e-108
UniRef100_Q5XF57 At5g57670 [Arabidopsis thaliana] 394 e-108
UniRef100_Q6YW24 SERK1 protein-like [Oryza sativa] 387 e-106
UniRef100_O64619 Hypothetical protein At2g18890 [Arabidopsis tha... 330 9e-89
UniRef100_Q653Y9 Putative receptor protein kinase PERK1 [Oryza s... 316 1e-84
UniRef100_Q653Y7 Putative receptor protein kinase PERK1 [Oryza s... 316 1e-84
UniRef100_Q5Z8L3 Putative benzothiadiazole-induced somatic embry... 313 1e-83
UniRef100_Q7XTD9 OSJNBb0022F16.6 protein [Oryza sativa] 307 5e-82
UniRef100_Q8GXZ5 Hypothetical protein At5g18910/F17K4_160 [Arabi... 306 8e-82
UniRef100_Q9LSM2 Similarity to lectin-like protein kinase [Arabi... 302 1e-80
UniRef100_Q8GWR7 Hypothetical protein At5g65530/K21L13_3 [Arabid... 302 1e-80
UniRef100_Q9LXA4 Pto kinase interactor-like protein [Arabidopsis... 300 7e-80
UniRef100_Q93ZZ8 Putative Pto kinase interactor [Arabidopsis tha... 300 7e-80
UniRef100_Q8H1D6 Putative Pto kinase interactor [Arabidopsis tha... 300 7e-80
UniRef100_Q8RXC8 Putative serine/threonine protein kinase [Arabi... 295 2e-78
UniRef100_Q9FGC3 Serine/threonine protein kinase-like [Arabidops... 294 4e-78
UniRef100_Q682Z0 Hypothetical protein At2g18890 [Arabidopsis tha... 268 2e-70
UniRef100_Q9MAA5 Putative serine/threonine protein kinase [Arabi... 251 4e-65
UniRef100_Q9LVN9 Similarity to protein kinase [Arabidopsis thali... 221 3e-56
UniRef100_Q8VZG4 At1g21590/F24J8_9 [Arabidopsis thaliana] 218 3e-55
>UniRef100_Q9FHH7 Similarity to unknown protein [Arabidopsis thaliana]
Length = 416
Score = 394 bits (1011), Expect = e-108
Identities = 185/257 (71%), Positives = 219/257 (84%), Gaps = 2/257 (0%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGYSEVY+GDL +G IAVKRLAK++ D NKEKEFL ELGII HV HPNTA LLG C E
Sbjct: 113 GGYSEVYRGDLWDGRRIAVKRLAKESGDMNKEKEFLTELGIISHVSHPNTALLLGCCVEK 172
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
GLYL+F +S+NG L +ALH SLDWP+RYKIA+GVARGLHYLHK C HRIIHRDI
Sbjct: 173 GLYLVFRFSENGTLYSALHE--NENGSLDWPVRYKIAVGVARGLHYLHKRCNHRIIHRDI 230
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K+SNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPE M G +DEKTDI+
Sbjct: 231 KSSNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPESLMQGTIDEKTDIY 290
Query: 474 AFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTA 533
AFG+LLLEI+TGRRPV+ ++++ILLWAKP ME+GN +EL DP+++ +YD +Q+ ++VLTA
Sbjct: 291 AFGILLLEIITGRRPVNPTQKHILLWAKPAMETGNTSELVDPKLQDKYDDQQMNKLVLTA 350
Query: 534 SYCVRQTAIWRPAMTEV 550
S+CV+Q+ I RP MT+V
Sbjct: 351 SHCVQQSPILRPTMTQV 367
>UniRef100_Q5XF57 At5g57670 [Arabidopsis thaliana]
Length = 579
Score = 394 bits (1011), Expect = e-108
Identities = 185/257 (71%), Positives = 219/257 (84%), Gaps = 2/257 (0%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGYSEVY+GDL +G IAVKRLAK++ D NKEKEFL ELGII HV HPNTA LLG C E
Sbjct: 276 GGYSEVYRGDLWDGRRIAVKRLAKESGDMNKEKEFLTELGIISHVSHPNTALLLGCCVEK 335
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
GLYL+F +S+NG L +ALH SLDWP+RYKIA+GVARGLHYLHK C HRIIHRDI
Sbjct: 336 GLYLVFRFSENGTLYSALHE--NENGSLDWPVRYKIAVGVARGLHYLHKRCNHRIIHRDI 393
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K+SNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPE M G +DEKTDI+
Sbjct: 394 KSSNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPESLMQGTIDEKTDIY 453
Query: 474 AFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTA 533
AFG+LLLEI+TGRRPV+ ++++ILLWAKP ME+GN +EL DP+++ +YD +Q+ ++VLTA
Sbjct: 454 AFGILLLEIITGRRPVNPTQKHILLWAKPAMETGNTSELVDPKLQDKYDDQQMNKLVLTA 513
Query: 534 SYCVRQTAIWRPAMTEV 550
S+CV+Q+ I RP MT+V
Sbjct: 514 SHCVQQSPILRPTMTQV 530
Score = 53.9 bits (128), Expect = 1e-05
Identities = 25/51 (49%), Positives = 38/51 (74%)
Query: 6 SSTILIGLSLDPNDSKELLSWAIRVLANPNDTIVAVHVLGEEDNHNLGKQK 56
S+ IL+ +SLD ++S+ +LSWAI VLA P+DT+VA+H+L E+ L +K
Sbjct: 9 SNKILVAISLDRDESQNVLSWAINVLAKPSDTVVALHLLVGEEPRKLPMKK 59
>UniRef100_Q6YW24 SERK1 protein-like [Oryza sativa]
Length = 601
Score = 387 bits (993), Expect = e-106
Identities = 181/261 (69%), Positives = 216/261 (82%), Gaps = 2/261 (0%)
Query: 290 LSPRGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGY 349
++ RGGY+EVYKG L +G+ +AVKRLA+ KEKEFL ELGI GHVCHPNTA LLG
Sbjct: 305 MAGRGGYAEVYKGILSDGQCVAVKRLAQGKPTEQKEKEFLTELGIQGHVCHPNTAYLLGC 364
Query: 350 CFENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRII 409
C ENGLYL+F + +NG L++ALHG K+ L+WP+RYKIA+GVARGL YLH C+HRII
Sbjct: 365 CVENGLYLVFEFCENGTLASALHG--KSAKILEWPLRYKIAVGVARGLQYLHMFCRHRII 422
Query: 410 HRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEK 469
HRDIKASNVLLG D+EPQI+DFGLAKWLP +WTHH+VIP+EGTFGYLAPE FMHGIVDEK
Sbjct: 423 HRDIKASNVLLGDDFEPQISDFGLAKWLPKQWTHHSVIPIEGTFGYLAPEYFMHGIVDEK 482
Query: 470 TDIFAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRV 529
TDIFAFGVLLLEIVTGRRP+D SK ++L WAKPL+E+G + ELADP + G YD +QL R+
Sbjct: 483 TDIFAFGVLLLEIVTGRRPIDCSKLSLLQWAKPLLEAGQVTELADPNLGGDYDKDQLKRM 542
Query: 530 VLTASYCVRQTAIWRPAMTEV 550
V AS C+ + A+WRP+M EV
Sbjct: 543 VAVASRCIMRPAMWRPSMAEV 563
Score = 60.8 bits (146), Expect = 1e-07
Identities = 49/181 (27%), Positives = 76/181 (41%), Gaps = 49/181 (27%)
Query: 7 STILIGLSLDPNDSKELLSWAIRVLANPNDTIVAVHVLGEEDNHNLGKQKH--MLYLCLE 64
S IL+G+ + ELLSWAIRV+A PND++VAVHVLG N ++ + ++Y+ E
Sbjct: 20 SRILVGVPNNSRGCSELLSWAIRVVARPNDSVVAVHVLGGRGRKNRLQKANAFVIYMLGE 79
Query: 65 ----------NLHRLVGPNRLIWRQKWH*VPQ*VVV*WRKPSQFLLIFFFFLAEQETKQL 114
NL V + IWR QE
Sbjct: 80 FVETCEAKQVNLEAKVVCSPSIWR---------------------------ALTQEATLT 112
Query: 115 SKNSSVIGTSKG--------ITNYCFEHVHEGCTIVSVGRN--TKTEQNINSTDFQGNSV 164
N ++G S + NYC+ + C++++VGR+ ++ S F +S+
Sbjct: 113 DANFLIVGRSGNAYRRNHFEVANYCYMNAPRNCSVIAVGRDGLPQSAARFKSRSFDDSSI 172
Query: 165 F 165
F
Sbjct: 173 F 173
>UniRef100_O64619 Hypothetical protein At2g18890 [Arabidopsis thaliana]
Length = 392
Score = 330 bits (845), Expect = 9e-89
Identities = 153/260 (58%), Positives = 205/260 (78%), Gaps = 4/260 (1%)
Query: 293 RGGYSEVYKGDLC-NGETIAVKRLAKDNKDPNK-EKEFLMELGIIGHVCHPNTASLLGYC 350
RGG++EVYKG L NGE IAVKR+ + +D + EKEFLME+G IGHV HPN SLLG C
Sbjct: 76 RGGFAEVYKGILGKNGEEIAVKRITRGGRDDERREKEFLMEIGTIGHVSHPNVLSLLGCC 135
Query: 351 FENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
+NGLYL+F +S G+L++ LH +A L+W RYKIAIG A+GLHYLHK C+ RIIH
Sbjct: 136 IDNGLYLVFIFSSRGSLASLLHDLNQA--PLEWETRYKIAIGTAKGLHYLHKGCQRRIIH 193
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
RDIK+SNVLL D+EPQI+DFGLAKWLP++W+HH++ P+EGTFG+LAPE + HGIVDEKT
Sbjct: 194 RDIKSSNVLLNQDFEPQISDFGLAKWLPSQWSHHSIAPIEGTFGHLAPEYYTHGIVDEKT 253
Query: 471 DIFAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
D+FAFGV LLE+++G++PVD+S Q++ WAK +++ G I +L DPR+ +D++QL+R+
Sbjct: 254 DVFAFGVFLLELISGKKPVDASHQSLHSWAKLIIKDGEIEKLVDPRIGEEFDLQQLHRIA 313
Query: 531 LTASYCVRQTAIWRPAMTEV 550
AS C+R +++ RP+M EV
Sbjct: 314 FAASLCIRSSSLCRPSMIEV 333
>UniRef100_Q653Y9 Putative receptor protein kinase PERK1 [Oryza sativa]
Length = 550
Score = 316 bits (809), Expect = 1e-84
Identities = 143/258 (55%), Positives = 196/258 (75%), Gaps = 3/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG++EVYKG L +G+ +AVKRL K ++ +FL ELGII HV HPN A LLG+ E
Sbjct: 242 KGGHAEVYKGHLADGQFVAVKRLTKGGNKEDRISDFLSELGIIAHVNHPNAAQLLGFSVE 301
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
GL+L+ +S +G+L++ LHG +L W R+ IA+G+A GL YLH+ C RIIHRD
Sbjct: 302 GGLHLVLQFSPHGSLASVLHG---TKGALKWKARFNIALGIAEGLLYLHEGCHRRIIHRD 358
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL DY+PQI+DFGLAKWLP+KWTHH V P+EGTFGY++PE FMHGI++EKTD+
Sbjct: 359 IKASNILLTEDYQPQISDFGLAKWLPDKWTHHVVFPIEGTFGYMSPEYFMHGIINEKTDV 418
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FA+GVLLLE+VTGR+ VDSS+Q++++WAKPL++S N+ EL DP ++ YD E++ ++
Sbjct: 419 FAYGVLLLELVTGRKAVDSSRQSLVIWAKPLLDSNNMKELVDPSLDVGYDPEEMAHILAV 478
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+ ++ RP+M V
Sbjct: 479 ASMCIHHSSSSRPSMKSV 496
>UniRef100_Q653Y7 Putative receptor protein kinase PERK1 [Oryza sativa]
Length = 555
Score = 316 bits (809), Expect = 1e-84
Identities = 143/258 (55%), Positives = 196/258 (75%), Gaps = 3/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG++EVYKG L +G+ +AVKRL K ++ +FL ELGII HV HPN A LLG+ E
Sbjct: 247 KGGHAEVYKGHLADGQFVAVKRLTKGGNKEDRISDFLSELGIIAHVNHPNAAQLLGFSVE 306
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
GL+L+ +S +G+L++ LHG +L W R+ IA+G+A GL YLH+ C RIIHRD
Sbjct: 307 GGLHLVLQFSPHGSLASVLHG---TKGALKWKARFNIALGIAEGLLYLHEGCHRRIIHRD 363
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL DY+PQI+DFGLAKWLP+KWTHH V P+EGTFGY++PE FMHGI++EKTD+
Sbjct: 364 IKASNILLTEDYQPQISDFGLAKWLPDKWTHHVVFPIEGTFGYMSPEYFMHGIINEKTDV 423
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FA+GVLLLE+VTGR+ VDSS+Q++++WAKPL++S N+ EL DP ++ YD E++ ++
Sbjct: 424 FAYGVLLLELVTGRKAVDSSRQSLVIWAKPLLDSNNMKELVDPSLDVGYDPEEMAHILAV 483
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+ ++ RP+M V
Sbjct: 484 ASMCIHHSSSSRPSMKSV 501
>UniRef100_Q5Z8L3 Putative benzothiadiazole-induced somatic embryogenesis receptor
kinase 1 [Oryza sativa]
Length = 392
Score = 313 bits (801), Expect = 1e-83
Identities = 147/258 (56%), Positives = 190/258 (72%), Gaps = 1/258 (0%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG SEVY+G+L +G +AVKRL +E++FL ELG +GH HPN +LLG C +
Sbjct: 77 KGGSSEVYRGELPDGRAVAVKRLMGAWACERRERDFLAELGTVGHARHPNVCALLGCCVD 136
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
LYL+F++S G++S LH KA ++ W +R IA+G ARGL YLHK C+ RIIHRD
Sbjct: 137 RDLYLVFHFSGRGSVSANLHDEKKAP-AMGWAVRRAIAVGTARGLEYLHKGCQRRIIHRD 195
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASNVLL D++PQI+DFGLAKWLP++WTH A+ P+EGTFG LAPE + HGIVDEKTD+
Sbjct: 196 IKASNVLLTDDFQPQISDFGLAKWLPSEWTHRAIAPIEGTFGCLAPEYYTHGIVDEKTDV 255
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FAFGV LLEI+TGR+PVD S +++L WA+P + G I L DPR+ G YD E+ R+
Sbjct: 256 FAFGVFLLEIMTGRKPVDGSHKSLLSWARPFLNEGRIESLVDPRIGGDYDGEEARRLAFV 315
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+R +A WRP+MTEV
Sbjct: 316 ASLCIRSSAKWRPSMTEV 333
>UniRef100_Q7XTD9 OSJNBb0022F16.6 protein [Oryza sativa]
Length = 348
Score = 307 bits (787), Expect = 5e-82
Identities = 152/260 (58%), Positives = 188/260 (71%), Gaps = 4/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNK-DPNKEKEFLMELGIIGHVCHPNTASLLGYCF 351
RGGY EVY+G L +G +AVKRL+ D KEK+FL ELG +GHV HPN +LLG C
Sbjct: 67 RGGYGEVYRGVLEDGSAVAVKRLSPAAAADEKKEKDFLTELGTVGHVRHPNVTALLGCCV 126
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+LIF +S G++S LH + + W R+ IA+G ARGL YLHK C RIIHR
Sbjct: 127 DRGLHLIFEFSARGSVSANLHD--ERLPVMPWRRRHGIAVGTARGLRYLHKGCARRIIHR 184
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASNVLL DYEPQI+DFGLA+WLP++WTHHA+ P+EGTFG LAPE F HGIVDEKTD
Sbjct: 185 DIKASNVLLTADYEPQISDFGLARWLPSEWTHHAIAPIEGTFGCLAPEYFTHGIVDEKTD 244
Query: 472 IFAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRM-EGRYDVEQLYRVV 530
+FAFGV LLE+++GR+PVD S +++L WAKP + L DPR+ +G YD QL R++
Sbjct: 245 VFAFGVFLLELISGRKPVDGSHKSLLAWAKPYLNDCVAQGLVDPRLGDGGYDGAQLRRLM 304
Query: 531 LTASYCVRQTAIWRPAMTEV 550
AS CVR A WRP MT+V
Sbjct: 305 FVASLCVRPAAAWRPTMTQV 324
>UniRef100_Q8GXZ5 Hypothetical protein At5g18910/F17K4_160 [Arabidopsis thaliana]
Length = 511
Score = 306 bits (785), Expect = 8e-82
Identities = 141/257 (54%), Positives = 198/257 (76%), Gaps = 4/257 (1%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GGY+EVYKG + +G+ +A+K+L + + + ++L ELGII HV HPN A L+GYC E
Sbjct: 201 GGYAEVYKGQMADGQIVAIKKLTRGSAE-EMTMDYLSELGIIVHVDHPNIAKLIGYCVEG 259
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
G++L+ S NG+L++ L+ +A L+W +RYK+A+G A GL+YLH+ C+ RIIH+DI
Sbjct: 260 GMHLVLELSPNGSLASLLY---EAKEKLNWSMRYKVAMGTAEGLYYLHEGCQRRIIHKDI 316
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
KASN+LL ++E QI+DFGLAKWLP++WTHH V VEGTFGYL PE FMHGIVDEKTD++
Sbjct: 317 KASNILLTQNFEAQISDFGLAKWLPDQWTHHTVSKVEGTFGYLPPEFFMHGIVDEKTDVY 376
Query: 474 AFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTA 533
A+GVLLLE++TGR+ +DSS+ +I++WAKPL++ I +L DP +E YDVE+L R+V A
Sbjct: 377 AYGVLLLELITGRQALDSSQHSIVMWAKPLIKENKIKQLVDPILEDDYDVEELDRLVFIA 436
Query: 534 SYCVRQTAIWRPAMTEV 550
S C+ QT++ RP M++V
Sbjct: 437 SLCIHQTSMNRPQMSQV 453
>UniRef100_Q9LSM2 Similarity to lectin-like protein kinase [Arabidopsis thaliana]
Length = 418
Score = 302 bits (774), Expect = 1e-80
Identities = 145/260 (55%), Positives = 190/260 (72%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L +GET+A+K+L + K+ + +FL ELGII HV HPN A L G+
Sbjct: 114 KGGHAEVYKGVLPDGETVAIKKLTRHAKEVEERVSDFLSELGIIAHVNHPNAARLRGFSC 173
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + YS +G+L++ L G + LDW RYK+A+G+A GL YLH C RIIHR
Sbjct: 174 DRGLHFVLEYSSHGSLASLLFG---SEECLDWKKRYKVAMGIADGLSYLHNDCPRRIIHR 230
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEKTD
Sbjct: 231 DIKASNILLSQDYEAQISDFGLAKWLPEHWPHHIVFPIEGTFGYLAPEYFMHGIVDEKTD 290
Query: 472 IFAFGVLLLEIVTGRRPVDS-SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+TGRR VD+ S+Q+I++WAKPL+E N+ E+ DP++ +D ++ RV+
Sbjct: 291 VFAFGVLLLEIITGRRAVDTDSRQSIVMWAKPLLEKNNMEEIVDPQLGNDFDETEMKRVM 350
Query: 531 LTASYCVRQTAIWRPAMTEV 550
TAS C+ + RP M +
Sbjct: 351 QTASMCIHHVSTMRPDMNRL 370
>UniRef100_Q8GWR7 Hypothetical protein At5g65530/K21L13_3 [Arabidopsis thaliana]
Length = 456
Score = 302 bits (774), Expect = 1e-80
Identities = 145/260 (55%), Positives = 190/260 (72%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L +GET+A+K+L + K+ + +FL ELGII HV HPN A L G+
Sbjct: 152 KGGHAEVYKGVLPDGETVAIKKLTRHAKEVEERVSDFLSELGIIAHVNHPNAARLRGFSC 211
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + YS +G+L++ L G + LDW RYK+A+G+A GL YLH C RIIHR
Sbjct: 212 DRGLHFVLEYSSHGSLASLLFG---SEECLDWKKRYKVAMGIADGLSYLHNDCPRRIIHR 268
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEKTD
Sbjct: 269 DIKASNILLSQDYEAQISDFGLAKWLPEHWPHHIVFPIEGTFGYLAPEYFMHGIVDEKTD 328
Query: 472 IFAFGVLLLEIVTGRRPVDS-SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+TGRR VD+ S+Q+I++WAKPL+E N+ E+ DP++ +D ++ RV+
Sbjct: 329 VFAFGVLLLEIITGRRAVDTDSRQSIVMWAKPLLEKNNMEEIVDPQLGNDFDETEMKRVM 388
Query: 531 LTASYCVRQTAIWRPAMTEV 550
TAS C+ + RP M +
Sbjct: 389 QTASMCIHHVSTMRPDMNRL 408
>UniRef100_Q9LXA4 Pto kinase interactor-like protein [Arabidopsis thaliana]
Length = 464
Score = 300 bits (768), Expect = 7e-80
Identities = 144/260 (55%), Positives = 187/260 (71%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L NGET+A+K+L K+ + +FL ELGII HV HPN A L G+
Sbjct: 161 KGGHAEVYKGVLINGETVAIKKLMSHAKEEEERVSDFLSELGIIAHVNHPNAARLRGFSS 220
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + Y+ G+L++ L G + L+W IRYK+A+G+A GL YLH C RIIHR
Sbjct: 221 DRGLHFVLEYAPYGSLASMLFG---SEECLEWKIRYKVALGIADGLSYLHNACPRRIIHR 277
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEK D
Sbjct: 278 DIKASNILLNHDYEAQISDFGLAKWLPENWPHHVVFPIEGTFGYLAPEYFMHGIVDEKID 337
Query: 472 IFAFGVLLLEIVTGRRPVD-SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+T RR VD +S+Q+I+ WAKP +E ++ ++ DPR+ ++ ++ RV+
Sbjct: 338 VFAFGVLLLEIITSRRAVDTASRQSIVAWAKPFLEKNSMEDIVDPRLGNMFNPTEMQRVM 397
Query: 531 LTASYCVRQTAIWRPAMTEV 550
LTAS CV A RP MT +
Sbjct: 398 LTASMCVHHIAAMRPDMTRL 417
>UniRef100_Q93ZZ8 Putative Pto kinase interactor [Arabidopsis thaliana]
Length = 467
Score = 300 bits (768), Expect = 7e-80
Identities = 144/260 (55%), Positives = 187/260 (71%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L NGET+A+K+L K+ + +FL ELGII HV HPN A L G+
Sbjct: 161 KGGHAEVYKGVLINGETVAIKKLMSHAKEEEERVSDFLSELGIIAHVNHPNAARLRGFSS 220
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + Y+ G+L++ L G + L+W IRYK+A+G+A GL YLH C RIIHR
Sbjct: 221 DRGLHFVLEYAPYGSLASMLFG---SEECLEWKIRYKVALGIADGLSYLHNACPRRIIHR 277
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEK D
Sbjct: 278 DIKASNILLNHDYEAQISDFGLAKWLPENWPHHVVFPIEGTFGYLAPEYFMHGIVDEKID 337
Query: 472 IFAFGVLLLEIVTGRRPVD-SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+T RR VD +S+Q+I+ WAKP +E ++ ++ DPR+ ++ ++ RV+
Sbjct: 338 VFAFGVLLLEIITSRRAVDTASRQSIVAWAKPFLEKNSMEDIVDPRLGNMFNPTEMQRVM 397
Query: 531 LTASYCVRQTAIWRPAMTEV 550
LTAS CV A RP MT +
Sbjct: 398 LTASMCVHHIAAMRPDMTRL 417
>UniRef100_Q8H1D6 Putative Pto kinase interactor [Arabidopsis thaliana]
Length = 467
Score = 300 bits (768), Expect = 7e-80
Identities = 144/260 (55%), Positives = 187/260 (71%), Gaps = 5/260 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKE-KEFLMELGIIGHVCHPNTASLLGYCF 351
+GG++EVYKG L NGET+A+K+L K+ + +FL ELGII HV HPN A L G+
Sbjct: 161 KGGHAEVYKGVLINGETVAIKKLMSHAKEEEERVSDFLSELGIIAHVNHPNAARLRGFSS 220
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
+ GL+ + Y+ G+L++ L G + L+W IRYK+A+G+A GL YLH C RIIHR
Sbjct: 221 DRGLHFVLEYAPYGSLASMLFG---SEECLEWKIRYKVALGIADGLSYLHNACPRRIIHR 277
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASN+LL DYE QI+DFGLAKWLP W HH V P+EGTFGYLAPE FMHGIVDEK D
Sbjct: 278 DIKASNILLNHDYEAQISDFGLAKWLPENWPHHVVFPIEGTFGYLAPEYFMHGIVDEKID 337
Query: 472 IFAFGVLLLEIVTGRRPVD-SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVV 530
+FAFGVLLLEI+T RR VD +S+Q+I+ WAKP +E ++ ++ DPR+ ++ ++ RV+
Sbjct: 338 VFAFGVLLLEIITSRRAVDTASRQSIVAWAKPFLEKNSMEDIVDPRLGNMFNPTEMQRVM 397
Query: 531 LTASYCVRQTAIWRPAMTEV 550
LTAS CV A RP MT +
Sbjct: 398 LTASMCVHHIAAMRPDMTRL 417
>UniRef100_Q8RXC8 Putative serine/threonine protein kinase [Arabidopsis thaliana]
Length = 460
Score = 295 bits (755), Expect = 2e-78
Identities = 146/258 (56%), Positives = 184/258 (70%), Gaps = 3/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGGY++VY+G L G+ IAVKRL K D + EFL ELGII HV HPNTA +G C E
Sbjct: 151 RGGYADVYQGILPEGKLIAVKRLTKGTPD-EQTAEFLSELGIIAHVDHPNTAKFIGCCIE 209
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G++L+F S G+L + LHG K L W RY +A+G A GL YLH+ C+ RIIHRD
Sbjct: 210 GGMHLVFRLSPLGSLGSLLHGPSKY--KLTWSRRYNVALGTADGLVYLHEGCQRRIIHRD 267
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA N+LL D++PQI DFGLAKWLP + THH V EGTFGY APE FMHGIVDEKTD+
Sbjct: 268 IKADNILLTEDFQPQICDFGLAKWLPKQLTHHNVSKFEGTFGYFAPEYFMHGIVDEKTDV 327
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FAFGVLLLE++TG +D S+Q+++LWAKPL+E I EL DP + Y+ E+L R+ T
Sbjct: 328 FAFGVLLLELITGHPALDESQQSLVLWAKPLLERKAIKELVDPSLGDEYNREELIRLTST 387
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+ Q+++ RP M++V
Sbjct: 388 ASLCIDQSSLLRPRMSQV 405
>UniRef100_Q9FGC3 Serine/threonine protein kinase-like [Arabidopsis thaliana]
Length = 429
Score = 294 bits (753), Expect = 4e-78
Identities = 144/258 (55%), Positives = 187/258 (71%), Gaps = 4/258 (1%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GGY+EVYKG L NG+ +A+KRL + N + +FL E+GI+ HV HPN A LLGY E
Sbjct: 142 KGGYAEVYKGMLPNGQMVAIKRLMRGNSE-EIIVDFLSEMGIMAHVNHPNIAKLLGYGVE 200
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G++L+ S +G+L++ L+ + + W IRYKIA+GVA GL YLH+ C RIIHRD
Sbjct: 201 GGMHLVLELSPHGSLASMLYS---SKEKMKWSIRYKIALGVAEGLVYLHRGCHRRIIHRD 257
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA+N+LL D+ PQI DFGLAKWLP WTHH V EGTFGYLAPE HGIVDEKTD+
Sbjct: 258 IKAANILLTHDFSPQICDFGLAKWLPENWTHHIVSKFEGTFGYLAPEYLTHGIVDEKTDV 317
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FA GVLLLE+VTGRR +D SKQ+++LWAKPLM+ I EL DP + G Y+ Q+ V+L
Sbjct: 318 FALGVLLLELVTGRRALDYSKQSLVLWAKPLMKKNKIRELIDPSLAGEYEWRQIKLVLLA 377
Query: 533 ASYCVRQTAIWRPAMTEV 550
A+ ++Q++I RP M++V
Sbjct: 378 AALSIQQSSIERPEMSQV 395
>UniRef100_Q682Z0 Hypothetical protein At2g18890 [Arabidopsis thaliana]
Length = 277
Score = 268 bits (686), Expect = 2e-70
Identities = 126/196 (64%), Positives = 159/196 (80%), Gaps = 4/196 (2%)
Query: 293 RGGYSEVYKGDLC-NGETIAVKRLAKDNKDPNK-EKEFLMELGIIGHVCHPNTASLLGYC 350
RGG++EVYKG L NGE IAVKR+ + +D + EKEFLME+G IGHV HPN SLLG C
Sbjct: 76 RGGFAEVYKGILGKNGEEIAVKRITRGGRDDERREKEFLMEIGTIGHVSHPNVLSLLGCC 135
Query: 351 FENGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIH 410
+NGLYL+F +S G+L++ LH +A L+W RYKIAIG A+GLHYLHK C+ RIIH
Sbjct: 136 IDNGLYLVFIFSSRGSLASLLHDLNQA--PLEWETRYKIAIGTAKGLHYLHKGCQRRIIH 193
Query: 411 RDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
RDIK+SNVLL D+EPQI+DFGLAKWLP++W+HH++ P+EGTFG+LAPE + HGIVDEKT
Sbjct: 194 RDIKSSNVLLNQDFEPQISDFGLAKWLPSQWSHHSIAPIEGTFGHLAPEYYTHGIVDEKT 253
Query: 471 DIFAFGVLLLEIVTGR 486
D+FAFGV LLE+++G+
Sbjct: 254 DVFAFGVFLLELISGK 269
>UniRef100_Q9MAA5 Putative serine/threonine protein kinase [Arabidopsis thaliana]
Length = 476
Score = 251 bits (641), Expect = 4e-65
Identities = 131/258 (50%), Positives = 168/258 (64%), Gaps = 25/258 (9%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
RGGY++VY+G L G+ IAVKRL K D + EFL ELGII HV HPNTA +G C E
Sbjct: 189 RGGYADVYQGILPEGKLIAVKRLTKGTPD-EQTAEFLSELGIIAHVDHPNTAKFIGCCIE 247
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
G++L+F S G+L + LHG K L W RY +A+G A GL YLH+ C+ RIIHRD
Sbjct: 248 GGMHLVFRLSPLGSLGSLLHGPSKY--KLTWSRRYNVALGTADGLVYLHEGCQRRIIHRD 305
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKA N+LL D++PQI DFG Y APE FMHGIVDEKTD+
Sbjct: 306 IKADNILLTEDFQPQICDFG----------------------YFAPEYFMHGIVDEKTDV 343
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FAFGVLLLE++TG +D S+Q+++LWAKPL+E I EL DP + Y+ E+L R+ T
Sbjct: 344 FAFGVLLLELITGHPALDESQQSLVLWAKPLLERKAIKELVDPSLGDEYNREELIRLTST 403
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+ Q+++ RP M++V
Sbjct: 404 ASLCIDQSSLLRPRMSQV 421
>UniRef100_Q9LVN9 Similarity to protein kinase [Arabidopsis thaliana]
Length = 705
Score = 221 bits (564), Expect = 3e-56
Identities = 116/257 (45%), Positives = 165/257 (64%), Gaps = 8/257 (3%)
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG S VY+GDL +G +AVK L + KEF++E+ +I V H N SL G+CFEN
Sbjct: 371 GGNSYVYRGDLPDGRELAVKIL---KPCLDVLKEFILEIEVITSVHHKNIVSLFGFCFEN 427
Query: 354 G-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L L+++Y G+L LHG K W RYK+A+GVA L YLH +IHRD
Sbjct: 428 NNLMLVYDYLPRGSLEENLHGNRKDAKKFGWMERYKVAVGVAEALDYLHNTHDPEVIHRD 487
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K+SNVLL D+EPQ++DFG A + H A + GTFGYLAPE FMHG V +K D+
Sbjct: 488 VKSSNVLLADDFEPQLSDFGFASLASSTSQHVAGGDIAGTFGYLAPEYFMHGKVTDKIDV 547
Query: 473 FAFGVLLLEIVTGRRP--VDSSK--QNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
+AFGV+LLE+++GR+P VD SK ++++LWA P+++SG A+L DP +E + + +
Sbjct: 548 YAFGVVLLELISGRKPICVDQSKGQESLVLWANPILDSGKFAQLLDPSLENDNSNDLIEK 607
Query: 529 VVLTASYCVRQTAIWRP 545
++L A+ C+++T RP
Sbjct: 608 LLLAATLCIKRTPHDRP 624
>UniRef100_Q8VZG4 At1g21590/F24J8_9 [Arabidopsis thaliana]
Length = 756
Score = 218 bits (556), Expect = 3e-55
Identities = 110/260 (42%), Positives = 165/260 (63%), Gaps = 8/260 (3%)
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG S V++G L NG +AVK L + K+F+ E+ II + H N SLLGYCFE
Sbjct: 417 KGGSSRVFRGYLPNGREVAVKILKRTEC---VLKDFVAEIDIITTLHHKNVISLLGYCFE 473
Query: 353 NG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
N L L++NY G+L LHG K + W RYK+A+G+A L YLH +IHR
Sbjct: 474 NNNLLLVYNYLSRGSLEENLHGNKKDLVAFRWNERYKVAVGIAEALDYLHNDAPQPVIHR 533
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K+SN+LL D+EPQ++DFGLAKW T V GTFGYLAPE FM+G ++ K D
Sbjct: 534 DVKSSNILLSDDFEPQLSDFGLAKWASESTTQIICSDVAGTFGYLAPEYFMYGKMNNKID 593
Query: 472 IFAFGVLLLEIVTGRRPVDS----SKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLY 527
++A+GV+LLE+++GR+PV+S ++ ++++WAKP+++ ++L D ++ + +Q+
Sbjct: 594 VYAYGVVLLELLSGRKPVNSESPKAQDSLVMWAKPILDDKEYSQLLDSSLQDDNNSDQME 653
Query: 528 RVVLTASYCVRQTAIWRPAM 547
++ L A+ C+R RP M
Sbjct: 654 KMALAATLCIRHNPQTRPTM 673
Score = 44.7 bits (104), Expect = 0.007
Identities = 19/40 (47%), Positives = 29/40 (72%), Gaps = 1/40 (2%)
Query: 8 TILIGLSLDPNDSKELLSWAIRVLANPNDTIVAVHVLGEE 47
T+++G+ D + S ELL WA+ +A P DT++A+HVLG E
Sbjct: 13 TVMVGVKFDES-SNELLDWALVKVAEPGDTVIALHVLGNE 51
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.337 0.149 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 901,951,143
Number of Sequences: 2790947
Number of extensions: 37638948
Number of successful extensions: 165631
Number of sequences better than 10.0: 17294
Number of HSP's better than 10.0 without gapping: 6496
Number of HSP's successfully gapped in prelim test: 10799
Number of HSP's that attempted gapping in prelim test: 134478
Number of HSP's gapped (non-prelim): 19686
length of query: 550
length of database: 848,049,833
effective HSP length: 132
effective length of query: 418
effective length of database: 479,644,829
effective search space: 200491538522
effective search space used: 200491538522
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC127169.5


