

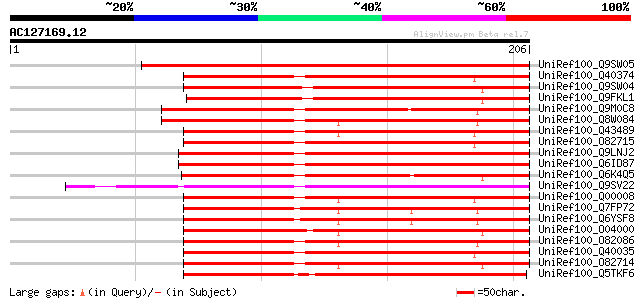
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.12 + phase: 0
(206 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SW05 Putative pathogenesis-related protein [Arabidop... 232 5e-60
UniRef100_Q40374 Pathogenesis-related protein PR-1 precursor [Me... 177 2e-43
UniRef100_Q9SW04 Putative pathogenesis-related protein [Arabidop... 172 4e-42
UniRef100_Q9FKL1 Similarity to pathogenesis-related protein [Ara... 166 3e-40
UniRef100_Q9M0C8 PR-1-like protein [Arabidopsis thaliana] 160 3e-38
UniRef100_Q8W084 Putative pathogenesis-related protein [Oryza sa... 156 4e-37
UniRef100_Q43489 PR-1a pathogenesis related protein (Hv-1a) prec... 155 5e-37
UniRef100_O82715 Pathogenisis-related protein 1.2 precursor [Tri... 154 1e-36
UniRef100_Q9LNJ2 F6F3.11 protein [Arabidopsis thaliana] 154 1e-36
UniRef100_Q6ID87 At1g01310 [Arabidopsis thaliana] 154 1e-36
UniRef100_Q6K4Q5 Putative Pathogenesis-related protein PR-1 [Ory... 154 2e-36
UniRef100_Q9SV22 Pathogenesis-related protein homolog [Arabidops... 152 4e-36
UniRef100_Q00008 Pathogenesis-related protein PRMS precursor [Ze... 152 4e-36
UniRef100_Q7FP72 PR1a protein [Oryza sativa] 152 7e-36
UniRef100_Q6YSF8 PR-1 type pathogenesis-related protein PR-1a [O... 152 7e-36
UniRef100_O04000 Pathogenesis-related protein class 1 [Oryza sat... 152 7e-36
UniRef100_O82086 Pathogenesis related protein-1 [Zea mays] 150 3e-35
UniRef100_Q40035 Type-1 pathogenesis-related protein [Hordeum vu... 148 8e-35
UniRef100_O82714 Pathogenisis-related protein 1.1 precursor [Tri... 148 8e-35
UniRef100_Q5TKF6 Hypothetical protein OSJNBa0030I14.10 [Oryza sa... 148 1e-34
>UniRef100_Q9SW05 Putative pathogenesis-related protein [Arabidopsis thaliana]
Length = 190
Score = 232 bits (591), Expect = 5e-60
Identities = 95/154 (61%), Positives = 126/154 (81%)
Query: 53 IYKVSKQLCWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEH 112
++++ K LC C +SL+FLFRHNLVRA+++E PL+WD +L+ YA+ WA+QR+ DC + H
Sbjct: 37 VFRICKNLCPGCDHDSLQFLFRHNLVRAARFEPPLIWDRRLQNYAQGWANQRRGDCALRH 96
Query: 113 SFPEDGFKLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKS 172
S F LGENIYWG G++W+P DAV AWA E+++Y Y +N+C +GQMCGHYTQIVWKS
Sbjct: 97 SVSNGEFNLGENIYWGYGANWSPADAVVAWASEKRFYHYGSNTCDAGQMCGHYTQIVWKS 156
Query: 173 TRRIGCARVVCDDGDVFMTCNYDPVGNYVGERPY 206
TRR+GCARVVCD+G +FMTCNYDP GNY+G++PY
Sbjct: 157 TRRVGCARVVCDNGGIFMTCNYDPPGNYIGQKPY 190
>UniRef100_Q40374 Pathogenesis-related protein PR-1 precursor [Medicago truncatula]
Length = 173
Score = 177 bits (449), Expect = 2e-43
Identities = 79/138 (57%), Positives = 97/138 (70%), Gaps = 5/138 (3%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL N+ RA+ PL+WD +L YA+W+A+QR+ DC +EHS GENI+WGS
Sbjct: 40 QFLIPQNIARAAVGLRPLVWDDKLTHYAQWYANQRRNDCALEHS----NGPYGENIFWGS 95
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGDV 188
G W P AV AW DE+++Y Y NSCV G+MCGHYTQ+VW ST ++GCA VVC DD
Sbjct: 96 GVGWNPAQAVSAWVDEKQFYNYWHNSCVDGEMCGHYTQVVWGSTTKVGCASVVCSDDKGT 155
Query: 189 FMTCNYDPVGNYVGERPY 206
FMTCNYDP GNY GERPY
Sbjct: 156 FMTCNYDPPGNYYGERPY 173
>UniRef100_Q9SW04 Putative pathogenesis-related protein [Arabidopsis thaliana]
Length = 210
Score = 172 bits (437), Expect = 4e-42
Identities = 78/138 (56%), Positives = 99/138 (71%), Gaps = 5/138 (3%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL HN VR PL+WD ++ YA WWA+QR+ DC + HS GEN++WGS
Sbjct: 77 QFLDPHNTVRGGLGLPPLVWDVKIASYATWWANQRRYDCSLTHSTGP----YGENLFWGS 132
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-DV 188
GSD+T T AV++W E K Y ++TN+C MCGHYTQIVW+ TRR+GCARVVC++G V
Sbjct: 133 GSDFTSTFAVESWTVEAKSYNHMTNTCEGDGMCGHYTQIVWRETRRLGCARVVCENGAGV 192
Query: 189 FMTCNYDPVGNYVGERPY 206
F+TCNYDP GNYVGE+PY
Sbjct: 193 FITCNYDPPGNYVGEKPY 210
>UniRef100_Q9FKL1 Similarity to pathogenesis-related protein [Arabidopsis thaliana]
Length = 207
Score = 166 bits (421), Expect = 3e-40
Identities = 75/137 (54%), Positives = 96/137 (69%), Gaps = 5/137 (3%)
Query: 71 FLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGSG 130
FL HN +R+ PL+WD +L YA WWA+QR+ DC + HS GEN++WGSG
Sbjct: 75 FLDPHNALRSGLGLPPLIWDGKLASYATWWANQRRYDCSLTHSTGP----YGENLFWGSG 130
Query: 131 SDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-DVF 189
S W P AV++W E + Y + TNSC MCGHYTQ+VW+ T+R+GCARVVC++G VF
Sbjct: 131 SSWAPGFAVQSWIVEGRSYNHNTNSCDGSGMCGHYTQMVWRDTKRLGCARVVCENGAGVF 190
Query: 190 MTCNYDPVGNYVGERPY 206
+TCNYDP GNYVGE+PY
Sbjct: 191 ITCNYDPPGNYVGEKPY 207
>UniRef100_Q9M0C8 PR-1-like protein [Arabidopsis thaliana]
Length = 161
Score = 160 bits (404), Expect = 3e-38
Identities = 72/147 (48%), Positives = 97/147 (65%), Gaps = 6/147 (4%)
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 120
C +C +F+ N RA PL WD +L +YA+WWA+QR+ DC + HS
Sbjct: 20 CCHCATYQEQFMGPQNAARAHLRLKPLKWDAKLARYAQWWANQRRGDCALTHS----NGP 75
Query: 121 LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 180
GEN++WGSG+ W P+ A W E + Y Y +NSC S +MCGHYTQIVWK+T++IGCA
Sbjct: 76 YGENLFWGSGNRWGPSQAAYGWLSEARSYNYRSNSCNS-EMCGHYTQIVWKNTQKIGCAH 134
Query: 181 VVCD-DGDVFMTCNYDPVGNYVGERPY 206
V+C+ G VF+TCNYDP GN++G +PY
Sbjct: 135 VICNGGGGVFLTCNYDPPGNFLGRKPY 161
>UniRef100_Q8W084 Putative pathogenesis-related protein [Oryza sativa]
Length = 167
Score = 156 bits (394), Expect = 4e-37
Identities = 69/148 (46%), Positives = 99/148 (66%), Gaps = 6/148 (4%)
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 120
C + ++L N R++ P+ W +L+ +A +A QRK DC+++HS G
Sbjct: 24 CSDAQNSPQDYLSPQNAARSAVGVGPMSWSTKLQGFAEDYARQRKGDCRLQHS----GGP 79
Query: 121 LGENIYWGS-GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCA 179
GENI+WGS G+DWT DAV++W DE+KYY Y +NSC +G++CGHYTQ+VW+ + +GCA
Sbjct: 80 YGENIFWGSAGADWTAADAVRSWVDEKKYYNYASNSCAAGKVCGHYTQVVWRDSTNVGCA 139
Query: 180 RVVCD-DGDVFMTCNYDPVGNYVGERPY 206
RV CD + +F+ CNY+P GN VG RPY
Sbjct: 140 RVRCDANRGIFIICNYEPRGNIVGRRPY 167
>UniRef100_Q43489 PR-1a pathogenesis related protein (Hv-1a) precursor [Hordeum
vulgare]
Length = 164
Score = 155 bits (393), Expect = 5e-37
Identities = 71/139 (51%), Positives = 99/139 (71%), Gaps = 6/139 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
++L HN RA+ + W +L+ YA+ +A+QR DCK++HS G GENI+WGS
Sbjct: 30 DYLSPHNAARAAVGVGAVSWSTKLQAYAQSYANQRIGDCKLQHS----GGPYGENIFWGS 85
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGD 187
G+DW DAVK W DE+K Y Y +N+C G++CGHYTQ+VW+++ IGCARVVC ++G
Sbjct: 86 AGADWKAADAVKLWVDEKKDYDYGSNTCAGGKVCGHYTQVVWRASTSIGCARVVCNNNGG 145
Query: 188 VFMTCNYDPVGNYVGERPY 206
VF+TCNY+P GN VG++PY
Sbjct: 146 VFITCNYEPAGNVVGQKPY 164
>UniRef100_O82715 Pathogenisis-related protein 1.2 precursor [Triticum aestivum]
Length = 173
Score = 154 bits (390), Expect = 1e-36
Identities = 69/138 (50%), Positives = 96/138 (69%), Gaps = 5/138 (3%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+F+ HN RA P+ WD + ++A+ WA+QR DC+++HS G GENI+WGS
Sbjct: 31 DFVNLHNRARAVDGVGPVAWDNNVARFAQDWAAQRAGDCRLQHS----GGPFGENIFWGS 86
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGDV 188
G WT DAVK W DE++ Y +N+C +G++CGHYTQ+VW+ + RIGCARVVC + V
Sbjct: 87 GQSWTAADAVKLWVDEKQNYHLDSNTCDAGKVCGHYTQVVWRKSTRIGCARVVCTGNRGV 146
Query: 189 FMTCNYDPVGNYVGERPY 206
F+TCNY+P GN+ GERP+
Sbjct: 147 FITCNYNPPGNFNGERPF 164
>UniRef100_Q9LNJ2 F6F3.11 protein [Arabidopsis thaliana]
Length = 283
Score = 154 bits (389), Expect = 1e-36
Identities = 69/139 (49%), Positives = 88/139 (62%), Gaps = 4/139 (2%)
Query: 68 SLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYW 127
S EFL HNLVRA E P WD +L YAR WA+QR DC++ HS GENI+W
Sbjct: 127 SREFLIAHNLVRARVGEPPFQWDGRLAAYARTWANQRVGDCRLVHS----NGPYGENIFW 182
Query: 128 GSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGD 187
++W+P D V WADE+K+Y N+C MCGHYTQIVW+ + ++GCA V C +G
Sbjct: 183 AGKNNWSPRDIVNVWADEDKFYDVKGNTCEPQHMCGHYTQIVWRDSTKVGCASVDCSNGG 242
Query: 188 VFMTCNYDPVGNYVGERPY 206
V+ C Y+P GNY GE P+
Sbjct: 243 VYAICVYNPPGNYEGENPF 261
>UniRef100_Q6ID87 At1g01310 [Arabidopsis thaliana]
Length = 241
Score = 154 bits (389), Expect = 1e-36
Identities = 69/139 (49%), Positives = 88/139 (62%), Gaps = 4/139 (2%)
Query: 68 SLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYW 127
S EFL HNLVRA E P WD +L YAR WA+QR DC++ HS GENI+W
Sbjct: 85 SREFLIAHNLVRARVGEPPFQWDGRLAAYARTWANQRVGDCRLVHS----NGPYGENIFW 140
Query: 128 GSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGD 187
++W+P D V WADE+K+Y N+C MCGHYTQIVW+ + ++GCA V C +G
Sbjct: 141 AGKNNWSPRDIVNVWADEDKFYDVKGNTCEPQHMCGHYTQIVWRDSTKVGCASVDCSNGG 200
Query: 188 VFMTCNYDPVGNYVGERPY 206
V+ C Y+P GNY GE P+
Sbjct: 201 VYAICVYNPPGNYEGENPF 219
>UniRef100_Q6K4Q5 Putative Pathogenesis-related protein PR-1 [Oryza sativa]
Length = 172
Score = 154 bits (388), Expect = 2e-36
Identities = 71/139 (51%), Positives = 93/139 (66%), Gaps = 6/139 (4%)
Query: 69 LEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWG 128
++FL + N RA+ L+WD ++ YARW+A R+ DC + HS GEN++WG
Sbjct: 39 MQFLGQQNAARAAMGLPALVWDERVAGYARWYAESRRGDCALVHS----SGPYGENLFWG 94
Query: 129 SGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-D 187
SG+ W+P AV AW E+ Y Y +NSC G MCGHYTQI+W++TRR+GCA V C +G
Sbjct: 95 SGTGWSPAQAVGAWLAEQPRYNYWSNSCYGG-MCGHYTQIMWRATRRVGCAMVACYNGRG 153
Query: 188 VFMTCNYDPVGNYVGERPY 206
F+TCNYDP GNYVG RPY
Sbjct: 154 TFITCNYDPPGNYVGMRPY 172
>UniRef100_Q9SV22 Pathogenesis-related protein homolog [Arabidopsis thaliana]
Length = 185
Score = 152 bits (385), Expect = 4e-36
Identities = 81/184 (44%), Positives = 102/184 (55%), Gaps = 14/184 (7%)
Query: 23 TSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNCMQESLEFLFRHNLVRASK 82
++P PS S P S + + I+ K L N +Q+ +FL HN++RA
Sbjct: 16 STPLPSLSFQIP--------SNRTPTTSTLIFSQDKALARNTIQQ--QFLRPHNILRAKL 65
Query: 83 WELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGSGSDWTPTDAVKAW 142
PL W L YA WA R+ DCK+ HS G GEN++WGSG WTP DAV AW
Sbjct: 66 RLPPLKWSNSLALYASRWARTRRGDCKLIHS----GGPYGENLFWGSGKGWTPRDAVAAW 121
Query: 143 ADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVFMTCNYDPVGNYVG 202
A E KYY T+ C + C HYTQ+VWK + RIGCA C GD F+ CNYDP GN VG
Sbjct: 122 ASEMKYYDRRTSHCKANGDCLHYTQLVWKKSSRIGCAISFCKTGDTFIICNYDPPGNIVG 181
Query: 203 ERPY 206
+ P+
Sbjct: 182 QPPF 185
>UniRef100_Q00008 Pathogenesis-related protein PRMS precursor [Zea mays]
Length = 167
Score = 152 bits (385), Expect = 4e-36
Identities = 69/139 (49%), Positives = 97/139 (69%), Gaps = 6/139 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
++L N RA+ P+ W +L+Q+A +A+QR DC+++HS G GENI+WGS
Sbjct: 33 DYLTPQNSARAAVGVGPVTWSTKLQQFAEKYAAQRAGDCRLQHS----GGPYGENIFWGS 88
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGD 187
G DW DAV++W DE+++Y Y TNSC +G++CGHYTQ+VW++T IGCARVVC D+
Sbjct: 89 AGFDWKAVDAVRSWVDEKQWYNYATNSCAAGKVCGHYTQVVWRATTSIGCARVVCRDNRG 148
Query: 188 VFMTCNYDPVGNYVGERPY 206
VF+ CNY+P GN G +PY
Sbjct: 149 VFIICNYEPRGNIAGMKPY 167
>UniRef100_Q7FP72 PR1a protein [Oryza sativa]
Length = 168
Score = 152 bits (383), Expect = 7e-36
Identities = 73/141 (51%), Positives = 95/141 (66%), Gaps = 6/141 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+F+ HN RA P+ WD + YA +A+QR+ DCK+EHS + G K GENI+WGS
Sbjct: 30 DFVDPHNAARADVGVGPVSWDDTVAAYAESYAAQRQGDCKLEHS--DSGGKYGENIFWGS 87
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVS--GQMCGHYTQIVWKSTRRIGCARVVCD-D 185
G DWT AV AW E+++Y + +NSC + G CGHYTQ+VW+ + IGCARVVCD D
Sbjct: 88 PGGDWTAASAVSAWVSEKQWYDHGSNSCSAPEGSSCGHYTQVVWRDSTAIGCARVVCDGD 147
Query: 186 GDVFMTCNYDPVGNYVGERPY 206
VF+TCNY P GN+VG+ PY
Sbjct: 148 LGVFITCNYSPPGNFVGQSPY 168
>UniRef100_Q6YSF8 PR-1 type pathogenesis-related protein PR-1a [Oryza sativa]
Length = 168
Score = 152 bits (383), Expect = 7e-36
Identities = 73/141 (51%), Positives = 95/141 (66%), Gaps = 6/141 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+F+ HN RA P+ WD + YA +A+QR+ DCK+EHS + G K GENI+WGS
Sbjct: 30 DFVDPHNAARADVGVGPVSWDDTVAAYAESYAAQRQGDCKLEHS--DSGGKYGENIFWGS 87
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVS--GQMCGHYTQIVWKSTRRIGCARVVCD-D 185
G DWT AV AW E+++Y + +NSC + G CGHYTQ+VW+ + IGCARVVCD D
Sbjct: 88 AGGDWTAASAVSAWVSEKQWYDHGSNSCSAPEGSSCGHYTQVVWRDSTAIGCARVVCDGD 147
Query: 186 GDVFMTCNYDPVGNYVGERPY 206
VF+TCNY P GN+VG+ PY
Sbjct: 148 LGVFITCNYSPPGNFVGQSPY 168
>UniRef100_O04000 Pathogenesis-related protein class 1 [Oryza sativa]
Length = 164
Score = 152 bits (383), Expect = 7e-36
Identities = 68/139 (48%), Positives = 95/139 (67%), Gaps = 4/139 (2%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+++ HN RA+ P+ WD ++ +A +ASQR DC + HS + LGEN++WGS
Sbjct: 28 DYVRLHNAARAAVGVGPVTWDTSVQAFAENYASQRSGDCSLIHSSNRNN--LGENLFWGS 85
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-D 187
G DWT AV++W E+ Y Y +NSC G++CGHYTQ+VW+++ IGCARVVC +G
Sbjct: 86 AGGDWTAASAVQSWVGEKSDYDYASNSCAQGKVCGHYTQVVWRASTSIGCARVVCSNGRG 145
Query: 188 VFMTCNYDPVGNYVGERPY 206
VF+TCNY P GN+VG+RPY
Sbjct: 146 VFITCNYKPAGNFVGQRPY 164
>UniRef100_O82086 Pathogenesis related protein-1 [Zea mays]
Length = 163
Score = 150 bits (378), Expect = 3e-35
Identities = 69/139 (49%), Positives = 96/139 (68%), Gaps = 6/139 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+++ HN RA P+ WD + YA+ +A+QR+ DCK+ HS G GEN++WGS
Sbjct: 29 DYVDPHNAARADVGVGPVSWDDTVAAYAQSYAAQRQGDCKLIHS----GGPYGENLFWGS 84
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCD-DGD 187
G+DW+ +DAV +W E++YY + TNSC GQ+CGHYTQ+VW+ + IGCARVVCD +
Sbjct: 85 AGADWSASDAVGSWVSEKQYYDHDTNSCAEGQVCGHYTQVVWRDSTAIGCARVVCDNNAG 144
Query: 188 VFMTCNYDPVGNYVGERPY 206
VF+ C+Y+P GN VGE PY
Sbjct: 145 VFIICSYNPPGNVVGESPY 163
>UniRef100_Q40035 Type-1 pathogenesis-related protein [Hordeum vulgare]
Length = 174
Score = 148 bits (374), Expect = 8e-35
Identities = 67/138 (48%), Positives = 95/138 (68%), Gaps = 5/138 (3%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+F+ HN RA P+ WD + ++A+ +A++R DC+++HS G GENI+WGS
Sbjct: 31 DFVNLHNRARAVDGVGPVAWDNNVARFAQNYAAERAGDCRLQHS----GGPFGENIFWGS 86
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVC-DDGDV 188
G WT DAVK W DE++ Y +N+C +G++CGHYTQ+VW+ + RI CARVVC + V
Sbjct: 87 GRSWTAADAVKLWVDEKQNYHLDSNTCNAGKVCGHYTQVVWRKSIRIACARVVCAGNRGV 146
Query: 189 FMTCNYDPVGNYVGERPY 206
F+TCNYDP GN+ GERP+
Sbjct: 147 FITCNYDPPGNFNGERPF 164
>UniRef100_O82714 Pathogenisis-related protein 1.1 precursor [Triticum aestivum]
Length = 164
Score = 148 bits (374), Expect = 8e-35
Identities = 68/139 (48%), Positives = 97/139 (68%), Gaps = 6/139 (4%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
++L HN RA+ + W +L+ +A+ +A+QR DCK++HS G GENI+W S
Sbjct: 30 DYLSPHNAARAAVGVGAVSWSTKLQGFAQSYANQRINDCKLQHS----GGPYGENIFWRS 85
Query: 130 -GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-D 187
G+DW DAVK W DE+K Y Y +N+C G++CGHYTQ+VW+++ IGCARVVC++
Sbjct: 86 AGADWKAADAVKLWVDEKKDYDYGSNTCAGGKVCGHYTQVVWRASTSIGCARVVCNNNRG 145
Query: 188 VFMTCNYDPVGNYVGERPY 206
VF+TCNY+P GN VG++PY
Sbjct: 146 VFITCNYEPAGNVVGQKPY 164
>UniRef100_Q5TKF6 Hypothetical protein OSJNBa0030I14.10 [Oryza sativa]
Length = 247
Score = 148 bits (373), Expect = 1e-34
Identities = 70/136 (51%), Positives = 88/136 (64%), Gaps = 3/136 (2%)
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
EFL HN VRA PL W +L +YAR W++ R+ DC + HS PE + GEN++WG+
Sbjct: 113 EFLDAHNKVRAQYGLQPLKWSNKLARYARRWSAARRFDCVMMHS-PESPY--GENVFWGT 169
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVF 189
G W TDAVK+WA E Y + SC GQMCGH+TQIVW T+ +GC R C G VF
Sbjct: 170 GWGWRATDAVKSWAGESSVYDWRGQSCNPGQMCGHFTQIVWNDTKLVGCGRSECVAGGVF 229
Query: 190 MTCNYDPVGNYVGERP 205
+TC+YDP GN+ GE P
Sbjct: 230 ITCSYDPPGNWKGEVP 245
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.133 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 385,970,215
Number of Sequences: 2790947
Number of extensions: 16332958
Number of successful extensions: 44984
Number of sequences better than 10.0: 514
Number of HSP's better than 10.0 without gapping: 380
Number of HSP's successfully gapped in prelim test: 134
Number of HSP's that attempted gapping in prelim test: 43890
Number of HSP's gapped (non-prelim): 561
length of query: 206
length of database: 848,049,833
effective HSP length: 121
effective length of query: 85
effective length of database: 510,345,246
effective search space: 43379345910
effective search space used: 43379345910
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC127169.12


